Open Access Article
Open Access ArticleSelenium-atom-modified thymidine enhances the specificity and sensitivity of DNA polymerization and detection†
Yang
Li
 a,
Yingying
Zhou
a,
Danyan
Luo
a,
Yingying
Zhou
a,
Danyan
Luo
 b,
Zhaoyi
Yang
b,
Lillian Ruoduo
Hu
b and
Zhen
Huang
*ab
b,
Zhaoyi
Yang
b,
Lillian Ruoduo
Hu
b and
Zhen
Huang
*ab
aKey Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610000, Sichuan, P. R. China
bSzostak-CDHT Large Nucleic Acids Institute & SeNA Research Institute, Chengdu 610000, Sichuan, P. R. China. E-mail: huang@senaresearch.org
First published on 4th May 2021
Abstract
Nucleobase mismatches can jeopardize DNA polymerization specificity, causing mutations and errors in DNA replication and detection. Herein we report the first synthesis of novel 2-Se-thymidine triphosphate (SeTTP), describe the single-selenium atom-specific modification strategy (SAM) against T/G mismatches, and demonstrate SAM-assisted polymerization and detection with much higher specificity and sensitivity. SAM can effectively suppress the formation of non-specific products in DNA polymerization and detection. Thus, SAM enhances the specificity of DNA synthesis by approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold, and in turn, it allows the detection of clinical COVID-19 viral RNA in low copy numbers (single-digit copies), while the conventional RT-qPCR does not.
000 fold, and in turn, it allows the detection of clinical COVID-19 viral RNA in low copy numbers (single-digit copies), while the conventional RT-qPCR does not.
Studying the mechanisms of nucleic acid polymerization and replication is important in molecular and cellular biology as well as nucleic acid detection and diagnostics. Detection of specific nucleic acids is essential for pathogen identification and epidemic control,1–4 while nucleic acids play vital roles in living systems as genetic materials.5 In addition to the enzymatic synthesis, many recent studies have demonstrated the non-enzymatic synthesis of nucleic acids.6,7 Since the base pairs are the basis of nucleic acid recognition,8,9 they are of great significance in studying the mechanisms of genetic information storage,10,11 sequence recognition, DNA and RNA diagnosis, DNA replication,12 RNA transcription13 and protein translation. Furthermore, the canonical pairing and stacking of DNA nucleobases (T/A and C/G) determine the structure and stability of DNA duplex.14,15 The non-canonical base pairs (e.g., U/G wobble pair) in RNAs (such as ribozymes and viral RNAs) diversify RNA structures and enhance RNA functions,16–18 while they (e.g., T/G wobble pair) are not desired in DNA recognition and synthesis, since they can cause mutations and reduce DNA polymerization specificity.19,20 This wobble pairing needs to be addressed in order to facilitate highly-specific DNA polymerization and accurate detection.
A T/G wobble pair is formed by the alternative hydrogen-bond (Fig. 1a) through T 2-exo-oxygen, which does not participate in the T/A base pairing.21 Since the atomic radius of selenium (1.16 Å) is much larger than that of oxygen (0.73 Å) in the same family group of elements, and Se has poor ability in hydrogen-bond formation,22 we have hypothesized that by substituting the 2-O atom with Se, thymidine triphosphate modified with the 2-Se atom (SeTTP) can be used to discriminate against the G template during DNA polymerization, significantly increasing T specificity to the A template. The incorporated Se at the 2-position of thymine discourages the T/G wobble pairing (Fig. 1b) while keeping SeT/A the same as canonical T/A without any significant difference, indicated by the UV-melting study.23 This Se atom can greatly increase the electronic and steric effects, discouraging the wobble pairing, and thereby the Se-atom substitution of thymine 2-oxygen enhances the SeT/A specificity.
 | ||
| Fig. 1 The base pair structures with or without the Se replacement. (a) T/A pair and T/G wobble pair; (b) SeT/A pair without the Se disruption, while SeT/G wobble pair with the Se disruption. | ||
Herein we report the first synthesis of SeTTP and its recognition-and-incorporation by DNA polymerase, offering much higher specificity and sensitivity for DNA polymerization and detection. Our synthesis started from 5′-trityl-2-thiothymidine 1 (Scheme 1), and CH3I was used to alkylate the 2-thio-functionality in order to activate the 2-thio-moiety and offer 2. Compound 2 was reacted with freshly prepared NaSeH in order to incorporate the Se functionality,23 synthesizing 3 in 80% yield, followed by detritylation with trichloracetic acid, offering 2-seleniothymidine 4 (SeT). The generated Se-nucleoside 4 was converted to its triphosphate 5via the one-pot and mild synthesis.24,25 The Se-triphosphate (5, SeTTP) was purified and characterized by NMR, MS and HPLC to confirm its integrity and purity, followed by DNA synthesis with SeTTP and DNA polymerase, such as Klenow and Taq.
To investigate the SeTTP recognition by DNA polymerases, we designed a DNA primer (10 nt, 5′ FAM) and the “standing-start” template (30 nt) that allow incorporation of one “T” on the underlined template “A” (Fig. 2a). Indeed, SeTTP offered the extension with only one “T”, while canonical TTP allowed the extension with up to three “T”s (Fig. 2b and c). In addition, our time-course experiments showed that in the presence of TTP or SeTTP and the other three dNTPs, DNA polymerase effectively synthesized the full-length products (Fig. 2e and f), while it didn’t synthesize the products in the absence of TTP or SeTTP under the same conditions. Furthermore, it was exciting to discover that the incorporation rate with SeTTP was approximately 95% of TTP (Fig. 2b–d). Furthermore, it is worthy to note that in the presence of SeTTP or TTP and the other three dNTPs (dATP, dCTP and dGTP), the yield of the full-length Se-DNA was approximately 90% of that of the canonical DNA (Fig. 2e–g). Obviously, SeTTP can be recognized almost as well as the canonical TTP by DNA polymerase and its incorporation specificity is much higher than that of canonical TTP.
To investigate the discrimination of SeTTP on wobble pairing, we designed DNA templates with G, GG and GGG (Fig. 3a), and included “TC” before the underlined template “G, GG or GGG” in order to allow a “running-start” wobble-pairing extension. We found that in the DNA polymerization with only dATP and dGTP (a negative control), the full-length product via the primer extension was not observed. When canonical TTP was added to the polymerization with only dATP and dGTP, the T/G wobble-pairing extension was observed (Fig. 3b). The formation of the wobble-extended full-length products was prevented only in the presence of multiple “G” on the templates (at least 3 “G”). However, when SeTTP was used to replace TTP (Fig. 3c), the wobble-pairing extended products were not observed. This surprising result indicated that SeTTP discouraged the T/G wobble pairing and polymerization, and the quantitative PAGE analysis is presented in Fig. 3d. In addition, to study the steady-state kinetics of SeTTP incorporation compared to TTP, the primer and template 1 were used for the investigation, in the presence of dATP and dGTP. We found that the wobble-pairing extension rate of SeTTP on the G-template was approximately 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 folds slower than that of canonical TTP (Fig. 3e–g), shedding light on the high specificity of SeTTP.
000 folds slower than that of canonical TTP (Fig. 3e–g), shedding light on the high specificity of SeTTP.
To further demonstrate the specificity value of SeTTP in DNA cloning and amplification, such as PCR, we designed two different target fragments (Fig. 4). We found that in the absence of SeTTP, the PCR nonspecific amplification was a serious problem (indicated by the smeared by-products), while in the presence of SeTTP (50%), the nonspecific products were almost completely suppressed, and the product yields were as high as the canonical. Further, the PCR products were analyzed by sequencing to confirm their integrity. The sequencing results (see ESI,† S15–S22) have indicated that the PCR products with TTP or SeTTP had identical sequences. Furthermore, SeT-DNA can be easily converted to the corresponding canonical DNA by a few cycles of PCR with canonical dNTPs. Our discovery may offer a new strategy for DNA amplification, gene cloning, reverse transcription and nucleic acid detection.
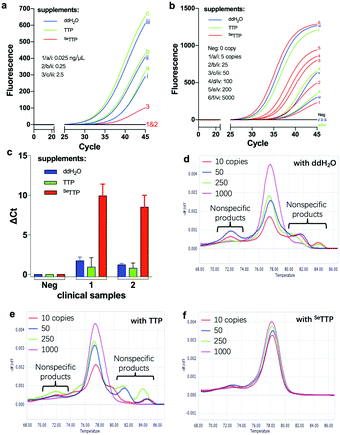
Nucleic acid detection is essential in the prevention and control of epidemics, such as the one caused by the COVID-19 virus, which has infected over 14 million people and resulted in six hundred thousand deaths worldwide. Unfortunately, it has been reported that nucleic acid detection of COVID-19 viral RNA offered a high false rate (approximately 20% false positive and negative per detection). In order to reduce the false diagnosis rate, we have decided to explore the COVID-19 detection with our unique SAM strategy (Fig. 5). First, we investigated its anti-background interference by including the viral background RNA (namely, human total RNA) in the detection. Since the typical concentration of background RNA in clinical COVID-19 detection is 0.25 ng mL−1, we set the detection concentration of background RNA as 0.025, 0.25, and 2.5 ng μL−1. Our experiment demonstrated that the Se-approach can successfully suppress the nonspecific amplification (Fig. 5a). No or low fluorescence signal was observed in the presence of supplementary SeTTP, while high signals were observed similarly with supplementary water or canonical TTP, and the high signals could be mis-interpreted as false positives.
Furthermore, we have found that the Se-strategy can detect viral RNA in up to single-digit 5 copies (Fig. 5b), while the conventional kit can detect up to 100 copies, thereby indicating the higher sensitivity of the Se-strategy (20 folds better) than the conventional one. Furthermore, we examined clinical COVID-19 viral RNA samples (Fig. 5c) and found that our SAM methodology detected 1000-fold-diluted viral RNA samples, while the commercial kits (or supplementary TTP) did not. Moreover, in order to investigate the non-specific product suppression, we performed RT-qPCR and melting-temperature studies26 and discovered that in the presence of SeTTP, the non-specific products were not observed, while they were when supplemented with water or TTP, indicated by multiple peaks (Fig. 5d–f). Our study indicated the severe non-specific amplification by RT-qPCR, especially when the copy number of viral RNAs was low (Fig. 5d and e). However, non-specific amplification in nucleic acid detection can be significantly inhibited by supplementary SeTTP (Fig. 5f).
In summary, we have first synthesized 2-seleno-thymidine triphosphate (SeTTP) and SeT-DNAs via DNA polymerization. We have found that DNA polymerase can effectively recognize the Se-triphosphate, almost as well as the canonical one. Furthermore, we have discovered high discrimination of the 2-Se-thymine against T/G mis-pairing and polymerization, and the SAM triphosphate is over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold more discriminative than the canonical one. Furthermore, we have demonstrated this unique SAM strategy for DNA polymerization, amplification and nucleic acid detection (such as PCR and RT-PCR) with high specificity, sensitivity and product purity. In addition, SAM provides a novel approach for investigating base-pair recognition, molecular interaction and nucleic acid polymerization, such as DNA replication, RNA transcription and reverse transcription. Moreover, this SAM strategy can be useful in nucleic acid detection, such as COVID-19 epidemic diagnostics, control and prevention by enhancing the sensitivity and accuracy of nucleic acid detection.
000 fold more discriminative than the canonical one. Furthermore, we have demonstrated this unique SAM strategy for DNA polymerization, amplification and nucleic acid detection (such as PCR and RT-PCR) with high specificity, sensitivity and product purity. In addition, SAM provides a novel approach for investigating base-pair recognition, molecular interaction and nucleic acid polymerization, such as DNA replication, RNA transcription and reverse transcription. Moreover, this SAM strategy can be useful in nucleic acid detection, such as COVID-19 epidemic diagnostics, control and prevention by enhancing the sensitivity and accuracy of nucleic acid detection.
This work was supported by the National Natural Science Foundation of China (21761132029 and 22077089), SeNA Research Institute, and the Fundamental Research Funds for the Central Universities.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J.-W. Ai, H.-C. Zhang, T. Xu, J. Wu, M. Zhu, Y.-Q. Yu, H.-Y. Zhang, Z. Shen, Y. Li, X. Zhou, G.-Q. Zang, J. Xu, W.-J. Chen, Y.-J. Li, D.-S. Xie, M.-Z. Zhou, J.-Y. Sun, J.-Z. Chen and W.-H. Zhang, medRxiv, 2020 DOI:10.1101/2020.02.13.20022673.
- L. M. Kucirka, S. A. Lauer, O. Laeyendecker, D. Boon and J. Lessler, Ann. Intern. Med., 2020, 173(4), 262–267 CrossRef PubMed.
- S. Ren, Q. Wang, P. Yang, S. Cui, T. Yuan, D. Zhang, L. Long and Y. Pan, Clin. Chem., 2020, 66, 794–801 CrossRef PubMed.
- S. Woloshin, N. Patel and A. S. Kesselheim, N. Engl. J. Med., 2020, 383(6), e38 CrossRef CAS PubMed.
- J. D. Watson and F. H. C. Crick, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- M. Sosson, D. Pfeffer and C. Richert, Nucleic Acids Res., 2019, 47, 3836–3845 CrossRef CAS PubMed.
- J. Niu, R. Hili and D. R. Liu, Nat. Chem., 2013, 5, 282–292 CrossRef CAS PubMed.
- A. M. Sismour and S. A. Benner, Nucleic Acids Res., 2005, 33, 5640–5646 CrossRef CAS PubMed.
- R. Laos, C. Lampropoulos and S. A. Benner, Acta Crystallogr., Sect. C: Struct. Chem., 2019, 75, 22–28 CrossRef CAS PubMed.
- G. Storz, Science, 2002, 296, 1260–1263 CrossRef CAS PubMed.
- Y. Zhang, J. L. Ptacin, E. C. Fischer, H. R. Aerni, C. E. Caffaro, K. S. Jose, A. W. Feldman, C. R. Turner and F. E. Romesberg, Nature, 2017, 551, 644–647 CrossRef CAS PubMed.
- Y. Gao, Y. Cui, T. Fox, S. Lin, H. Wang, N. de Val, Z. H. Zhou and W. Yang, Science, 2019, 363(6429), eaav7003 CrossRef CAS PubMed.
- S. Becker, J. Feldmann, S. Wiedemann, H. Okamura, C. Schneider, K. Iwan, A. Crisp, M. Rossa, T. Amatov and T. Carell, Science, 2019, 366, 76–82 CrossRef CAS PubMed.
- G. Ferry, Nature, 2019, 575, 35–36 CrossRef CAS PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2016, 533, 420–424 CrossRef CAS PubMed.
- Z. Lan and J. A. Doudna, Science, 2002, 295, 2084–2088 CrossRef PubMed.
- C. Roost, S. R. Lynch, P. J. Batista, K. Qu, H. Y. Chang and E. T. Kool, J. Am. Chem. Soc., 2015, 137(5), 2107–2115 CrossRef CAS PubMed.
- C. Schneider, S. Becker, H. Okamura, A. Crisp, T. Amatov, M. Stadlmeier and T. Carell, Angew. Chem., Int. Ed., 2018, 57, 5943–5946 CrossRef CAS PubMed.
- O. Kennard, J. Biomol. Struct. Dyn., 1985, 3, 205 CrossRef CAS PubMed.
- J. R. Kiefer, C. Mao, J. C. Braman and L. S. Beese, Nature, 1998, 391, 304–307 CrossRef CAS PubMed.
- L. E. Orgel, J. Mol. Biol., 1968, 38, 381–393 CrossRef CAS PubMed.
- H. Sun, S. Jiang, J. Caton-Williams, H. Liu and Z. Huang, RNA, 2013, 19, 1309–1314 CrossRef CAS PubMed.
- A. E. Hassan, J. Sheng, W. Zhang and Z. Huang, J. Am. Chem. Soc., 2010, 132, 2120–2121 CrossRef CAS PubMed.
- J. Caton-Williams, M. Smith, N. Carrasco and Z. Huang, Org. Lett., 2011, 13, 4156–4159 CrossRef CAS PubMed.
- B. Hu, Y. Wang, S. Sun, W. Yan, C. Zhang, D. Luo, H. Deng, L. R. Hu and Z. Huang, Angew. Chem., Int. Ed., 2019, 58, 7835–7839 CrossRef CAS PubMed.
- T. B. Morrison, J. J. Weis and C. T. Wittwer, Biotechniques, 1998, 24, 954–962 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, NMR, Maldi-Tof, sequencing data. See DOI: 10.1039/d0cc07922g |
| This journal is © The Royal Society of Chemistry 2021 |