 Open Access Article
Open Access ArticleSynthesis of redox-active fluorinated 5-hydroxytryptophans as molecular reporters for biological electron transfer†
Amanda
Ohler‡
a,
Hanna
Long‡
a,
Kei
Ohgo
a,
Kristin
Tyson
a,
David
Murray
a,
Amanda
Davis
a,
Chris
Whittington
a,
Eli G.
Hvastkovs
a,
Liam
Duffy
b,
Alice
Haddy
b,
Andrew L.
Sargent
a,
William E.
Allen
a and
Adam R.
Offenbacher
 *a
*a
aDepartment of Chemistry, East Carolina University, Greenville, NC, USA. E-mail: offenbachera17@ecu.edu
bDepartment of Chemistry and Biochemistry, University of North Carolina, Greensboro, NC, USA
First published on 11th February 2021
Abstract
Fluorinated 5-hydroxytryptophans (Fn-5HOWs) were synthesized in gram scale quantities and incorporated into a β-hairpin peptide and the protein azurin. The redox-active Fn-5HOWs exhibit unique radical spectroscopic signatures that expand the function of 5HOW as probes for biological electron transfer.
Proton-coupled electron transfer (PCET) is a ubiquitous and fundamental process in biological energy conversion and catalysis. In addition to metals, proteins exploit their natural amino acid building blocks, primarily tyrosine (Y) and tryptophan (W), to shuttle electrons, sometimes reversibly, over long distances.1,2 The mechanistic exploration of tyrosine-mediated PCET has been advanced by the advent of unnatural amino acids (UAAs), most notably fluoro- and nitro-tyrosine, that have altered redox properties to enable the ‘trapping’ of organic radicals associated with PCET reactions.3 Despite its low abundance in nature, tryptophan is particularly appealing because it has a chemically complex and electron rich ring structure that is expected to serve as a vital intermediary in long-range electron hopping pathways, including ribonucleotide reductase, cytochrome c oxidase, DNA photolyase, and peroxidases.2 Tryptophan wires, found abundantly in oxidoreductases, have been implicated in mitigating oxidative damage.4 Mechanistic details of Trp-mediated electron transfer in select systems are not well resolved, in part due to a lack of tractable UUAs. Further, quantum coherent electron spins between a flavin and tryptophan radical pair in cryptochromes might even be responsible for macroscopic properties including avian migration and plant circadian clocks.5,6 Thus, development of novel tryptophan-based UAAs, with distinct thermodynamic and spectroscopic properties, could have many potential applications, ranging from biophysical probes to protein engineering and peptide-based pharmaceuticals.7
Among available indole rings with subtle ring modifications, 5-hydroxytryptophan (5HOW) was selected for its desirable redox properties (Table S1, ESI†). In this report, we show that oxidation of 5HOW occurs at the hydroxy group, which enables manipulation of its indoxyl pKa and the spectral features of its radical state through introducing a flanking, nuclear spin active fluorine atom, similar to that reported for fluorotyrosines.9,10 Further, we present a facile synthetic approach that leads to high yields (80–90% conversion from the indole) of enantiomerically pure, monofluorinated L-5HOWs and their Fmoc derivatives using a one-pot reaction (Scheme 1). The strategy, which exploits an evolved TrpB synthase,11 marks a significant improvement over conventional synthetic methods for unnatural L-Trp derivatives that require multiple steps and are associated with low yields.12,13 While 5HOW is expected to have different electrostatic properties than Trp (see Fig. S1, ESI†), incorporation of 5HOW into different proteins has been performed using human and E. coli expression systems without significant deleterious effects on protein structure or function.14,15 In former applications, 5HOW has been utilized for its red-shifted electronic and fluorescence spectral features as well as its ability to perform bioconjugation. Results, presented herein, reveal unique thermodynamic and radical spectroscopic properties of Fn-5HOWs. These data implicate, for the first time, that 5HOW can be used as a biophysical probe for electron transfer.
