Open Access Article
Open Access ArticleCovalent capture and electrochemical quantification of pathogenic E. coli†
Sarah H.
Klass
 a,
Laura E.
Sofen
a,
Laura E.
Sofen
 a,
Zachary F.
Hallberg
a,
Zachary F.
Hallberg
 b,
Tahoe A.
Fiala
a,
Alexandra V.
Ramsey
a,
Nicholas S.
Dolan
a,
Matthew B.
Francis
b,
Tahoe A.
Fiala
a,
Alexandra V.
Ramsey
a,
Nicholas S.
Dolan
a,
Matthew B.
Francis
 ac and
Ariel L.
Furst
ac and
Ariel L.
Furst
 *d
*d
aDepartment of Chemistry, University of California, Berkeley, California 94720, USA
bDepartment of Plant and Microbial Biology, University of California, Berkeley, California 94720, USA
cMaterials Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA
dDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA. E-mail: afurst@mit.edu
First published on 15th February 2021
Abstract
Pathogenic E. coli pose a significant threat to public health, as strains of this species cause both foodborne illnesses and urinary tract infections. Using a rapid bioconjugation reaction, we selectively capture E. coli at a disposable gold electrode from complex solutions and accurately quantify the pathogenic microbes using electrochemical impedance spectroscopy.
Pathogenic strains of Escherichia coli are the most common cause of urinary tract infections (UTIs)1,2 and are a major contributor to foodborne diseases,3–6 both of which cause millions of illnesses each year. Unfortunately, current methods to identify bacterial pathogens are limited to techniques performed in centralized laboratory facilities that require trained personnel and specialized equipment. The most prevalent methods to detect bacteria in complex samples such as bodily fluids or foods are colony-based detection and the polymerase chain reaction (PCR).7–11 Despite the sensitivity of these methods, they cannot be deployed for point-of-contamination detection of foodborne pathogens, nor can they be used for the rapid point-of-care diagnosis of UTIs. Colony-based detection often requires several days of growth to determine accurate colony counts, and, though PCR can be used to identify bacteria, it cannot discern between living and dead cells. In order to combat the spread of these dangerous pathogens, the development of a rapid-readout, field-deployable test is paramount.
Electrochemical detection offers a rapid and inexpensive alternative to traditional methods for detection and quantitation of pathogenic bacteria. For E. coli sensing, antibody- and aptamer-based electrochemical sensors have been reported that enable highly sensitive monitoring of these microbes. Such technologies have enabled detection limits of 10 colony-forming units (CFU) mL−1 or lower.12–18 However, the incorporation of biorecognition elements increases the cost and complexity of these platforms and decreases their stability, often necessitating highly controlled storage and transportation conditions. Additionally, neither antibodies nor aptamers covalently capture bacteria, which can lead to variability in measurements due to washes or interactions with components of the analyte solution.
Here, we report an electrochemical sensor to detect E. coli from complex food samples and bodily fluids. By combining covalent capture and electrochemical impedance spectroscopy (EIS) for sensing,19 quantitative detection of the bacteria was achieved with a limit of detection (LOD) of 12 CFU mL−1. To accomplish this, a fast and selective bioorthogonal conjugation reaction between the E. coli cells and the electrochemical surface was needed. As demonstrated by Bandyopadhyay et al.,20 the synthetic amino acid 2-acetylphenylboronic acid can be incorporated into the E. coli cell wall via peptidoglycan remodelling, which can then be reacted with a fluorescently-labelled carbazide. The bioconjugation reaction proceeds rapidly, with rates faster than 103 M−1 s−1, to form a stable diazaborine linkage. Following the reported synthetic protocol,21 the synthesis of the amino acid was completed, albeit with significant racemization of the final product, as observed and quantified by chiral HPLC (ESI,† Fig. S5). The enantiomeric ratio is important to note as the D-enantiomer has been shown to incorporate into the E. coli peptidoglycan at higher levels than the L-enantiomer. However, specificity over other bacterial species was retained for both configurations of the synthetic amino acid.20 To capture the cell at the gold electrode surface, a PEGylated thiol with a terminal carbazide group was synthesized in four steps (Fig. 1a). The reactivity of this terminal carbazide with 2-acetylphenylboronic acid incorporated in the E. coli peptidoglycans was confirmed by flow cytometry (ESI,† Fig. S7).
