Specific enrichment and glycosylation discrepancy profiling of cellular exosomes using a dual-affinity probe†
Ning
Zhang
ab,
Xufang
Hu
b,
Haolin
Chen
b,
Chunhui
Deng
 *ab and
Nianrong
Sun
*ab and
Nianrong
Sun
 *a
*a
aDepartment of Gastroenterology and Hepatology, Zhongshan Hospital, Fudan University, Shanghai, 200032, China
bDepartment of Chemistry, Fudan University, Shanghai, 200433, China. E-mail: chdeng@fudan.edu.cn; sunnianrong@fudan.edu.cn
First published on 24th May 2021
Abstract
A hydrophilic probe is employed to enrich exosomes from three kinds of cancer cells by TiO2–phosphate interaction and exosomal glycoproteins by hydrophilic interaction in succession. The probe performs efficiently in both the enrichment processes. And the analytical results confirm that unique exosomal glycoproteins can distinguish parent exosomes from others.
Exosomes are 40–160 nm vesicles with a phospholipid bilayer membrane secreted by nearly all component cells.1 Recently, exosomes have remarkably shown their unique function in human physiological reactions, especially in the occurrence and deterioration of diseases.2–4 It has been strongly indicated that exosomal contents present high information consistency with parental cells like tumour cells, and thus have great potential in disease diagnosis and prognosis.5–8 For exploiting this potential, exosome separation from complex samples becomes the key procedure. On the other hand, some conventional separation methods confront many problems including long-time consumption and high cost. Moreover, the acquired exosomes are normally impure and disruptive. All these extremely restrict the high-throughput analysis of exosomes to a large extent, let alone exosomal contents. As a consequence, it's a critical to exploit new methods with low cost and high efficiency for separation of intact exosomes.
Studies have pointed out that more than half of the proteins in the human body are glycosylated, which are involved in numerous physical processes such as human immune response, cell recognition, and tumor invasion.9–12 More importantly, most of the disease markers with great potential are glycoproteins.13 For the importance of glycosylation, glycans attached to proteins take the lion's share of credit. Analogically, it has been revealed that glycoproteins are important components of exosomes especially their membrane surface, leading exosomes to participate in multitudinous physical activities. In addition, glycoproteins are also involved in the regulation of secretion and uptake of exosomes.14 Therefore, exosomal glycoprotein profiling is significant for disclosing the biological function of exosomes in organisms and promoting their clinical application. However, the absence of exosome separation methods with low cost and high efficiency results in the difficulty of profiling exosomal glycoproteins. As a result, particular information about exosomal glycoproteins has scarcely been acquainted so far.
In this study, a new strategy which can go straight to meet head-on the above problem including exosome separation and exosomal glycoprotein analysis was proposed. A dual-affinity probe (denoted Fe3O4@TiO2-GSH) was first fabricated by briefly coating titanium oxide on magnetic nanoparticles and grafting hydrophilic glutathione (GSH) in order. Titanium oxide is a widely applied nanomaterial in phosphoprotemics research, adopting the separation mechanism of metal oxide affinity chromatography. In this work, in view of its excellent separation performance for phosphoproteomes, titanium oxide was expected as the functional unit to separate exosomes by binding the phosphate group of the exosomal phospholipid bilayer. Moreover, GSH was expected to exert a subsidiary separation effect through its hydrophilic interactions with the phosphate group. And magnetic nanoparticles were expected to shorten the separation time, ensuring the reduction of the possibility of damage to exosomes. Further, the dual-affinity probe was employed to enrich glycopeptides from exosome lysate digests based on hydrophilic interactions between GSH and glycopeptides, followed by LC-MS/MS analysis. In order to test its availability, the exosomes secreted by different cells were separated and the related glycoproteins were analyzed. As expected, the dual-affinity probe exhibited good performance and achieved a “one for two” goal of separation of exosomes derived from different cells and analysis of exosomal glycoproteins. Furthermore, glycosylation discrepancy profiling between exosomes derived from these different cells reveals the significance of exosomes and exosomal glycoproteins in distinguishing their parent cells.
