Open Access Article
Open Access ArticleIon mobility-mass spectrometry shows stepwise protein unfolding under alkaline conditions†
Cagla
Sahin‡
 ab,
Nicklas
Österlund‡
ab,
Nicklas
Österlund‡
 cd,
Axel
Leppert
e,
Jan
Johansson
e,
Erik G.
Marklund
cd,
Axel
Leppert
e,
Jan
Johansson
e,
Erik G.
Marklund
 f,
Justin L. P.
Benesch
*g,
Leopold L.
Ilag
*d,
Timothy M.
Allison
f,
Justin L. P.
Benesch
*g,
Leopold L.
Ilag
*d,
Timothy M.
Allison
 *h and
Michael
Landreh
*h and
Michael
Landreh
 *a
*a
aDepartment of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Stockholm 171 65, Sweden. E-mail: Michael.Landreh@ki.se
bDepartment of Biology, University of Copenhagen, Ole Maaløes vej 5, Copenhagen N, 2200, Denmark
cDepartment of Biochemistry and Biophysics, Stockholm University, Stockholm 106 91, Sweden
dDepartment of Materials and Environmental Chemistry, Stockholm University, Stockholm 106 91, Sweden. E-mail: leopold.ilag@mmk.su.se
eDepartment of Biosciences and Nutrition, Karolinska Institutet, Neo, Huddinge 141 83, Sweden
fDepartment of Chemistry – BMC, Uppsala University, Box 576, Uppsala, 751 23, Sweden
gDepartment of Chemistry, University of Oxford, South Parks Road, Oxford OX1 3QZ, UK. E-mail: justin.benesch@chem.ox.ac.uk
hBiomolecular Interaction Centre and School of Physical and Chemical Sciences, University of Canterbury, Christchurch 8140, New Zealand. E-mail: timothy.allison@canterbury.ac.nz
First published on 6th January 2021
Abstract
Although native mass spectrometry is widely applied to monitor chemical or thermal protein denaturation, it is not clear to what extent it can inform about alkali-induced unfolding. Here, we probe the relationship between solution- and gas-phase structures of proteins under alkaline conditions. Native ion mobility-mass spectrometry reveals that globular proteins are destabilized rather than globally unfolded, which is supported by solution studies, providing detailed insights into alkali-induced unfolding events. Our results pave the way for new applications of MS to monitor structures and interactions of proteins at high pH.
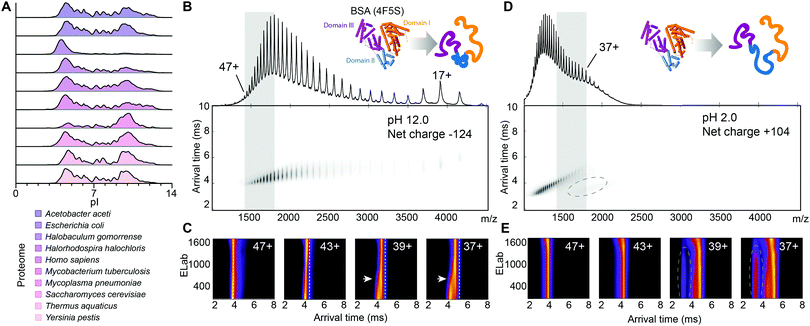
Understanding the folded states and interactions of proteins under destabilizing conditions is of importance for basic science as well as biotechnological and pharmacological applications. For example, enzymes can display activity optima in very high or low pH ranges.1 Several methods are available to study protein folding under alkaline and acidic conditions, including high-resolution strategies like NMR, and ensemble methods such as circular dichroism (CD) spectroscopy. Mass spectrometry (MS) is a robust and time-efficient method to analyse the conformational landscapes of proteins. By measuring the molecular weight, charge state distribution (CSD), and, by ion mobility (IM) spectrometry, collision cross section (CCS) of a protein, it is possible to draw conclusions about its structure and interactions.2 Nano-electrospray ionization (nESI)-MS from solutions that mimic physiological conditions can preserve the native fold during transfer to the gas-phase, resulting in lowly charged, compactly folded ions. Denaturing conditions that destabilize non-covalent interactions yield unfolded, highly charged species.3 nESI-MS is most efficient in positive polarity, and as a result, MS studies are usually performed on positively charged protein ions. Analysis of the distribution of isoelectric points across ten genomes shows that there is a comparable number of acidic proteins with a pI < 7 as basic proteins with pI > 7 (Fig. 1A and Tables S1, S2, ESI†), giving them a negative net charge under physiological conditions.4 During MS analysis, acidic proteins have to traverse their pI due to ion-pairing reactions in the electrospray droplet and are additionally subjected to protonation. While native MS has been found to preserve protein structures, the switch from solution to gas-phase charge can affect their fold and stability.