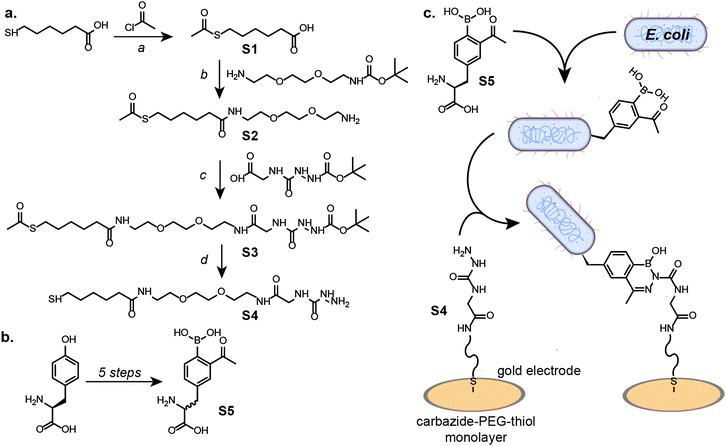 | ||
| Fig. 1 Synthetic route for E. coli capture at a modified electrode. (a) The carbazide-PEG-thiol small molecule (S4) was synthesized in 4 steps (a–d). Acetyl chloride was reacted with 6-mercaptohexanoic acid, generating S1 (a). To S1 was added tert-butyl (2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamate, followed by deprotection, generating S2 (b). S2 was reacted with (2-(tert-butoxycarbonyl)hydrazine-1-carbonyl)glycine to generate S3 (c), followed by reaction with K2CO3 and deprotection to generate S4 (d). (b) 2-Acetylphenylboronic acid (S5) was synthesized in five steps following a previously-published protocol21 to yield a mixture of the enantiomers. (c) The reaction between 2-acetylphenylboronic acid-labelled E. coli and the carbazide-PEG-thiol monolayer self-assembled on the gold surface yields a covalent diazaborine linkage to capture cells on the electrode. | ||
For electrochemical detection, E. coli cells were labelled through the initial incorporation of 2-acetylphenylboronic acid into the peptidoglycan cell wall. As the synthetic amino acid must be enzymatically incorporated into the peptidoglycan, only viable cells of specific species are able to incorporate it.20 Further specificity for the labelling of E. coli over other bacterial species (S. aureus) is due to the rate of incorporation of the 2-acetylphenylboronic acid, as was previously demonstrated.20 To capture the 2-acetylphenylboronic acid-labelled E. coli cells on the electrode surface, the carbazide-PEG-thiol was self-assembled on the gold electrode, forming a monolayer through gold-thiol bonds. When exposed to the reactive monolayer, the labelled E. coli were covalently captured by the rapid bioconjugation reaction, resulting in stable diazaborine linkages (Fig. 1c).
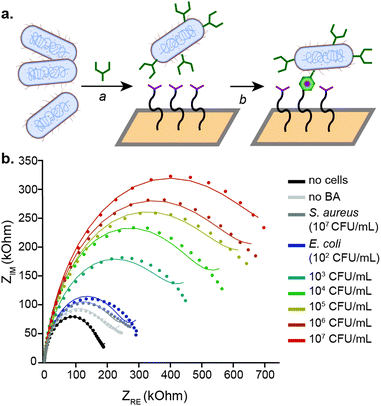
Combining the covalent capture of cells with an EIS-based readout, which is a facile and highly sensitive electrochemical sensing technique, enabled the quantification of E. coli in multiple complex solutions.19 As the platform is intended for point-of-contamination and point-of-care measurements, the capture and detection strategy was developed using inexpensive, disposable gold electrodes, which necessitate only 10 μL of sample volume for sensing (ESI,† Fig. S6).22 The workflow for labelling, capture, and quantification requires less than two hours. The cells are initially incubated with the synthetic amino acid for one hour at 37 °C, followed by washing and a five-minute heat killing step. The labelled cells are then incubated on the electrode for 30 minutes, followed by electrochemical detection, which requires less than five minutes (Fig. 2a).
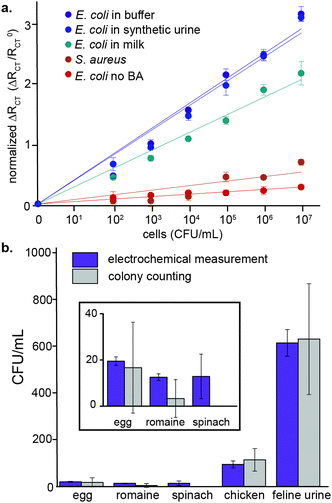
For sensitive electrochemical detection, faradaic EIS in the presence of ferricyanide and ferrocyanide was performed. Example Nyquist plots of the detection of known E. coli concentrations in buffer are shown (Fig. 2b). The data were fit with a Warburg impedance model, which is overlaid with the raw plots. From the resulting model fits, the charge transfer resistance (RCT) was extrapolated and correlated to the number of cells in samples of known concentration. E. coli were measured and quantified over a range of 102–107 CFU mL−1. The RCT increased by an average of 46.5% in the presence of 100 cells, with an order of magnitude larger increase for 107 cells (315% change). From these data, the LOD was calculated to be 12 CFU mL−1 in buffer. Based on the measured range of bacterial concentrations, cells can be detected and quantified over a range of 102–107 CFU mL−1, which can be seen in the normalized RCT values for E. coli quantification in buffer, shown in (Fig. 3a). This range is well within what is necessary for both foodborne pathogen detection and UTI diagnosis.23
The specificity of the platform was then evaluated for E. coli quantification. As can be seen in Fig. 3a, the sensor does not respond to the presence of S. aureus, independent of the concentration of cells. Even at the highest concentration of S. aureus evaluated, 107 CFU mL−1, the change in the RCT for these cells was only 69%, as compared to 315% for E. coli. Similarly, if the E. coli are not exposed to the 2-acetylphenyloronic acid prior to incubation with the electrode surface, non-specific adhesion is not observed. Only a 28% change in the normalized RCT occurred, even upon treatment with 107 unlabelled cells mL−1. The lack of electrochemical response from S. aureus as well as from unlabelled E. coli confirms the specificity of the platform for cells that rapidly incorporate 2-acetylphenylboronic acid into their peptidoglycan cell wall. One limitation of the platform, though, is that it cannot discern between E. coli strains. The electrochemical results were further confirmed by flow cytometry. E. coli and S. aureus were exposed to varying concentrations of the synthetic amino acid, followed by secondary labelling with a fluorescent semicarbazide. Incubating with 2-acetylphenylboronic acid concentrations between 20–100 μM showed labelling of E. coli with minimal background labelling of S. aureus (ESI,† Fig. S7). Consistent with prior literature,20 the phenylboronic acid labelling is specific for viable E. coli, which are capable of rapidly integrating the synthetic amino acid through peptidoglycan remodelling.
Following the initial sensor characterization with pathogens detected from buffer, the ability of the sensor to detect E. coli in complex solutions was further investigated. E. coli were detected and quantified in artificial urine (obtained from Innovating Science™) with similar sensitivity to buffer; the LOD was also 12 CFU mL−1 in this matrix. Although the percent change in the RCT was slightly lower for detection of these bacteria in milk (216% change in the normalized RCT for 107 cells), a similar LOD and range of detection is maintained (Fig. 3a). This slightly smaller change is likely due to proteins from the milk adhering to the electrode surface or sugars in solution interfering with the coupling, as a slight decrease in signal is similarly observed by flow cytometry.
Finally, detection of endogenous E. coli was performed with samples commonly contaminated with this pathogen. Rinsings from the surface of eggs, raw chicken, spinach, and romaine lettuce24–26 were measured electrochemically and compared to colony counts following growth on agar plates. Additionally, a sample of UTI-contaminated feline urine was obtained. As can be seen in Fig. 3b, CFU counts of E. coli measured using the electrochemical platform were consistent with values determined by colony counting, but with significantly lower error. The bacteria present in the samples were confirmed to be of the Escherichia genus by 16S sequencing. Thus, the electrochemical impedance sensor is capable of competing with established technologies to detect contaminants from complex samples.
E. coli pose a serious threat to public health, both as a foodborne pathogen and a leading cause of UTIs. Accepted methods to detect these microbes generally require centralized laboratory facilities with specialized equipment, trained personnel, and hours or days to complete. A sensitive electrochemical sensor has been developed to covalently capture, detect, and quantify E. coli from complex sample matrices with an LOD of 12 CFU mL−1 and a linear range of detection up to 107 CFU mL−1. The technology enables quantification of these pathogens within two hours on disposable electrodes, even from complex matrices including milk and artificial urine. Detection of endogenous E. coli was successful in commonly-contaminated samples, including eggs, raw chicken, spinach, and romaine lettuce, in addition to an infected urine sample. The quantification of E. coli from these samples tracked well with the current gold-standard of colony counting on agar plates. This platform represents significant progress in the development of field-deployable sensors to detect these dangerous pathogenic microbes.
We would like to acknowledge the University of California Cancer Research Coordinating Committee predoctoral fellowship for supporting S. H. K., the National Science Foundation Graduate Research Fellowship under grant No. DGE1752814 for supporting L. E. S., the Chemical Biology Graduate Program at UC Berkeley (NIH T32-GM066698) for supporting A. V. R., and the Stanley G. Thompson Memorial Scholarship for supporting T. A. F. A. L. F. acknowledges the Arnold and Mabel Beckman Foundation for funding. Some figure elements were constructed using the freely available online platform biorender.com.
Conflicts of interest
There are no conflicts to declare.Notes and references
- W. E. Stamm and S. R. Norrby, J. Infect. Dis., 2001, 183(Suppl), S1–S4 CrossRef.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269–284 CrossRef CAS.
- J. Meng, J. T. LeJeune, T. Zhao and M. P. Doyle, Food Microbiol., 2012, 287–309 Search PubMed.
- E. Scallan, R. M. Hoekstra, F. J. Angulo, R. V. Tauxe, M.-A. Widdowson, S. L. Roy, J. L. Jones and P. M. Griffin, Emerg. Infect. Dis., 2011, 17, 7–15 CrossRef.
- S. Viazis and F. Diez-Gonzalez, The Twentieth Century’s Emerging Foodborne Pathogen: A Review, in Advances in Agronomy, ed. D. L. Sparks, Elsevier Inc, New York, 1st edn, 2011, vol. 111, pp. 1–50 Search PubMed.
- S. Park, R. W. Worobo and R. A. Durst, Crit. Rev. Food Sci. Nutr., 1999, 39, 481–502 CrossRef CAS.
- A. C. G. Foddai and I. R. Grant, Appl. Microbiol. Biotechnol., 2020, 104, 4281–4288 CrossRef CAS.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, Analyst, 2019, 144, 396–411 RSC.
- J. Li, Y. Zhu, X. Wu and M. R. Hoffmann, Clin. Infect. Dis., 2020, 71, S84–S90 CrossRef.
- J. W.-F. Law, N.-S. Ab Mutalib, K.-G. Chan and L.-H. Lee, Front. Microbiol., 2015, 5, 770 Search PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Muñoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS.
- M. Amiri, A. Bezaatpour, H. Jafari, R. Boukherroub and S. Szunerits, ACS Sens., 2018, 3, 1069–1086 CrossRef CAS.
- Y. Wang and E. C. Alocilja, J. Biol. Eng., 2015, 9, 16 CrossRef.
- M. Barreiros dos Santos, S. Azevedo, J. P. Agusil, B. Prieto-Simón, C. Sporer, E. Torrents, A. Juárez, V. Teixeira and J. Samitier, Bioelectrochemistry, 2015, 101, 146–152 CrossRef CAS.
- V. Soheili, S. M. Taghdisi, K. Abnous and M. Ebrahimi, Iran J. Basic Med. Sci., 2020, 23, 901–908 Search PubMed.
- H. Jayamohan, B. K. Gale, B. Minson, C. J. Lambert, N. Gordon and H. J. Sant, Sensors, 2015, 15, 12034–12052 Search PubMed.
- H. Kaur, M. Shorie, M. Sharma, A. K. Ganguli and P. Sabherwal, Biosens. Bioelectron., 2017, 98, 486–493 CrossRef CAS.
- S. L. Burrs, M. Bhargava, R. Sidhu, J. Kiernan-Lewis, C. Gomes, J. C. Claussen and E. S. McLamore, Biosens. Bioelectron., 2016, 85, 479–487 CrossRef CAS.
- A. L. Furst and M. B. Francis, Chem. Rev., 2019, 119, 700–726 CrossRef CAS.
- A. Bandyopadhyay, S. Cambray and J. Gao, J. Am. Chem. Soc., 2017, 139, 871–878 CrossRef CAS.
- A. Bandyopadhyay and J. Gao, J. Am. Chem. Soc., 2016, 138, 2098–2101 CrossRef CAS.
- A. L. Furst, A. C. Hoepker and M. B. Francis, ACS Cent. Sci., 2017, 3, 110–116 CrossRef CAS.
- N. A. Cornick and A. F. Helgerson, Appl. Environ. Microbiol., 2004, 70, 5331–5335 CrossRef CAS.
- O. A. Oyarzabal, K. S. Macklin, J. M. Barbaree and R. S. Miller, Appl. Environ. Microbiol., 2005, 71, 3351–3354 CrossRef CAS.
- N. E. Secretariat, J. Food Prot., 2007, 70, 241–250 CrossRef.
- K. M. Leach, J. M. Stroot and D. V. Lim, Appl. Environ. Microbiol., 2010, 76, 8044–8052 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc08420d |
| This journal is © The Royal Society of Chemistry 2021 |