The synthesis procedure of Fe3O4@TiO2-GSH is shown in Fig. 1a. A TiO2 shell wrapped around a magnetic core (Fe3O4) through hydrolysis reaction of titanium butoxide. And GSH was modified onto the TiO2 shell through the Ti–S bond. The morphological structure of Fe3O4@TiO2-GSH was characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). From Fig. S1a and b (ESI†), with excellent uniformity and dispersion of materials, the diameter of the nanoparticles is found to be approximately 300 nm constituted by a 250 nm magnetic core and a 50 nm TiO2 shell. The zeta potential values of Fe3O4, Fe3O4@TiO2 and Fe3O4@TiO2-GSH changed obviously (Fig. S2, ESI†), ascribed to the different metal surface charges and the negative groups of GSH modification. The X-ray photoelectron spectroscopy (XPS) spectra in Fig. S1c (ESI†) disclosed that the peaks of N 1s and S 2p3 of Fe3O4@TiO2-GSH were not present in Fe3O4@TiO2, and the high-resolution XPS spectrum of Ti 2p from Fe3O4@TiO2-GSH in Fig. S1d (ESI†) also indicated the bonding mode of GSH and the nanoparticles (Ti-S at 456.6 eV), indicating the successful modification of GSH. Besides, the magnetic saturation value of Fe3O4@TiO2-GSH was tested to be 66.0 emu g−1 (Fig. S3, ESI†), indicating its excellent magnetic responsiveness which can guarantee rapid separation in a solid–liquid system. The contact angles of Fe3O4@TiO2 and Fe3O4@TiO2-GSH were 29.2° and 20.8°, respectively (Fig. S4, ESI†), in which the obvious improvement is attributed to the modification of hydrophilic GSH. The great hydrophilicity is beneficial to capture exosomes from biological samples, and is also essential for glycopeptide enrichment.
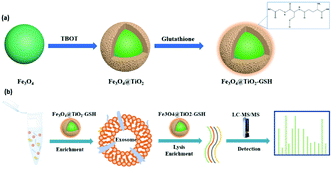 | ||
| Fig. 1 (a) Synthetic route to Fe3O4@TiO2-GSH; and (b) the workflow of exosome enrichment and glycopeptide enrichment using Fe3O4@TiO2-GSH. | ||
With incubation in conditioned medium without fetal bovine serum for 24 h, the late supernatants of MCF-7 cells, MDA-MB-231 cells and HeLa cells were collected as exosome samples. Following a previous report,15 the collected exosome samples were filtered using 0.22 μm filters and concentrated with 100 kDa ultrafiltration tubes to remove cells, cell debris and apoptotic blebs. Then according to the abstract workflow in Fig. S5 (ESI†), pre-treated samples were incubated with Fe3O4@TiO2-GSH at 4 °C for 10 min, during which exosomes were captured by Fe3O4@TiO2-GSH via the interaction between titanium dioxide and the phosphate groups of the phospholipid bilayer on exosomes. Subsequently, exosomes were eluted from nanoparticles using 0.4 M ammonia aqueous buffer followed by rapid solid–liquid magnetic separation. Then buffer substitution with PBS via ultrafiltration was implemented with the aim of decreasing the structural destruction caused by high pH.
Many necessary characterization techniques were employed to evaluate the feasibility of Fe3O4@TiO2-GSH for exosome isolation. For instance, the typical morphological structure of exosomes after negative staining, extracted from HeLa cells, MDA-MB-231 cells and MCF-7 cells, were characterized visually by TEM. As shown in Fig. 2a–c, apart from the background difference that may be caused by instrument error or uneven dyeing, there is no obvious difference in terms of flat-cup appearance among the three kinds of exosomes. The sizes of the HeLa cell and MDA-MB-231 cell derived exosomes were similar, but MCF-7 cell-derived exosomes were slightly smaller. The above results preliminarily proved the stable performance of Fe3O4@TiO2-GSH in exosome separation. Then, western blot analysis of the TSG101 protein (a common antigen marker of exosomes) was carried out for exosome identification. As shown in Fig. S6 (ESI†), the TSG101 protein was present in all three kinds of exosomes, and the concentration of the TSG101 protein in MDA-MB-231-derived exosomes was slightly lower than those of the other two. It can be seen that the discrepancy among the three kinds of exosomes began to appear to a certain extent.