We therefore asked if the change from negative to positive charge during transfer to the gas-phase affects the folded state of proteins. The most extreme case is nESI-MS of proteins subjected to alkaline-induced unfolding, where a protein with highly negative solution charge is converted into a positively charged protein ion. Basic conditions are employed very infrequently in nESI-MS, but some studies suggest alkaline conditions (pH 9–11) result in the generation of unfolded protein ions.5–9
To investigate the effect of alkaline conditions in nESI-MS, we prepared a variety of proteins in aqueous ammonia, i.e. ammonium hydroxide, solutions, and recorded mass spectra in positive ionization mode using instrument parameters designed to minimize disruption of the native fold.10 We analysed bovine serum albumin (BSA) and found that 7 M ammonia, which results in a pH of 12.0, produces the highest average ion charge, indicating unfolding (Fig. S1, ESI†). Inspection of the CSDs obtained at pH 6.9 and 12.0 shows that high pH results in two major CSD envelopes and loss of dimers (Fig. 1B and Fig. S1A, ESI†). These findings suggest that MS captures a folded-to-unfolded transition between pH 11.5 and 13.0.11
To probe the structures of the different BSA populations observed at pH 12.0, we employed IM-MS to measure their respective folded state(s) in the gas phase. By activating the ions in the trap region of the mass spectrometer and measuring the resulting change in arrival time distribution (ATD), it is possible to induce and monitor collision-induced unfolding (CIU) of the desolvated protein ions. Plotting the arrival time of an ion population as a function of activation energy (ELab) yields CIU profiles that show how a folded state changes in response to thermal activation in the gas-phase.12 It has been proposed that unfolding steps are correlated with the number of domains in a complex.13 The energies at which folded-to-unfolded transitions occur inform about the stabilities of protein domains and complexes, and can serve as fingerprints for complex macromolecules.14,15
We obtained CIU profiles for BSA at pH 12.0, as well as at pH 2.0, where BSA is completely unfolded in solution. BSA yields a CSD ranging from 47+ to 15+ at pH 12, in contrast to approximately 60+ to 31+ at pH 2.0 (Fig. 1B–E). The higher charges at acidic pH likely reflect a more unfolded population, where BSA has transitioned through its pI of 4.8, resulting in global destabilization, yet there is a significant overlap between the CSDs at pH 2.0 and 12.0. CIU profiles show that at pH 2.0, increasing collisional activation did not affect the ATDs of the 47+ to 37+ ions. CIU of the same charge states at pH 12.0 revealed an unfolding step for ions with charges below 43+, and lower arrival times even after gas-phase unfolding. These observations indicate that at pH 12.0, BSA ions have a more compact structure than ions with the same charge produced at pH 2.0.
We then recorded IM-mass spectra of BSA at pH 6.9 and 11.0, where the protein exhibits native-like CSDs, and compared CIU profiles to those recorded at pH 12.0 (Fig. S2, ESI†). We found two clear unfolding steps for the 16+ and 17+ charge states of BSA at pH 6.9 and 11.0, as well as an additional unfolding step for both charge states at pH 12.0 (Fig. S2, ESI†). These data indicate that the BSA ions at pH 12.0 have different structural features than those ions of identical charge produced at pH 6.9 or 11.0. Indeed, base-induced unfolding of BSA has been found to occur in a step-wise fashion, with domain II retaining its fold up to a pH of 12.5.11 The presence of partially folded segments likely accounts for the unfolding step observed in the CIU experiments. The different arrival times at pH 2.0 and 12.0 suggest additional conformational restraints that are not present in ions produced from globally unfolded protein.
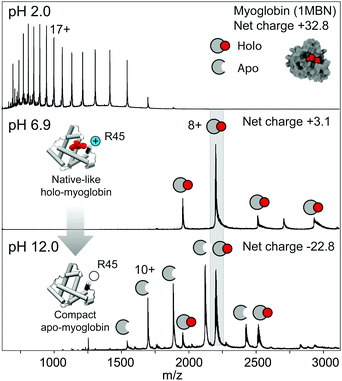
Next, we asked to what extent the alkali unfolding observed in nESI-MS correlates with solution folded states. For this purpose, we selected horse heart myoglobin, which in the native state contains a non-covalently bound heme co-factor that is lost if the protein unfolds, reporting on the structural integrity of the complex in solution.16 Mass spectra of myoglobin at pH 2.0 show only apo-myoglobin with a broad CSD centred around the 17+ charge state, in line with complete unfolding (Fig. 2). At pH 6.9, we detected four charge states centred around 8+, and complete retention of the heme group. Increasing the pH to 12.0 resulted in the appearance of 11+, 10+, and 9+ charge states for apo-myoglobin, but not the highly charged ions (>13+) observed at acidic pH (Fig. 2), as well as peaks corresponding to holo-myoglobin with 7, 8, or 9 positive charges, consistent with partial unfolding. It is tempting to speculate that the low degree of unfolding observed for myoglobin may be related to its higher pI of 7.2, compared to 4.8 for BSA, resulting in a smaller shift in net charge at basic pH.
To probe the structural integrity of apo- and holo-myoglobin, we recorded CIU profiles of the 8+ charge states at pH 6.9 and 12.0 (Fig. S3, ESI†). Interestingly, the profiles of holo-myoglobin reveal virtually identical single-step unfolding trajectories at the same activation energy, suggesting similar gas-phase structures in both solution conditions. The CIU profile of the 8+ charge state of apo-myoglobin at pH 12.0 also shows a single unfolding step, albeit at lower activation energy, which is consistent with a compact but destabilized structure (Fig. 2 inset). To compare the MS data to the folded state of the protein in solution, we recorded absorbance spectra at pH 2.0, 6.9, and 12.0. We found that the absorbance peak at 409 nm, which indicates heme bound to myoglobin, is not affected at pH 6.9 or 12.0, but completely abolished at pH 2.0 (Fig. S3, ESI†). Similarly, CD spectroscopy showed that myoglobin retains its native secondary structure content at pH 12.0 (Fig. S3, ESI†). Taken together, these observations agree with previous reports that myoglobin is destabilized at pH 12 but does not undergo unfolding.17 During nESI-MS under alkaline conditions, myoglobin destabilization could cause a subpopulation to unfold during desolvation and undergo co-factor dissociation. In fact, the heme group of horse heart myoglobin directly interacts with Arg45, which has a predicted pKa of 11.5, at the entrance to the co-factor binding site. It therefore appears possible that deprotonation of this residue could promote heme dissociation, giving rise to partially folded apo-protein as observed here. In the remaining holo-protein population, the destabilizing effect is mitigated upon desolvation, giving rise to a native-like CIU profile (Fig. S3, ESI†).
Having established that we can observe alkali-induced unfolding in IM-MS, we selected two proteins whose conformational preferences should be insensitive to alkaline pH: the intrinsically disordered α-synuclein, and the disulfide-stabilized lysozyme. Mass spectra of α-synuclein18 showed a broad CSD at pH 6.9 that was similar when the pH was decreased to 2.0 or increased to 12.0 (Fig. S4, ESI†). Likewise, CIU profiles did not reveal change in arrival times or any unfolding steps, suggesting that alkaline pH has no significant effect on the ionization or gas-phase conformations of the protein (Fig. S4, ESI†). Next, we investigated lysozyme, which contains a high number of basic residues and is resistant to unfolding at alkaline pH.19 The CSDs of lysozyme at pH 6.9 and 12.0 show no pronounced differences, suggesting that it does not undergo unfolding at pH 12.0 (Fig. S4, ESI†). The CSDs, as well as the CIU profiles, are consistent with the high pH stability of lysozyme and are reflected by the near-identical CD spectra at neutral and alkaline pH (Fig. S4, ESI†). We conclude that the observed CSDs and CIU profiles at alkaline pH accurately reflect the solution unfolding behaviour of BSA and myoglobin.
The generally good agreement between solution- and gas-phase protein folded states at alkaline pH led us to reason that IM-MS could be employed to probe specific conformational changes. The native trimer of the BRICHOS domain of the lung surfactant peptide C protein (proSP-C) is stabilized by an inter-subunit salt bridge between Arg139 and Glu191 on neighbouring protomers, with a predicted pKa of 11.0 (Fig. 3A inset and Fig. S5, ESI†).20 We speculated that de-protonation of this salt bridge would de-stabilize the trimer and result in monomer formation. In line with previous studies, mass spectra of BRICHOS in pure water showed mostly monomeric protein (Fig. 3A).21 Addition of 1 M AmAc, pH 6.9, resulted in a shift towards the trimeric form, likely because higher AmAc concentrations help to preserve a neutral droplet pH during the ionization process.22 However, raising the pH to 11.0 mostly reverted BRICHOS to its monomeric state (Fig. 3A). To investigate whether trimer dissociation is consistent with the release of folded monomers, we employed CIU of the BRICHOS trimers and monomers observed at pH 6.9 and 11.0 (Fig. 3B and C). We then generated difference plots for each species at neutral and high pH (Fig. S5, ESI†). Although the CIU profiles of monomers and trimers are quite similar, the gas-phase unfolded trimer at neutral pH shows a lower arrival time and an additional unfolding step at the highest activation energies. We suggest that the lower coulombic repulsion and the intact Arg139–Glu191 salt bridge could restrict unfolding at neutral pH and allow the trimer to withstand higher activation energies without dissociating. The observations from IM-MS are consistent with the release of folded monomers via de-protonation of the Arg139–Glu191 salt bridge. CD spectroscopy and size exclusion chromatography of proSP-C BRICHOS support this conclusion, showing on one hand highly similar secondary structures and on the other hand extensive monomerization going from pH 7.0 to 11.0 (Fig. S5, ESI†).
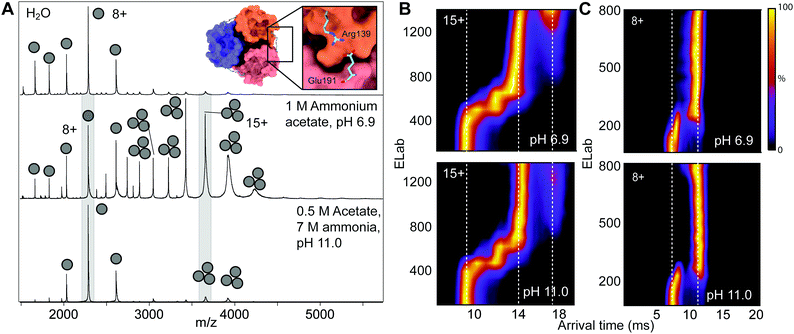 | ||
| Fig. 3 pH-Dependent dissociation of the proSP-C BRICHOS domain trimer. (A) nESI-MS spectra of proSP-C BRICHOS in pure water (top) shows mostly monomeric protein, and only a minor fraction of the native trimer. In 1 M AmAc, pH 6.9 (middle), most of the protein is trimeric. Increasing the pH to 11.0 (bottom) results in mostly monomeric protein. No MS settings were changed when recording the water and the AmAc spectra. Inset: The proSP-C trimer (PDB 2YAD) contains an inter-molecular salt bridge between Arg139 and Glu191. (B and C) CIU profiles of the 15+ charge state of the proSP-C BRICHOS trimer and 8+ charge state of the monomer show minor changes in the unfolding pattern of the trimer at high pH. | ||
Considering the behaviour of globular proteins in this study, we observe that high pH induces mild to moderate destabilization of the native structure, as opposed to the complete unfolding at low pH. This effect could be due to protein-specific differences such as pI, but can even be considered in the context of desolvation during ionisation. For example, a superoxide dismutase mutant containing an additional arginine exhibits altered stability in IM-MS due to the formation of an additional salt bridge after desolvation.23 In solution, alkaline pH unfolds proteins by breaking salt bridges and H-bonds and causing coulombic repulsion due to the high density of negative charges. In positive ionization mode, the negative charges are neutralized mitigating their repulsive effect. This process would in principle enable the protein to adopt a partially compact structure, which in turn would acquire fewer charges than a fully unfolded protein. At low pH, on the other hand, coulombic repulsion between positive charges is instead exacerbated during nESI, giving rise to fully unfolded protein, consistent with lower charges at alkaline than at acidic pH.9 This process is best illustrated by the unfolding of BSA: in solution, the protein has a similar secondary structure content at pH 2.0 and 12.0, as evident from their near-identical CD spectra, as its predicted net charge moves by +110 or −110 from the net charge at neutral pH (−10). In nESI-MS, the acid-unfolded protein is completely unfolded, whereas the alkali-unfolded protein remains partially compact and can undergo CIU steps. We suggest that for some protein systems, positive-mode nESI may artificially increase the degree of acid-induced unfolding but provide a relatively accurate picture of alkali-induced unfolding. In summary, we have explored alkaline-conditions that are compatible with IM-MS and reveal specific unfolding events during alkali-unfolding of these proteins, expanding its applicability to protein structural biology.
Conflicts of interest
There are no conflicts to declare.References
- K. Talley and E. Alexov, Proteins: Struct., Funct., Bioinf., 2010, 78, 2699–2706 CAS.
- P. Lössl, M. van de Waterbeemd and A. J. Heck, EMBO J., 2016, 35, 2634–2657 CrossRef.
- Z. Hall and C. V. Robinson, J. Am. Soc. Mass Spectrom., 2012, 23, 1161–1168 CrossRef CAS.
- J. Kiraga, P. Mackiewicz, D. Mackiewicz, M. Kowalczuk, P. Biecek, N. Polak, K. Smolarczyk, M. R. Dudek and S. Cebrat, BMC Genomics, 2007, 8, 163 CrossRef.
- L. Konermann and D. J. Douglas, J. Am. Soc. Mass Spectrom., 1998, 9, 1248–1254 CrossRef CAS.
- G. Invernizzi, M. Šamalikova, S. Brocca, M. Lotti, H. Molinari and R. Grandori, J. Mass Spectrom., 2006, 41, 717–727 CrossRef CAS.
- M. Bruneaux, P. Terrier, E. Leize, J. Mary, F. H. Lallier and F. Zal, Proteins: Struct., Funct., Bioinf., 2009, 77, 589–601 CrossRef CAS.
- P. Liuni, B. Deng and D. J. Wilson, Analyst, 2015, 140, 6973–6979 RSC.
- C. J. Hogan, J. A. Carroll, H. W. Rohrs, P. Biswas and M. L. Gross, Anal. Chem., 2009, 81, 369–377 CrossRef CAS.
- F. Sobott, H. Hernández, M. G. McCammon, M. A. Tito and C. V. Robinson, Anal. Chem., 2002, 74, 1402–1407 CrossRef CAS.
- B. Ahmad, M. Kamal and R. Khan, Protein Pept. Lett., 2005, 11, 307–315 CrossRef.
- J. T. S. Hopper and N. J. Oldham, J. Am. Soc. Mass Spectrom., 2009, 20, 1851–1858 CrossRef CAS.
- Y. Zhong, L. Han and B. T. Ruotolo, Angew. Chem., Int. Ed., 2014, 53, 9209–9212 CrossRef CAS.
- S. M. Dixit, D. A. Polasky and B. T. Ruotolo, Curr. Opin. Chem. Biol., 2018, 42, 93–100 CrossRef CAS.
- A. J. Borysik and C. V. Robinson, Phys. Chem. Chem. Phys., 2012, 14, 14439 RSC.
- V. Katta and B. T. Chait, J. Am. Chem. Soc., 1991, 113, 8534–8535 CrossRef CAS.
- S. Uppal, S. Salhotra, N. Mukhi, F. K. Zaidi, M. Seal, S. G. Dey, R. Bhat and S. Kundu, J. Biol. Chem., 2015, 290, 1979–1993 CrossRef CAS.
- A. S. Phillips, A. F. Gomes, J. M. D. Kalapothakis, J. E. Gillam, J. Gasparavicius, F. C. Gozzo, T. Kunath, C. MacPhee and P. E. Barran, Analyst, 2015, 140, 3070–3081 RSC.
- F. Murakami, T. Sasaki and T. Sugahara, Cytotechnology, 1997, 24, 177–182 CrossRef CAS.
- H. Willander, G. Askarieh, M. Landreh, P. Westermark, K. Nordling, H. Keranen, E. Hermansson, A. Hamvas, L. M. Nogee, T. Bergman, A. Saenz, C. Casals, J. Aqvist, H. Jornvall, H. Berglund, J. Presto, S. D. Knight and J. Johansson, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2325–2329 CrossRef CAS.
- M. Fitzen, G. Alvelius, K. Nordling, H. Jörnvall, T. Bergman and J. Johansson, Rapid Commun. Mass Spectrom., 2009, 23, 3591–3598 CrossRef CAS.
- L. Konermann, J. Am. Soc. Mass Spectrom., 2017, 28, 1827–1835 CrossRef CAS.
- L. McAlary, J. A. Harrison, J. A. Aquilina, S. P. Fitzgerald, C. Kelso, J. L. P. Benesch and J. J. Yerbury, Anal. Chem., 2020, 92, 1702–1711 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental methods, Fig. S1–S5. See DOI: 10.1039/d0cc08135c |
| ‡ Equal Contribution. |
| This journal is © The Royal Society of Chemistry 2021 |