Open Access Article
Open Access ArticleRNA diversification by a self-reproducing ribozyme revealed by deep sequencing and kinetic modelling†
Cyrille
Jeancolas‡
 ab,
Yoshiya J.
Matsubara‡
c,
Mykhailo
Vybornyi
a,
Camille N.
Lambert
a,
Alex
Blokhuis
ab,
Yoshiya J.
Matsubara‡
c,
Mykhailo
Vybornyi
a,
Camille N.
Lambert
a,
Alex
Blokhuis
 ad,
Thomas
Alline
a,
Andrew D.
Griffiths
a,
Sandeep
Ameta
ad,
Thomas
Alline
a,
Andrew D.
Griffiths
a,
Sandeep
Ameta
 c,
Sandeep
Krishna
c and
Philippe
Nghe
*a
c,
Sandeep
Krishna
c and
Philippe
Nghe
*a
aLaboratoire de Biochimie, UMR CNRS-ESPCI 8231, Chimie Biologie Innovation, PSL University, ESPCI Paris, 10 rue Vauquelin, Paris 75005, France. E-mail: philippe.nghe@espci.psl.eu
bLaboratoire d’Anthropologie Sociale, Collège de France, 52 rue du Cardinal Lemoine, Paris 75005, France
cSimons Centre for the Study of Living Machines, National Centre for Biological Sciences, Bellary Road, Bangalore 560 065, Karnataka, India
dGroningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen 9747 AG, The Netherlands
First published on 6th July 2021
Abstract
We demonstrate that a recombinase ribozyme achieves multiple functions in the same reaction network: self-reproduction, iterative elongation and circularization of other RNAs, leading to synthesis of diverse products predicted by a kinetic model. This shows that key mechanisms can be integrated and controlled toward Darwinian evolution in RNA reaction networks.
The “RNA world” hypothesis surmises RNA as a primordial biopolymer capable of evolution due to its ability to store information and catalyze reactions.1 An RNA polymerase ribozyme that catalyzes its own copying and the copying of other RNAs would support this hypothesis but has not yet been found.2 Alternatively, ribozymes can make copies of themselves from fragments via energy-rich3 or energy-neutral4 chemistries. Autocatalytic reproduction has been engineered using the ∼200 nt long Azoarcus group I intron ribozyme via recombination reactions between hybridized RNA fragments.5,6 Such catalysis requires the binding of a 3 nt long internal guide sequence (IGS) of the ribozyme to a 3 nt long ‘tag’ sequence of the substrates.7 Different IGS-tag combinations result in autocatalytic sets that differ in their dynamics and ability to reproduce.5 However, open-ended evolution further requires self-reproducing RNAs to generate novel sequences. Group I intron ribozymes are promising candidates8–10 for sequence diversification as they catalyze the recombination of other RNAs in addition to their own reproduction.
Here, we characterize sequence variation occurring through the combination of two mechanisms catalyzed by the self-reproducing Azoarcus ribozyme. A first mechanism, here called repeated transfer (RT), allows covalent additions of multiple RNA fragments to the 3′-end of an RNA substrate (RNA1, Fig. 1), leading to RNA elongation from 26 nt to at least 136 nt. A second mechanism, here called terminal strand attack (TSA), results in the ligation of hybridized RNAs (RNA2, Fig. 1) into hairpin and circular products. RT and TSA can be combined (RNA3, Fig. 1), leading to diversification of the RNA products together with ribozyme self-reproduction. For further mechanistic details, see ESI.† Furthermore, the reaction network dynamics parameterized from gel data can be integrated in a kinetic model that predicts species distributions revealed by sequencing (ESI†).