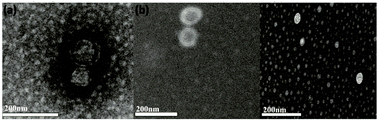 | ||
| Fig. 2 TEM images of HeLa cell (a), MDA-MB-231 cell (b) and MCF-7 cell (c) derived exosomes isolated with Fe3O4@TiO2-GSH. | ||
Western blotting was also used for semi-quantitative analysis of HeLa cell derived exosomes to assess the exosome isolation efficiency of Fe3O4@TiO2-GSH. As illustrated in Fig. S7a (ESI†), the bands of TSG101 were almost invisible in the supernatant and the first washing with buffer after enrichment, while the other two bands of the original culture medium and eluent presented similar densities, indicating a high capture efficiency of Fe3O4@TiO2-GSH. Furthermore, the reusability of the probe was investigated through continuous enrichment application. In Fig. S7b (ESI†), there was almost no difference between the freshly prepared probe and that recycled five times in the western blot result of MCF-7-exosomes. In addition, fluorescence detection was used for evaluating exosome recovery according to a previous report.16 Namely, the lipid bilayers of exosomes were tagged with fluorescent carbocyanine dye (DiI), then enriched with Fe3O4@TiO2-GSH. By comparing the relative fluorescence values in Fig. S8 (ESI†), the separation efficiencies of the exosomes increased first and then remained constant with increasing time or material dosage. The inflection point of the curve was regarded as the optimal conditions (40 μg exosomes incubated with 2.5 mg Fe3O4@TiO2-GSH for 10 min). With the optimal conditions, the recovery of exosomes reached up to 90.8% (nine parallel experiments, Fig. S8d, ESI†) by calculating the ratio of the fluorescence intensities at 570 nm between the initial sample and eluent.
In this work, we planned to enrich and analyze glycopeptides belonging to exosomes via a bottom-up strategy; therefore, the enrichment ability of Fe3O4@TiO2-GSH for glycopeptides was also tested. We first adopted a standard glycoprotein (horseradish peroxidase, HRP, 100 fmol μL−1) digest as a sample to optimize the incubation conditions for glycopeptides. A loading buffer of 90%ACN/3%TFA/7%H2O (v/v/v), an elution buffer of 50% ACN aqueous solution, 30 min incubation and 45 min elution were finally selected, see Fig. S9 (ESI†). Compared with Fe3O4@TiO2, more peaks with higher signal to noise ratios (SNRs) were detected after enrichment with Fe3O4@TiO2-GSH (Fig. S10, ESI†), which indicated the significance of GSH modification. Totally, 22 glycopeptides from the HRP tryptic digest were detected (Table S1, ESI†). Additionally, when the concentration of the HRP digest was as low as 0.5 fmol μL−1, five target peaks still showed significant signal intensities (Fig. S11b, ESI†). When using a tryptic digest mixture of HRP and non-glycoprotein BSA (albumin from bovine serum) as a sample (HRP/BSA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), there were 17 target peaks detected with high SNR levels after enrichment with Fe3O4@TiO2-GSH (Fig. S12, ESI†). These results indicated that Fe3O4@TiO2-GSH exhibited high sensitivity and selectivity. Furthermore, the reusability of the dual-affinity probe was also investigated during the glycopeptide enrichment process, see Fig. S13 (ESI†). After exosome isolation, the reused materials showed an excellent result of glycopeptide enrichment. And the intensities of the goal peaks with the materials recycled third times were lower than the former but still remained at good levels, while several MS peaks were missed with the particles in the fifth recycle whose affinity decreased a bit after four adsorption–elution recycles.
100), there were 17 target peaks detected with high SNR levels after enrichment with Fe3O4@TiO2-GSH (Fig. S12, ESI†). These results indicated that Fe3O4@TiO2-GSH exhibited high sensitivity and selectivity. Furthermore, the reusability of the dual-affinity probe was also investigated during the glycopeptide enrichment process, see Fig. S13 (ESI†). After exosome isolation, the reused materials showed an excellent result of glycopeptide enrichment. And the intensities of the goal peaks with the materials recycled third times were lower than the former but still remained at good levels, while several MS peaks were missed with the particles in the fifth recycle whose affinity decreased a bit after four adsorption–elution recycles.
Inspired by the above results, Fe3O4@TiO2-GSH was further applied to enrich different cell-derived exosomes and N-glycopeptides from exosome digests. As a result, 531 N-glycopeptides corresponding to 186 N-glycoproteins from HeLa-exosomes, 659 N-glycopeptides corresponding to 242 N-glycoproteins from MDA-MB-231-exosomes and 754 N-glycopeptides corresponding to 256 N-glycoproteins from MCF-7-exosomes were identified using LC-MS/MS (listed in the ESI†). And from Fig. S14 (ESI†), it is clear that the commonly involved KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways of the three exosomes were ECM–receptor interaction and the PI3K-Akt signaling pathway. As is well known, the occurrence and metastasis of malignant tumors are often accompanied by abnormal changes in the expression of the extracellular matrix (ECM) and its cell surface receptors.17 The PI3K-Akt signaling pathway is also closely related to tumor occurrence, malignant transformation of cells, and even tumor angiogenesis.18 Therefore, the study on exosomal glycoproteins is quite significant for providing key information about the tumor mechanism.
Additionally, the glycoproteins presented in the results of the three experiments with 100% probability were chosen to perform further discrepancy analysis. As shown in Fig. 3a, three obviously segregated clusters were displayed in the results of principal component analysis (PCA) based on label-free quantification (LFQ) results and statistical analysis, and there are significant differences among glycoproteins expressed in the three kinds of exosomes. Also, the heatmap of exosomal glycoproteins in Fig. 3b demonstrated intuitively the heterogeneous abundance from different sources. Each row represents the expression level of each glycosylated protein in the different samples, and each column represents the expression level of each protein in a sample. Different shades of color represent protein contents in an exosome sample. Whether belonging to the same disease or not, there were obvious clusters among parts of N-glycoproteins. The quantified comparison in our result showed that CD9 are widely presented on nearly all kinds of exosomes surface but performed a big differentiation of expression among the three kinds of exosomes. Thus it can be seen that even the common glycoproteins also showed quantification differences in different diseases, the deep research into which can act as signs directed to specific physiological environments. The unique N-glycoproteins from different cell-exosomes were further compared. The results showed that the majority of unique glycoproteins from HeLa-exosomes are involved in cell migration and transport, and 80% are membrane-related. The unique N-glycoproteins from MCF-7-exosomes are mainly involved in cell communication and signal transduction, and more than 90% are membrane-related. However, the glycoproteins from MDA-MB-231-exosomes that are membrane related are much fewer, most of which belong to the cytoplasm and vesicles, and the unique N-glycoproteins from MDA-MB-231-exosomes are mainly involved in the protein metabolic process and macromolecule metabolic process. These unique glycoproteins tend to promote the corresponding exosomes to perform different physiological functions, which are likely to be the specific markers that distinguish different parent cells. Subsequently, the relationship between exosomes and their sources was explored by comparing these unique N-glycoproteins from different exosomes. Among the highly expressed unique N-glycoproteins, epidermal growth factor receptor (EGFR) only presented in HeLa-exosomes, and it is closely associated with human papillomavirus (HPV) infection which is considered an etiological factor in the development of cervical cancer.19 This result indicated that exosomal glycoproteins can exactly reflect the information about parent cells, which will be promising candidates for identifying cancer patients. Similar to EGFR, the tissue factor pathway inhibitor and folate receptor α associated with cervical carcinogenesis in previous reports also only presented in HeLa-exosomes, based on which we can distinguish HeLa-exosomes from MDA-MB-231-exosomes and MCF-7-exosomes.20,21 In addition, many exosomal glycoproteins such as neural cell adhesion molecule 2 are observed both in MDA-MB-231 cells and in MCF-7 cells; this is supposed to be because they belong to the same disease.22 However, some N-glycoproteins, such as different types of integrin and factor receptors, display obvious quantitative differentiation in their exosomes, indicating that different exosomal glycoproteins can accurately match with different tumor cells of the same cancer. These results suggested that it's possible to realize precise classification of tumor cells based on exosomal glycoproteins. In summary, huge differences of exosomes can be obtained through exosomal N-glycoprotein profiling, which may contribute to identifying exosomes from specific sources and exploring the physical mechanism by monitoring the respective abnormal glycosylation.
In conclusion, a dual-affinity probe was prepared to realize efficient separation of exosomes and exosomal glycopeptides consecutively, in which the interaction between TiO2 and the phosphate groups of the exosomal membrane and the excellent hydrophilicity introduced by GSH are the main contributors. With great superparamagnetism, high affinity and stable properties, Fe3O4@TiO2-GSH presented good exosome recovery in three kinds of cancer cell samples. By combining the bottom-up strategy with LC-MS/MS and enrichment with the probe, 186 N-glycoproteins from HeLa-exosomes, 242 N-glycoproteins from MDA-MB-231-exosomes and 256 N-glycoproteins from MCF-7-exosomes were identified. Three types of exosomes can be clearly distinguished based on the analysis of N-glycoproteins. And some unique glycoproteins involving different signaling pathways show potential in performing different biological functions. The analysis of these N-glycoproteins can reflect the pathological states of their parent cells. Owing to its low cost and stable properties, our strategy for consecutive enrichment of exosomes and glycopeptides is likely to be applied in complicated biological samples to analyze specific exosomes and contribute to studying the physical mechanism in depth.
This work was financially supported by the National Key R&D Program of China (2018YFA0507501), the National Natural Science Foundation of China (22074019, 21425518, 22004017), and the Shanghai Sailing Program (20YF1405300).
Conflicts of interest
There are no conflicts to declare. All the experiments in this work were carried out in compliance with the ethical standards, and conducted according to the Declaration of Helsinki and approved by the Ethics Committee of Fudan University. All volunteers gave their consent.Notes and references
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, 640–655 CrossRef PubMed.
- S. Yoon, A. Kovalenko, K. Bogdanov and D. Wallach, Immunity, 2017, 47, 51–65 CrossRef CAS PubMed.
- B. Y. Shen, Y. Fang, N. Wu and S. J. Gould, J. Biol. Chem., 2011, 286, 44162–44176 CrossRef CAS PubMed.
- N. Iraci, T. Leonardi, F. Gessler, B. Vega and S. Pluchino, Int. J. Mol. Sci., 2016, 17, 171 CrossRef PubMed.
- M. Alexander, R. Z. Hu, M. C. Runtsch, D. A. Kagele, T. L. Mosbruger, T. Tolmachova, M. C. Seabra, J. L. Round, D. M. Ward and R. M. O'Connell, Nat. Commun., 2015, 6, 7321 CrossRef CAS PubMed.
- T. Skotland and K. Sandvig, Nat. Commun., 2019, 10, 2752 CrossRef PubMed.
- C. Thery, L. Zitvogel and S. Amigorena, Nat. Rev. Immunol., 2002, 2, 569–579 CrossRef CAS PubMed.
- D. D. Taylor and C. Gercel-Taylor, Semin. Immunopathol., 2011, 33, 441–454 CrossRef CAS PubMed.
- N. R. Sun, H. L. Yu, H. Wu, X. Z. Shen and C. H. Deng, TrAC, Trends Anal. Chem., 2021, 135, 116168 CrossRef CAS.
- C. C. Xu and D. T. W. Ng, Nat. Rev. Mol. Cell Biol., 2015, 16, 742–752 CrossRef CAS PubMed.
- R. G. Spiro, Glycobiology, 2002, 12, 43R–56R CrossRef CAS PubMed.
- P. M. Rudd, T. Elliott, P. Cresswell, I. A. Wilson and R. A. Dwek, Science, 2001, 291, 2370–2376 CrossRef CAS PubMed.
- R. Huettenhain, M. Choi, L. M. de la Fuente, K. Oehl, C. Y. Chang, A. K. Zimmermann, S. Malander, H. Olsson, S. Surinova, T. Clough, V. Heinzelmann-Schwarz, P. J. Wild, D. M. Dinulescu, E. Nimeus, O. Vitek and R. Aebersold, Mol. Cell. Proteomics, 2019, 18, 1836–1850 CrossRef PubMed.
- H. L. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed.
- N. Zhang, N. R. Sun and C. H. Deng, Talanta, 2021, 221, 121571 CrossRef CAS PubMed.
- N. Zhang, N. R. Sun and C. H. Deng, Chem. Commun., 2020, 56, 13999–14002 RSC.
- V. Luga, L. Zhang, A. M. Viloria-Petit, A. A. Ogunjimi, M. R. Inanlou, E. Chiu, M. Buchanan, A. N. Hosein, M. Basik, J. L. T. Wrana, V. Heinzelmann-Schwarz, P. J. Wild, D. M. Dinulescu, E. Nimeus, O. Vitek and R. Aebersold, Mol. Cell. Proteomics, 2019, 18, 1836–1850 CrossRef PubMed.
- B. M. Slomovitz and R. L. Coleman, Clin. Cancer Res., 2012, 18, 5856–5864 CrossRef CAS PubMed.
- T. Soonthornthum, H. Arias-Pulido, N. Joste, L. Lomo, C. Muller, T. Rutledge and C. Verschraegen, Ann. Oncol., 2011, 22, 2166–2178 CrossRef CAS PubMed.
- Q. Zhang, Y. Zhang, S. Wang, N. Wang, W. Jiang, Y. Ji and S. Zhang, J. Exp. Clin. Cancer Res., 2012, 31, 1 CrossRef PubMed.
- C. Liu, L. Ding, L. Bai, X. Chen, H. Kang, L. Hou and J. Wang, Biochem. Biophys. Res. Commun., 2017, 491, 1083–1091 CrossRef CAS PubMed.
- S. Takahashi, K. Kato, K. Nakamura, R. Nakano, K. Kubota and H. Hamada, Cancer Sci., 2011, 102, 808–814 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc01530c |
| This journal is © The Royal Society of Chemistry 2021 |