 | ||
| Scheme 1 One-pot chemoenzymatic synthetic strategy to prepare L-Fn-5HOW UAAs and their Fmoc-protected derivatives. The model of the β-hairpin peptide was generated from PepFold.8 The site of 5HOW is shown by cyan sticks. For the peptide sequence, X represents the Fn-5HOWs and O refers to L-ornithine. The azurin structure is from the PDB: 1E67. | ||
We began assessment of the spectroscopic and electrochemical properties of the 5-hydroxyindole (5HOI) functional group. The reduction potential of the 5HOI˙/5HOI couple from square wave voltammetry (SWV) was found to be 565 ± 5 mV at pH 7 (Fig. S2 and Table S2, ESI†). The value is ca. 300 mV lower than Y (928 ± 4 mV) or W (983 ± 5 mV) at the same pH. Mono-fluorination of the 5HOI ring at either the 4 or 6 position (Scheme 1) produces E0 values of 579 ± 6 and 595 ± 4 mV, respectively. The trends, though not the magnitude, of these potentials are consistent with observations from fluorotyrosine9 and track the DFT-calculated ionization potentials (IPs) of Fn-5HOI derivatives (Table S2, ESI†). To explain these trends, the HOMOs, calculated from the geometry optimized structure of 5HOI (Fig. 1), reveal that the positions 4 and 6 in the 5-hydroxyindole ring have unequal π contributions. This potentially serves as a framework to understand the different IPs emerging for the 4F- and 6F-substituted 5HOI species, since the impact from the halide's inductive withdrawing and resonance donating character will be communicated to the π system to differing degrees depending on position.
 | ||
| Fig. 1 HOMO of the ground state for indole and 5HOI (A and B) and Mulliken spin densities for Fn-5HOW radicals (D–F). The spin densities were calculated for (C) W˙, (D) 5HOW˙, (E) 4F-5HOW˙, and (F) 6F-5HOW˙ and the positive Mulliken spin density charges are displayed. | ||
Within the pH dependent range, E0 values for Fn-5HOI show the expected pH dependence of ∼59 mV per pH unit, consistent with loss of a proton coupled with side chain oxidation (Fig. S2, ESI†). At elevated pH values (≥10), the potentials are no longer dependent on pH, consistent with the pKa values of the ground state species. To corroborate this behaviour, the pKa values were determined using UV-visible spectroscopy. The UV-visible spectra of aqueous solutions of Fn-5HOI are shown in Fig. 2A–C. Their electronic absorbance spectra are significantly red-shifted relative to and distinct from Y and W (Fig. 2D and E). The features are dependent upon the presence and positioning of the fluorine atom. Deprotonation of the indoxyl proton causes a ca. 30 nm red shift of the 290 nm feature in the electronic absorption spectrum (Fig. S3, ESI†). From this shifted band, the pKa values of the Fn-5HOI species were then estimated by pH titrations. The pKa of 5HOI was determined as 10.90 ± 0.02, comparable to the literature value (11.1).16 As expected, mono-fluorination of 5HOI led to one-unit decreases in the pKa (Table S2, ESI†).
 | ||
| Fig. 2 UV-vis (A–E) and EPR (F–J) spectra of Fn-5HOI (A–C and F–H), tryptophan (D and I), and tyrosine (E and J). EPR spectra were collected at 77 K post UV photolysis from a pulsed Nd-YAG laser. EPR parameters are presented in the ESI.† For UV, the solutions were prepared at 200 μM in 50 mM potassium phosphate, pH 7. For EPR, the solutions were prepared at 50 mM in 50 mM sodium borate, pH 11. A pre-flash spectrum is shown as a dashed line in (F). | ||
Fig. 2F–J presents X-band EPR spectra at 77 K of photolysis-generated radical species of Y, W, 5HOI, 4F-5HOI, and 6F-5HOI. The 5HOI˙ EPR spectrum (Fig. 2F) exhibits a typical line shape for a g = 2 organic radical with little-to-no hyperfine interaction. Importantly, the EPR spectra of 4F-5HOI˙ (Fig. 2G) and 6F-5HOI˙ (Fig. 2H) are quite distinct from 5HOI˙ and the characteristic features of natural Y and W radicals (Fig. 2I and J). These distinguishing EPR features establish that Fn-5HOIs could serve as useful reporters of Trp radicals. Note that the hyperfine interaction from the 19F nucleus at position 4 (Fig. 2G) with the delocalized electron spin for the 5HOI radical is more prominent than from the fluorine at the 6 position (Fig. 2H). To provide a physical basis for these spectra, DFT calculated spin densities for the 5HOW derivatives (Fig. 1D–F) reveal larger spin density character at the four position of the 5-hydroxyindole ring which, when coupled to the nuclear moment of a fluorine atom substituted at this position, provides a rationale for the observed hyperfine structure in the EPR spectrum for the 4F-5HOI radical system relative to that of 6F-5HOI.