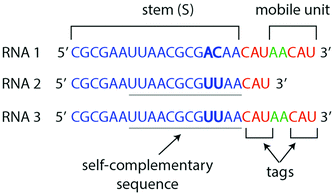
First, we show that RNA1 is elongated by multiple additions of small fragments in the presence of the ribozyme. RNA1 is composed of an 18 nt “stem” (S), a 3 nt tag, and a 5 nt “mobile unit” (Fig. 1). We observe a variety of products denoted Sn, which length corresponds to the addition of n mobile units appended to the stem. Polyacrylamide gel images reveal four bands (Fig. 2a) after 1 hour of reaction, corresponding to S0 (21 nt), S1 (26 nt), S2 (31 nt) and S3 (36 nt). The concentrations of putative S0, S1 and S2 species were measured over 7 h by gel electrophoresis, showing the transient formation of S2 during the first 4 h (Fig. 2b). Sequencing after 45 min of incubation confirmed that RNA sequences result from iterative additions of mobile units (Fig. 2c and see Fig. S2 for all Sn species detected, ESI†). Such additions can be explained by formerly identified reactions catalyzed by the Azoarcus ribozyme,7 here performed in 2 steps: first forward, then reverse after substrate exchange (Fig. 2d and see ESI† for details). Concentrations could be quantified by sequencing up to S7, and up to S23 sequences were detected (Fig. S2, ESI†). Sn concentration decreases exponentially with n (Fig. 2c) consistent with earlier theoretical predictions.11 We devised a kinetic model comprising the transfer reactions Sn + Sm → Sn+p + Sm−p, using the same rate k for all n, m and p, p ≤ m, and the degradation rates of stems and of mobile units (see “Model for RT” in ESI†). These 3 parameters were fitted using S0, S1 and S2 concentration at 0, 1, 3, 5 and 7 h from PAGE analysis (ESI,†Fig. 2b). The model reproduced the concentration time courses: S1 concentration decreases rapidly, S2 concentration reaches a maximum at around 45 min and decreases after 3 h of reaction, and S0 concentration increases until reaching a plateau after 3 h and subsequently slowly decreases. Without further fitting, the model predicted an exponential decrease of product frequencies as a function of their length at 45 min of reaction, as observed experimentally (Fig. 2c). From S1 to S7, the measured and predicted decay factor ln(Sn/Sn+1), with Sn being the fraction of Sn, agree quantitatively (−1.569 ± 0.057 and −1.589, respectively), with a correlation R2 = 0.988 (p < 10−5) between measured and predicted frequencies (Fig. S3, ESI†).
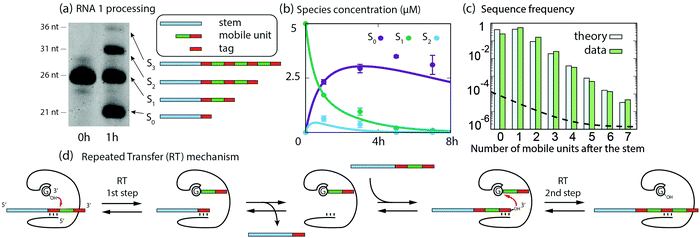 | ||
| Fig. 2 Ribozyme-based RNA elongation by RT. (a) Gel image showing the length diversity of products after 1 h of reaction (RNA1 with Azoarcus ribozyme). (b) Evolution of the concentrations of RNA1 (S1), products without a mobile unit (S0), and products with two mobile units (S2): experimental values from gel band intensities (dots) and kinetic model predictions (solid lines). Error bars are standard deviations from triplicates. (c) Fraction of products versus number of mobile units, after 45 min of reaction: RNA sequencing data (green bars) and kinetic model (blue bars, ESI†) fitted to gel electrophoresis data. The sequencing data is calibrated for length biases due to sequencing library preparation (see Fig. S1, ESI†). The dashed horizontal line indicates the calibrated limit of detection (materials and methods, ESI†). (d) Schematic of the RT mechanism accounting for the elongation of RNA1. | ||
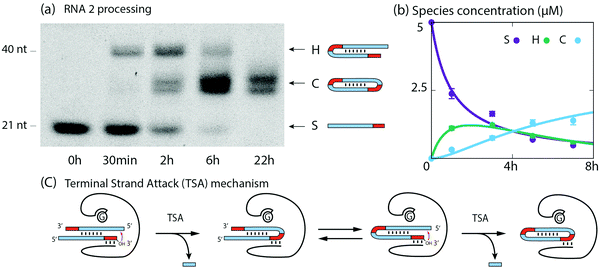
Next, we show that RNA2 undergoes catalytic dimerization and circularization in the presence of the ribozyme via the TSA mechanism. RNA2 is composed of an 18 nt long stem (S) with a a 12 nt self-complementary region and a 3 nt tag (Fig. 1). Over 22 h, polyacrylamide gels (PAGE) indicate the transient formation of almost twice longer RNAs and the appearance of lower bands assigned to circular RNAs (Fig. 3a). The formation of hairpins (H) was confirmed by sequencing and of circular RNA (C) by differential mobility assays with 12% and 18% polyacrylamide gels12 (Fig. S4, ESI†). Hairpins and cyclic RNAs result from the hybridization of two RNA2 molecules via their self-complementary region and by the ribozyme catalyzed TSA reaction, enabled by the presence of 3′ tags (Fig. 3c, see ESI† for details). We devised a kinetic model of the two TSA reactions S + S → H and H → C with rate constants k1 and k2. We fitted these 2 parameters to the concentration of S, H and C at 0, 1, 3, 5 and 7 h of reaction from PAGE gel data. Theoretical and experimental concentration time courses of the three species agree over 7 h of reaction (Fig. 3b): S is roughly halved in the first hour, then decreases more slowly, H increases to a maximum after 2 h and then decreases to reach the same concentration as S, while C continuously increases during the 7 h of reaction.
Thereafter, we show the combination of ribozyme catalyzed elongation, dimerization, and circularization using RNA3 as a substrate. RNA3 combines RNA2 self-complementary stem with RNA1 mobile unit (Fig. 1), thus can undergo both RT and TSA. Over 22 h, species S0 and S2 were produced transiently along with hairpin RNAs, followed by the production of circular RNAs (Fig. 4a) confirmed by RNA sequencing and gel mobility assay (Fig. S4, ESI†). Sequencing revealed a diversity of RNAs after 45 min: Hm,n, standing for hairpin (H) products with m and n mobile units after the first and the second stems, respectively (Fig. 4b and see Fig. S2 for all Sn species detected, ESI†). The 5 parameters of the combined models described above for RNA1 and RNA2 were fitted (Table S2, ESI†) to the concentration of S0, S1, S2 and H (the sum of all Hm,n) measured on gel over 7 h of reaction starting with RNA3 (Fig. S5, ESI†). The model was parameterized only with the gel data, which allowed quantification of circular RNAs absent from the sequencing data. The predicted proportions of Sn (with 0 ≤ n ≤ 5) and Hm,n (with 0 ≤ m, n ≤ 3) at 45 min reproduce the trends of each category of products (separated by horizontal dotted lines in Fig. 4b). Like for RNA1, Sn species decrease exponentially with their size, the measured and predicted decay factor being −1.74 ± 0.09 and −1.65, respectively. This trend is conserved with Hm,n species: at a given m (from 0 to 3), Hm,n species also decrease exponentially with n (from 1 to 3) with a similar exponential decay factor as Sn species (see “Model for RT + TSA” in ESI†). The correlation factor between measured and predicted frequencies is R2 = 0.940 (p < 10−5, Fig. S3, ESI†).
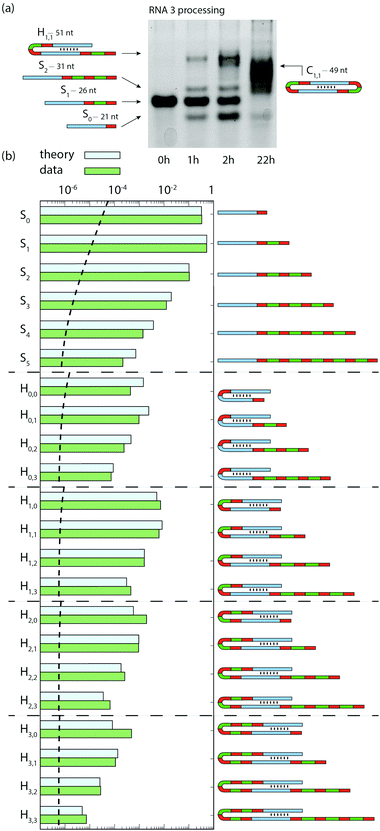 | ||
| Fig. 4 Combination of RT and TSA reactions. (a) Gel image showing the bands corresponding to the addition of one mobile unit and hairpin formation (from RNA3 substrate). At 22 h, diverse products longer than the substrate appear, among which hairpins and cyclic RNAs. (b) Relative proportion of expected products at 45 min measured by RNA sequencing compared to the predictions from the model, independently parameterized with gel data. Measured frequencies are calibrated for length biases (Fig. S1, ESI†). The dashed horizontal line indicates the calibrated limit of detection (materials and methods, ESI†). | ||
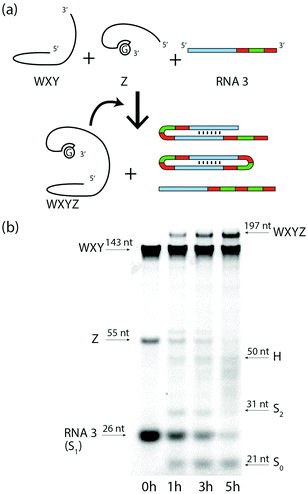
Finally, we show that the Azoarcus ribozyme (noted WXYZ, where the letters refer to 4 contiguous parts, Fig. S6 ESI†)4 self-reproduces from two fragments (WXY and Z) and simultaneously processes the RNA3 substrate through RT and TSA (Fig. 5a). We observed by PAGE the appearance of the band corresponding to the covalent Azoarcus ribozyme as well as those corresponding to the products of TSA and RT mechanisms on RNA3 (mainly S1, S2 and H) over 5 h of reaction (Fig. 5b). As bands characteristic of cyclic RNA could not be confidently detected in the product smear, we confirmed separately the ability of a WXY and Z mixture to generate a cyclic RNA from RNA2 (Fig. S7, ESI†). The concentration dynamics of S0, S1, S2 and H are well described by the same model as for RNA3, and the same fitting procedure results in slightly reduced parameter values, which reflects the inhibitory effect of free Z strands and the lower catalytic efficiency of the WXY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z non-covalent complex5 (Fig. S8, ESI†). The concentrations of WXY, Z and WXYZ were used to fit a set of kinetic equations for Azoarcus self-reproduction, comprising formation of the WXY
Z non-covalent complex5 (Fig. S8, ESI†). The concentrations of WXY, Z and WXYZ were used to fit a set of kinetic equations for Azoarcus self-reproduction, comprising formation of the WXY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z complex and covalent bond formation WXY
Z complex and covalent bond formation WXY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z → WXYZ catalyzed by both WXY
Z → WXYZ catalyzed by both WXY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Z and WXYZ.13 The kinetics show that recombination reactions involving RNA3 reduce the yield of self-reproduction of only ∼10% at 7 h, despite RNA3 being initially 10 times more concentrated than ribozyme fragments (Fig. S9, ESI†).
Z and WXYZ.13 The kinetics show that recombination reactions involving RNA3 reduce the yield of self-reproduction of only ∼10% at 7 h, despite RNA3 being initially 10 times more concentrated than ribozyme fragments (Fig. S9, ESI†).
This study shows mechanisms for ribozyme-mediated RNA sequence diversification, hairpin formation, circularization, extension, self-reproduction, and their integration in reaction networks. We have furthermore shown that the dynamics of these networks can be modeled quantitatively and that such a model predicts the diversity and concentration of products from the knowledge of substrate sequences. The ability of the Azoarcus ribozyme to catalyze its own formation from smaller fragments, diversify and elongate other fragments in an energy-neutral fashion is a notable advantage in the origin of life context. Furthermore, mechanisms for RNA elongation and structure formation are crucial to generate functional RNAs. Circularizing RNA could have been a way to store primordial genetic information,13 thus favoring heredity as in viroids,14 also of interest for the synthesis of cyclic siRNA for interfering therapies.15 This study overall paves the way to engineer evolvable RNA systems by combining mechanisms enabling reproduction, diversification, and structure formation.
Funding from CEFIPRA, HFSP GRY0077/2019, ANR-10-IDEX-0001-02 PSL, IPGG ANR-10-LABX-31, ANR-10-EQPX-34, U. Paris, ED FIRE-Bettencourt, ERC grant agreement No. [101002075].
Conflicts of interest
There are no conflicts to declare.References
- M. P. Robertson and G. F. Joyce, Cold Spring Harbor Perspect. Biol., 2012, 4, a003608 CrossRef PubMed.
- J. Attwater, A. Raguram, A. S. Morgunov, E. Gianni and P. Holliger, eLife, 2018, 7, 1–25 CrossRef PubMed.
- N. Paul and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12733–12740 CrossRef CAS PubMed.
- E. J. Hayden and N. Lehman, Chem. Biol., 2006, 13, 909–918 CrossRef CAS PubMed.
- S. Ameta, S. Arsène, S. Foulon, B. Saudemont, B. E. Clifton, A. D. Griffiths and P. Nghe, Nat. Commun., 2021, 12, 842 CrossRef CAS PubMed.
- E. J. Hayden, G. Von Kiedrowski and N. Lehman, Angew. Chem., Int. Ed., 2008, 47, 8424–8428 CrossRef PubMed.
- W. E. Draper, E. J. Hayden and N. Lehman, Nucleic Acids Res., 2008, 36, 520–531 CrossRef CAS PubMed.
- M. D. Been and T. R. Cech, Science, 1988, 239, 1412–1416 CrossRef CAS PubMed.
- B. M. Chowrira, A. Berzal-Herranz and J. M. Burke, EMBO J., 1993, 12, 3599–3605 CrossRef CAS PubMed.
- A. J. Zaug and T. R. Cech, Science, 1985, 229, 1060–1064 CrossRef CAS PubMed.
- A. Blokhuis and D. Lacoste, J. Chem. Phys., 2017, 147, 094905 CrossRef PubMed.
- T. R. Cech, A. J. Zaug and P. J. Grabowski, Cell, 1981, 27, 487–496 CrossRef CAS PubMed.
- J. Demongeot and A. Moreira, J. Theor. Biol., 2007, 249, 314–324 CrossRef CAS PubMed.
- M. C. Maurel, F. Leclerc, J. Vergne and G. Zaccai, Viruses, 2019, 11, 1–17 CrossRef PubMed.
- N. Abe, H. Abe and Y. Ito, Curr. Protoc. Nucleic Acid Chem., 2012, 1, 1–11 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc02290c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |