 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical probes reveal the timing of early chlorination in vancomycin biosynthesis†
Daniel J.
Leng
a,
Anja
Greule
b,
Max J.
Cryle
 bc and
Manuela
Tosin
bc and
Manuela
Tosin
 *a
*a
aDepartment of Chemistry, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK. E-mail: M.Tosin@warwick.ac.uk
bMonash Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria 3800, Australia
cARC Centre of Excellence for Innovations in Peptide and Protein Science, Monash University, Clayton, Victoria 3800, Australia
First published on 26th January 2021
Abstract
Glycopeptides such as vancomycin are antibiotics of last resort whose biosynthetic pathways still hold undefined details. Chemical probes were used to capture biosynthetic intermediates generated in the nonribosomal peptide formation of vancomycin in vivo. The putative intercepted intermediates were characterised via HR-LC-MS2. These species provided insights into the timing of the first chlorination of the peptide backbone by the halogenase VhaA: this holds significant interest for enzyme engineering towards the making of novel glycopeptides.
Nonribosomal peptides (NRPs) are a major class of secondary metabolites with a broad range of medicinally relevant biological activities.1 NRPs are assembled by the multifunctional enzymes nonribosomal peptide synthetases (NRPSs), which are able to utilise proteinogenic and non-proteinogenic amino acids to give structural variety.2 Each NRPS ‘module’ catalyses the addition of a single amino acid through the action of multiple domains (Scheme 1A and Fig. 1A). Specific amino acids are activated as adenosine monophosphate (AMP) esters by the NRPS adenylation domains.3 From AMP esters, amino acids are loaded onto a post-translationally modified peptidyl carrier protein (PCP) domain, as phosphopantetheine thioesters.4 Peptide bond formation is catalysed by the condensation (C) domain, whereby the upstream peptide is joined with a downstream aminoacyl moiety bound to a downstream PCP domain (Scheme 1A).5 The growing peptide intermediates can be further diversified by additional domains within the NRPS, including epimerisation, methyltransferase and cyclisation domains.6 Throughout peptide chain assembly, the intermediates remain covalently bound to the NRPS as thioesters, before chain release, generally catalysed by a terminal thioesterase domain promoting peptide hydrolysis or cyclisation, occurs.7 Tailoring enzymes can also act on the aminoacyl growing peptide either in trans or post-NRPS assembly, modifying the natural product through oxidation, halogenation and glycosylation.
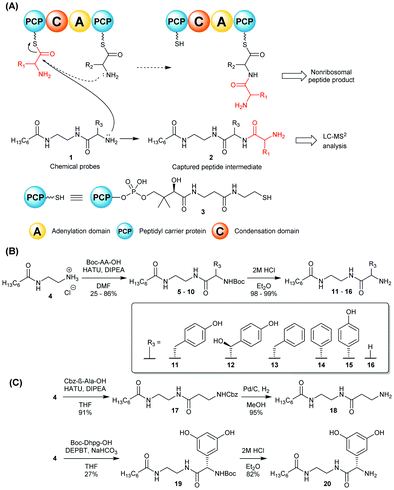 | ||
| Scheme 1 (A) Nonhydrolysable chemical probes 1 mimic PCP-bound amino acids and compete with them for C-catalysed peptide bond formation, intercepting and off-loading enzyme-bound peptide species (in red) for LC-MS2 analyses.8 (B) and (C): Synthesis of chemical probes 11–16, 18 and 20 utilised in this study. | ||
Vancomycin (21, Fig. 1) is glycopeptide antibiotic (GPA) that has been in clinical use for resistant Gram-positive bacterial infections, notably methicillin-resistant Staphylococcus aureus (MRSA). Vancomycin (21) is produced by the soil bacterium Amycolatopsis orientalis NRRL 2452, and it binds the D-Ala-D-Ala motif of the peptidoglycan precursor, thus inhibiting cell wall biosynthesis.9
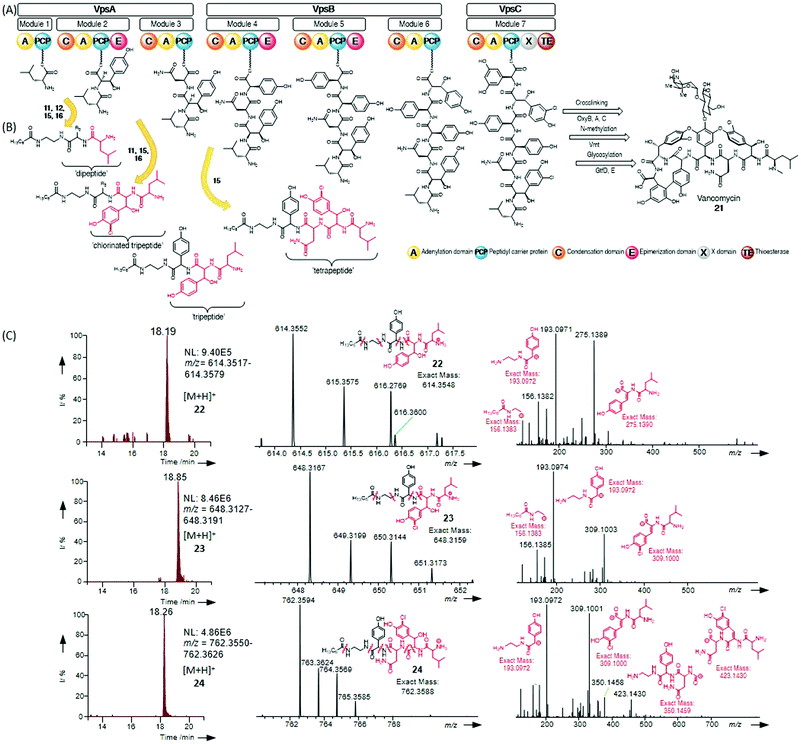 | ||
| Fig. 1 (A) Postulated vancomycin nonribosomal peptide biosynthesis. (B) In vivo offloading and capture of early vancomycin biosynthetic intermediates by chemical probes 11–16, 18, and 20, fully detailed in Table 1. Only structures related to α-amino acid based probes are shown (structures of off-loaded intermediates deriving from β-alanine probe 18 are shown in the ESI†). (C) Extracted ion chromatograms and HR-MSn analysis of the non-chlorinated and chlorinated peptide intermediates intercepted by the Hpg-based probe 15, and HR-MS2 analyses of these compounds containing diagnostic fragments related to probe and peptide intermediates. | ||
The heptapeptide backbone of vancomycin is constructed by a seven module NRPS (Fig. 1A).10 It is further modified by the action of three cytochrome P450s: OxyB, OxyA and Oxy C to install the phenolic and aryl crosslinks that impart a rigid three-dimensional structure crucial for bioactivity.11–14 Additionally, the vancomycin β-hydroxytyrosine residues are proposed to be chlorinated by the halogenase VhaA.15 The aglycone is methylated at the N-terminal leucine, and glycosylated with the addition of glucose and vancosamine.16,17
The rising resistance of Enterococci such as E. faecalis and E. faecium to glycopeptide antibiotics drives the urgent need for new derivatives of this vital therapeutic class. As the tailoring enzymes represent appealing tools for the preparation of novel glycopeptides, knowledge of the biosynthetic intermediates acting as substrates for tailoring enzymes is crucial.
Chemical probes of generic structure 1 have been developed in our group to enable the interrogation of nonribosomal peptide assembly lines in vivo (Scheme 1A).8 These mimic PCP-bound amino acids that act as acceptors during peptide bond formation catalysed by C domains, enabling the capture of nonhydrolysable biosynthetic intermediates for analysis by LC-MSn. The presence of chlorine atoms enhances the antibiotic activity of vancomycin in vivo, however the timing of peptide halogenation during its biosynthesis remains undetermined.18–20 The flavin-dependent (FADH2) halogenase VhaA present in the cluster is thought to be responsible for both vancomycin chlorinations, as shown by production of a non-chlorinated peptide in a ΔbhaA mutant of the analogous balhimycin biosynthetic pathway.15,21 Most halogenases of this class act on free substrates, but examples of PCP-bound substrates have also been reported as in the pyoluteorin pathway.22,23 For the balhimycin producer Amycolatopsis balhimycina it has been previously shown that supplementation of chloro-β-hydroxytyrosine was not able to restore antibiotic production in the halogenase deficient ΔbhaA mutant, strongly suggesting that halogenation of the free amino acid does not occur.24 Therefore chlorination events may occur at PCP-bound amino acid level, or on the PCP-bound peptide chain, or on the crosslinked heptapeptide aglycone. In vitro studies of Type IV glycopeptide teicoplanin biosynthesis has shown that the halogenase Tcp21 uses aminoacyl-PCP species as a substrate;25 however, it has also been shown that VhaA in vitro has limited activity towards PCP-bound vancomycin hexapeptide, suggesting the chlorination of the peptide is possible.21 The presence of chlorinated peptides in mutant strains with inactivated Oxy enzymes suggests the halogenation of the aglycone after crosslinking is unlikely,26 and Oxy activity was greater for the cyclisation of chlorinated peptides versus non-chlorinated ones.11
A. orientalis is a largely genetically intractable strain, which has impaired efforts to obtain information on vancomycin through direct study in vivo. The insolubility or instability of a number of GPA biosynthetic enzymes expressed recombinantly has also posed a significant challenge.27
In this work we report the use of chemical probes to investigate the timing of halogenation during vancomycin biosynthesis in its natural producer A. orientalis NRRL 2452. The probes utilised for this study were prepared as shown in Scheme 1B–C and detailed in the ESI.† The side chains (R3) of their amino acid components were based on 4-hydroxyphenylglycine (Hpg) (15), 3,5-dihydroxyphenylglycine (Dpg) (20), and β-hydroxytyrosine (βHT) (12), which are cognate amino acids to the vancomycin NRPS, and also on non-cognate amino acids tyrosine (11), phenylalanine (13), and phenylglycine (14). Achiral tools based on glycine (16) and β-alanine (18) were also utilised as ‘universal’ probes, as previously reported for the investigation of echinomycin assembly.8 The probes were supplemented to solid and liquid cultures of A. orientalis, both singly and portionwise over 4 days respectively, to a final concentration of 2.0 mM. No cytotoxicity to A. orientalis was observed. Organic extracts of the cultures were analysed by LC-HRMS2. Putative biosynthetic intermediate species intercepted by the chemical probes were identified by high-resolution mass detection and characteristic MS2 fragments. These included ions resulting from peptide bond cleavages, in addition to neutral loss of water and ammonia. No obvious differences in species detected were observed between solid and liquid cultures, and a range of species ranging from dipeptides to tetrapeptides were characterised (Fig. 1 and Table 1). The ability to effectively capture intermediate species from the biosynthetic pathway varied between the probes tested, providing insights into the substrate specificity of condensation domain acceptor sites. Offloading of leucine from the first NRPS module to obtain dipeptides could be observed using non-cognate probes based upon Gly 16, β-Ala 18, Tyr 11 and Hpg 15 as well as the cognate βHT substrate 12. Tripeptides were generated from intermediate offloading from the second module by Gly 16, Tyr 11 and 4-Hpg 15-based probes, whereas a tetrapeptide species 24 could be obtained only via the 4-Hpg probe 15 acting on module 4. No intercepted species were observed using Phg 13 or Phe 14-based substrates, suggesting the phenolic groups of Tyr and 4-Hpg are important for substrate recognition by the NRPS.
| Probe | R3 | Captured species |
|---|---|---|
| 11 | CH2C6H4OH | Di-, tripeptide |
| 12 | CH(OH)C6H4OH | Dipeptide |
| 13 | CH2C6H5 | n.d. |
| 14 | C6H5 | n.d. |
| 15 | C6H4OH | Di-, tri-, tetrapeptide |
| 16 | H | Di-, tripeptide |
| 18 | — | Dipeptide |
| 20 | C6H3(OH)2 | n.d. |
The second module of the NRPS VpsA is responsible for the addition of β-hydroxytyrosine, which is halogenated during vancomycin biosynthesis. As illustrated in Fig. 1C, a chemical probe based on Hpg 15 was able to intercept both chlorinated (23) and non-chlorinated (22) peptides, with the isotopic distributions supporting the proposed intermediate structures. This suggests that halogenation occurs at the latest at the dipeptide level. This agrees with the isolation of a chlorinated dipeptide from the fermentation of engineered A. balhimycina bearing a truncated form of the NRPS BpsA.25 Investigation of teicoplanin biosynthesis in vitro has also shown that hydroxylation of tyrosine is the primary factor in C-domain selectivity, accounting for the non-chlorinated peptide observed.28 A chlorinated species deriving from the tripeptide offloading was also identified and characterised (24). A probe based upon Dpg was prepared (20) and envisaged to intercept intermediates bound to module 6; however, this was unsuccessful and prevented the characterisation of the second chlorination event during vancomycin biosynthesis in the conditions explored here.
The identification and characterisation of a range of intercepted chlorinated peptides suggests that the donor substrate for VpsA module three is halogenated, which is important for the rational redesign of the NRPS. Species corresponding to condensation products of the C domain of module three were notably more abundant than those deriving from the condensation catalysed by module two, suggesting a broader substrate tolerance for the third condensation domain of the vancomycin NRPS. To unequivocally support the outcomes from our in vivo studies, further investigations will be necessary, e.g. involving aminoacyl-S-PCP and dipeptidyl-S-PCP constructs operating in the presence of the halogenase VhaA in vitro. Quantitative mass spectrometry tools to investigate the differing ratio of intercepted species from each module would also be helpful to better rationalise C domain selectivity and kinetics throughout glycopeptide assembly in vivo.
Nonetheless we have here shown that chemical probes are useful tools to investigate the timing of halogenation during glycopeptide bioassembly, with focus on vancomycin and its genetically intractable producer. Future work will focus on the elucidation of the timing of the second chlorination event and of other transformation events related to glycopeptide formation.
We gratefully acknowledge BBSRC (project grant BB/J007250/1 to M. T. and MIBTP studentship to D. J. L.); the Monash Warwick Alliance (Seed Fund Award to M. T. and M. C.); Dr Cleidiane Zampronio (School of Life Sciences, Warwick) for assistance with LC-HRMSn Orbitrap Fusion analyses, and Dr Lijiang Song (Warwick Chemistry) for preliminary MS data acquired on a BrukerMaXis Impact instrument.
Conflicts of interest
There are no conflicts to declare.Notes and references
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS.
- S. Caboche, M. Pupin, V. Leclère, A. Fontaine, P. Jacques and G. Kucherov, Nucleic Acids Res., 2008, 36, 326–331 CrossRef.
- T. Stachelhaus, H. D. Mootz and M. A. Marahiel, Chem. Biol., 1999, 6, 493–505 CrossRef CAS.
- Ø. Frøshov, T. L. Zimmer and S. G. Laland, FEBS Lett., 1970, 7, 68–71 CrossRef.
- V. De Crecy-Lagard, P. Marliere and W. Saurin, C. R. Acad. Sci., Ser. III, 1995, 318, 927–936 CAS.
- C. T. Walsh, H. Chen, T. A. Keating, B. K. Hubbard, H. C. Losey, L. Luo, C. G. Marshall, D. A. Miller and H. M. Patel, Curr. Opin. Chem. Biol., 2001, 5, 525–534 CrossRef CAS.
- C. A. Shaw-Reid, N. L. Kelleher, H. C. Losey, A. M. Gehring, C. Berg and C. T. Walsh, Chem. Biol., 1999, 6, 385–400 CrossRef CAS.
- Y. T. C. Ho, D. J. Leng, F. Ghiringhelli, I. Wilkening, D. P. Bushell, O. Kostner, E. Riva, J. Havemann, D. Passarella and M. Tosin, Chem. Commun., 2017, 53, 7088–7091 RSC.
- A. N. Chatterjee and H. R. Perkins, Biochem. Biophys. Res. Commun., 1966, 24, 489–494 CrossRef CAS.
- J. Recktenwald, R. Shawky, O. Puk, F. Pfennig, U. Keller, W. Wohlleben and S. Pelzer, Microbiology, 2002, 148, 1105–1118 CrossRef CAS.
- M. Peschke, C. Brieke, R. J. A. Goode, R. B. Schittenhelm and M. J. Cryle, Biochemistry, 2017, 56, 1239–1247 CrossRef CAS.
- C. C. Forneris and M. R. Seyedsayamdost, Angew. Chem., Int. Ed., 2018, 57, 8048–8052 CrossRef CAS.
- K. Woithe, N. Geib, K. Zerbe, B. L. Dong, M. Heck, S. Fournier-Rousset, O. Meyer, F. Vitali, N. Matoba, K. Abou-Hadeed and J. A. Robinson, J. Am. Chem. Soc., 2007, 129, 6887–6895 CrossRef CAS.
- K. Haslinger, M. Peschke, C. Brieke, E. Maximowitsch and M. J. Cryle, Nature, 2015, 521, 105–109 CrossRef CAS.
- O. Puk, P. Huber, D. Bischoff, J. Recktenwald, G. Jung, R. D. Süßmuth, K. H. Van Pée, W. Wohlleben and S. Pelzer, Chem. Biol., 2002, 9, 225–235 CrossRef CAS.
- D. P. O’Brien, P. N. Kirkpatrick, S. W. O’Brien, T. Staroske, T. I. Richardson, D. A. Evans, A. Hopkinson, J. B. Spencer and D. H. Williams, Chem. Commun., 2000, 103–104 RSC.
- H. C. Losey, M. W. Peczuh, Z. Chen, U. S. Eggert, S. D. Dong, I. Pelczer, D. Kahne and C. T. Walsh, Biochemistry, 2001, 40, 4745–4755 CrossRef CAS.
- U. Gerhard, J. P. Mackay, R. A. Maplestone and D. H. Williams, J. Am. Chem. Soc., 1993, 115, 232–237 CrossRef CAS.
- J. R. Pinchman and D. L. Boger, J. Med. Chem., 2013, 56, 4116–4124 CrossRef CAS.
- J. R. Pinchman and D. L. Boger, Bioorg. Med. Chem. Lett., 2013, 23, 4817–4819 CrossRef CAS.
- P. C. Schmartz, K. Zerbe, K. Abou-Hadeed and J. A. Robinson, Org. Biomol. Chem., 2014, 12, 5574–5577 RSC.
- P. C. Dorrestein, E. Yeh, S. Garneau-Tsodikova, N. L. Kelleher and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13843–13848 CrossRef CAS.
- S. Lin, S. G. Van Lanen and B. Shen, J. Am. Chem. Soc., 2007, 129, 12432–12438 CrossRef CAS.
- O. Puk, D. Bischoff, C. Kittel, S. Pelzer, S. Weist, E. Stegmann, R. D. Süssmuth and W. Wohlleben, J. Bacteriol., 2004, 186, 6093–6100 CrossRef CAS.
- T. Kittilä, C. Kittel, J. Tailhades, D. Butz, M. Schoppet, A. Büttner, R. J. A. Goode, R. B. Schittenhelm, K.-H. van Pee, R. D. Süssmuth, W. Wohlleben, M. J. Cryle and E. Stegmann, Chem. Sci., 2017, 8, 5992–6004 RSC.
- R. D. Süssmuth, S. Pelzer, G. Nicholson, T. Walk, W. Wohlleben and G. Jung, Angew. Chem., Int. Ed., 1999, 38, 1976–1979 CrossRef.
- J. W. Trauger and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2012, 97, 3112–3117 CrossRef.
- M. Kaniusaite, J. Tailhades, E. A. Marschall, R. J. A. Goode, R. B. Schittenhelm and M. J. Cryle, Chem. Sci., 2019, 10, 9466–9482 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Contains methods for the synthesis of chemical probes and LC-HRMS analyses of the biosynthetic intermediates captured from A. orientalis NRRL 2452. See DOI: 10.1039/d0cc07421g |
| This journal is © The Royal Society of Chemistry 2021 |