Oxidation of tryptophan occurs by loss of a hydrogen atom (H˙) from the indole N–H bond in a PCET reaction. The spin density of the resulting unpaired electron is primarily spread across the indole ring with some spin density delocalized into the backbone (Fig. 1C). The EPR detected hyperfine interactions and DFT calculations support that oxidation of Fn-5HOIs by photolysis occurs through loss of H˙ at the indole OH bond. Unlike W˙, there is no predicted delocalization of spin density into the backbone for 5HOW˙ (Fig. 1D–F and Fig. S4, ESI†). Consistent with this, EPR spectra of 5HOI and L-5HOW neutral radicals are identical (Fig. S5, ESI†). Therefore, the EPR spectra of Fn-5HOI˙ in Fig. 2H–J are reasonable model spectra.
The Fn-5HOW amino acids were synthesized from Fn-5HOI and L-serine using an engineered, pyridoxal phosphate (PLP)-dependent tryptophan B synthase (TrpB) variant from Thermotoga maritima, Tm9D8*. This TrpB variant was previously found to be highly efficient for the conversion of substituted, noncanonical indole rings and was functionally optimized for 37 °C,17 which provides the ability to prepare redox-active side chains in bacterial cultures. Reversed-phase HPLC analysis was used to determine the conversion of 5HOI (RT, 11 min) to L-5HOW (RT, 5.7 min) (Fig. S6A–F, ESI†). Previous reactions with engineered TrpB variants were carried out for extended times (≥24 h) and with excess L-serine. Herein, we show that with sub-stoichiometric L-serine (0.9 eq.), near complete conversion of 5HOI to 5HOW can be attained within 4 h. At longer reaction times, unwanted biproducts emerged. Using these conditions, high conversions of 4F- and 6F-5HOW were achieved (cf. Fig. S7A–E, ESI†). Thus, we demonstrate an expanded scope of this TrpB variant to these di-substituted indoles.
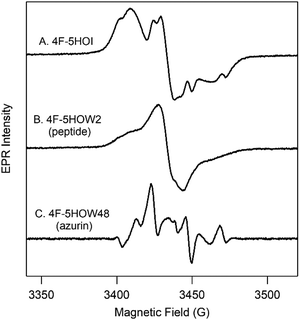 | ||
| Fig. 3 EPR spectra of 4F-5HOI/4F-5HOW radicals in solution (A), peptide (B), and azurin (C). Spectrum in (C) was calculated from background subtraction (Fig. S17, ESI†). | ||
From the optimized Tm9D8* enzymatic reaction, the amino groups of Fn-5HOW were Fmoc protected in the same reaction vessel through the addition of dioxane as a co-solvent (see SI for more details). Prior to adding Fmoc-succinimide (1 eq.), Tm9D8* was removed by Ni-NTA resin and filtration. The as-isolated, Fmoc-protected Fn-5HOW products (RT, 13 min) were obtained in good yield (≥ 90% crude mass) and at ca. 80% purity as judged by HPLC integrations (Fig. S6H–J, ESI†). The final products of newly synthesized Fmoc-4F-5HOW and Fmoc-6F-5HOW were confirmed by 1H and 13C NMR (Fig. S8 and S9, ESI†) and HRMS (Fig. S10, ESI†). To demonstrate the scalability of the two-step synthesis, near quantitative yield of Fmoc-L-5HOW (0.85 g crude; 96%) was isolated from 0.27 g of 5HOI starting material. The crude reaction product was sticky until purified. After purification, the product became an off-white powder and was confirmed by MS and NMR. Similar yields were determined for the fluorinated derivatives.
As proof-of-principle, L-5HOW and L-4F-5HOW were inserted into a designed β-hairpin peptide18 (cf.Scheme 1) using Fmoc solid-state chemistry. One notable exception was the substitution of leucine at position 11 for phenylalanine to model the cross-strand, staggered Trp-Phe π–π stacking interaction in the protein, azurin (see below). The HPLC-purified 5HOW2, 4F-5HOW2, and 6F-5HOW2 peptides are highly soluble in aqueous solutions (≥10 mM) and confer β-hairpin structure at room temperature based on characteristic CD spectra (Fig. S11, ESI†). These peptides exhibit reversible folding as indicated by the reproducible room temperature CD spectra before and after thermal denaturation. The incorporations of Fn-5HOWs are confirmed by HRMS and the UV-visible spectra (Fig. S12A–C, ESI†). It is important to note that the electronic spectra are slightly influenced by the secondary structure, consistent with the observation seen with tryptophan and tyrosine at position 2 in the peptide (Fig. S12D and E, ESI†). The light-generated neutral radical species were characterized by EPR spectroscopy (cf. Fig. S12F–J, ESI†). Qualitatively, the EPR signals of Fn-5HOW radicals in the β-hairpin peptides are comparable to their sidechains (cf.Fig. 2, 3A and B and Fig. S12, ESI†), though the hyperfine interactions are less defined. Similar observations were reported for tyrosyl radicals in other β-hairpins, and have been attributed to the dynamic interactions within the peptide.19,20
To further demonstrate the functionality of Fn-5HOWs as amino acids, these UAAs were incorporated into the robust model protein, azurin. Azurin was chosen because it contains only a single tryptophan residue and only two tyrosines; the latter were mutated to phenylalanine to generate a tyrosine-deficient variant (Y72F and Y108F) to enable direct spectroscopic and electrochemical characterization of the UAAs. Further, the tryptophan (W48) is buried and completely solvent excluded,21 enabling us to test the impact that these more polar 5HOW sidechains have on protein stability. 5HOW was incorporated into azurin using AMBER stop codon suppression at position 48 with an engineered tryptophanyl–tRNA synthetase (TrpRS)/tRNA pair. This strategy resulted in protein yields of 2–5 mg L−1 of pure azurin (Fig. S13, ESI†) with L-5HOW selectively incorporated at position 48 (80–90% incorporation assessed by LC-MS/MS; Table S3, ESI†). Exclusion of either 5HOW or the TrpRS/tRNA pair to the expression system resulted in truncated protein as assessed by SDS-PAGE (not shown). CD spectra of 5HOW48 azurin is nearly identical to the wild-type protein (Fig. S14, ESI†); the CD-derived melting temperature (Tm) of 5HOW48 is 71.9 ± 0.2 °C versus 74.0 ± 0.7 °C for WT azurin (Table S4, ESI†). Contrary to 5HOW solutions, the UV-visible spectrum of 5HOW48 shows some fine structure (cf. Fig. S15, ESI†), consistent with a low dielectric environment (cf. Fig. S16, ESI†).21
Extending this biosynthetic method for the insertion of 4F- and 6F-5HOW in azurin resulted in low yields (1 mg mL−1). For EPR characterization, the Fn-5HOW derivatives were incorporated using a tryptophan auxotroph (20 mg mL−1; see Methods for details); this has been described for 5HOW previously.22 CD and UV spectra demonstrate that these proteins are folded with comparable structures and thermostabilities to that of W48 (Fig. S14, S15 and Table S4, ESI†). Note that 4F-5HOW presents a slightly destabilizing effect, though the protein is still well folded. The corresponding EPR spectra of the light-induced neutral radical species of Fn-5HOW48 are presented in Fig. S15 (ESI†). The EPR spectra of W48˙ has previously been characterized by 9.4 and 700 GHz frequencies.23,24 Compared to W48˙, 4F-5HOW48˙ is also well-resolved but has a distinct EPR line shape (cf.Fig. 3C and Fig. S15, ESI†).
SWV of Fn-5HOW2 peptides and Fn-5HOW48 azurins exhibited reversible voltammograms with comparable reduction potentials to that of 5HOI˙/5HOI couple (Table S5, ESI†). These data validate the redox properties of Fn-5HOWs.
In conclusion, we report a facile, chemoenzymatic synthesis strategy to produce novel, fluorinated 5HOWs on the gram scale. The light-generated radicals of redox-active Fn-5HOWs, especially 4F-5HOW, exhibit compelling spectroscopic features that can be readily distinguished from canonical amino acids. With these unique spectroscopic and thermodynamic properties presented herein, Fn-5HOWs have great potential for the study of Trp-mediated biological PCET and protein engineering of Trp ‘wires’. While 5HOW works well with azurin and other proteins,14,15 careful control experiments to ensure structural and functional integrity are required for future applications.
A. O., K. O., K. T., D. M., A. D., C. W., L. D., and A. H. performed experiments. E. H., A. H., W. A., and A. R. O. analysed data. H. L. and A. S. performed DFT calculations. A. O., H. L., and A. R. O. wrote the text.
The authors thank Prof. Frances Arnold for the plasmids encoding evolved TrpB, Prof. Peter Schultz for plasmids encoding the orthogonal TrpRS/tRNA pair, and Prof. John Latham for help with HRMS data collection. This work was supported by the National Science Foundation (REU 1851844) for A. Ohler and (20-03956) to A. Offenbacher.
Conflicts of interest
There are no conflicts to declare.References
- J. Stubbe and W. A. van der Donk, Chem. Rev., 1998, 98, 705–762 CrossRef CAS.
- J. L. Dempsey, J. R. Winkler and H. B. Gray, Chem. Rev., 2010, 110, 7024–7039 CrossRef CAS.
- E. C. Minnihan, D. G. Nocera and J. Stubbe, Acc. Chem. Res., 2013, 46, 2524–2535 CrossRef CAS.
- J. R. Winkler and H. B. Gray, Q. Rev. Biophys., 2015, 48, 411–420 CrossRef CAS.
- I. Chaves, R. Pokorny, M. Byrdin, N. Hoang, T. Ritz, K. Brettel, L.-O. Essen, G. T. J. van der Horst, A. Batschauer and M. Ahmad, Annu. Rev. Plant Biol., 2011, 62, 335–364 CrossRef CAS.
- P. J. Hore and H. Mouritsen, Annu. Rev. Biophys., 2016, 45, 299–344 CrossRef CAS.
- A. Rezhdo, M. Islam, M. Huang and J. A. Van Deventer, Curr. Opin. Biotechnol., 2019, 60, 168–178 CrossRef CAS.
- P. Thevenet, Y. Shen, J. Maupetit, F. Guyon, P. Derreumaux and P. Tuffery, Nucleic Acids Res., 2012, 40, W288–W293 CrossRef CAS.
- M. R. Seyedsayamdost, S. Y. Reece, D. G. Nocera and J. Stubbe, J. Am. Chem. Soc., 2006, 128, 1569–1579 CrossRef CAS.
- P. H. Oyala, K. R. Ravichandran, M. A. Funk, P. A. Stucky, T. A. Stich, C. L. Drennan, R. D. Britt and J. Stubbe, J. Am. Chem. Soc., 2016, 138, 7951–7964 CrossRef CAS.
- E. J. Watkins-Dulaney, S. Straathof and F. H. Arnold, ChemBioChem, 2020, 22, 5–16 CrossRef.
- C.-H. Chen, S. Genapathy, P. M. Fischer and W. C. Chan, Org. Biomol. Chem., 2014, 48, 9764–9768 RSC.
- K. Boknevitz, J. S. Italia, A. Chatterjee and S.-Y. Liu, Chem. Sci., 2019, 10, 4994–4998 RSC.
- Z. Zhang, L. Alfonta, F. Tian, B. Bursulaya, S. Uryu, D. S. King and P. G. Schultz, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8882–8887 CrossRef CAS.
- P. Addy, S. B. Erickson, J. S. Italia and A. Chatterjee, J. Am. Chem. Soc., 2017, 139, 11670–11673 CrossRef CAS.
- S. V. Jovanovic, S. Steenken and M. G. Simic, J. Phys. Chem., 1990, 94, 3583–3588 CrossRef CAS.
- C. E. Boville, D. K. Romney, P. J. Almhjell, M. Sieben and F. H. Arnold, J. Org. Chem., 2018, 83, 7447–7452 CrossRef CAS.
- C. D. Tatko and M. L. Waters, J. Am. Chem. Soc., 2004, 126, 2028–2034 CrossRef CAS.
- R. Sibert, M. Joscowicz, F. Porcelli, G. Veglia, K. Range and B. A. Barry, J. Am. Chem. Soc., 2007, 129, 4393–4400 CrossRef CAS.
- R. Sibert, M. Joscowicz and B. A. Barry, ACS Chem. Biol., 2010, 5, 1157–1168 CrossRef CAS.
- G. Gilardi, G. Mei, N. Rosato, G. W. Canters and A. Finazzi-Agro, Biochemistry, 1994, 33, 1425–1432 CrossRef CAS.
- C. W. Hogue, I. Rasquinha, A. G. Szabo and J. P. MacManus, FEBS J., 1992, 310, 269–272 CrossRef CAS.
- H. S. Shafaat, B. S. Leigh, M. J. Tauber and J. E. Kim, J. Am. Chem. Soc., 2010, 132, 9030–9039 CrossRef CAS.
- S. Stoll, H. S. Shafaat, J. Krzystek, A. Ozarowski, M. J. Tauber, J. E. Kim and R. D. Britt, J. Am. Chem. Soc., 2011, 133, 18098–18101 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc00187f |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |
