Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity†
Yong
Sha
ab,
Ru
Huang
*ab,
Mengqi
Huang
ab,
Huahua
Yue
ab,
Yuanyue
Shan
ab,
Jiaming
Hu
ab and
Da
Xing
 *ab
*ab
aMOE Key Laboratory of Laser Life Science & Institute of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, P. R. China
bGuangdong Provincial Key Laboratory of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, China. E-mail: xingda@scnu.edu.cn; huangru@scnu.edu.cn
First published on 30th November 2020
Abstract
Existing CRISPR/cas-based biosensors usually improve sensitivity by target amplification, which is time-consuming and susceptible to impurities in complex biofluid. Herein, this is the first time a cascade CRISPR/cas (casCRISPR) system has been developed, which can provide a detection limit of 1.33 fM (∼1000 times lower than direct Cas13a-based miRNA detection) and single-base resolution for miR-17 detection without resorting to target amplification. casCRISPR can also be applied to detect miRNA in complicated cell extracts and serum samples. Overall, casCRISPR will provide a heuristic idea for CRISPR/cas based biosensing, and could be a promising tool for miRNA diagnostics.
Clustered regularly interspaced short palindromic repeats (CRISPR) and the corresponding CRISPR-associated (cas) protein derive from adaptive immune systems of bacteria and archaea against the invading nucleic acid components.1,2 The programmable guide RNA (gRNA) guided DNA/RNA cleavage activity enables CRISPR/cas system to be developed into a versatile gene editing tool.3–5 Furthermore, CRISPR/cas system also holds great potential in the biological detection field.6–8
Cas9 is one of the most widely applied cas protein and has been confirmed to possess a certain ability for biosensing.9–11 Although Cas9 has excellent selectivity, Cas9 recognizes double-stranded (ds-) DNA with a unique protospacer adjacent motif (PAM) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and generates a break with blunt ends, which limit its exploitability in biosensing. Moreover, low-efficient RNA cleavage also restricted its application in RNA detection.12 Coincidentally, Cas12, Cas13, and Cas14a were found to have target-activated trans-cleavage activity that can efficiently cleave random ssDNA/ssRNA,13–17 which makes them suitable for signal amplified nucleic acid detection.18 However, the detection limit of CRISPR/cas mediated amplification-free nucleic acid detection is usually at the pM level,19 which could not meet the requirement of real sample detection. Numerous CRISPR/cas-based biosensors improve the sensitivity to aM, or even zM by target amplification.20–22 However, these methods are often time-consuming, and could produce false positive amplification due to the effect of impurities in a complex sample. Therefore, developing a sensitive and specific CRISPR/cas biosensor without resorting to target amplification could be valuable for rapid and accurate miRNA detection.
1 ratio and generates a break with blunt ends, which limit its exploitability in biosensing. Moreover, low-efficient RNA cleavage also restricted its application in RNA detection.12 Coincidentally, Cas12, Cas13, and Cas14a were found to have target-activated trans-cleavage activity that can efficiently cleave random ssDNA/ssRNA,13–17 which makes them suitable for signal amplified nucleic acid detection.18 However, the detection limit of CRISPR/cas mediated amplification-free nucleic acid detection is usually at the pM level,19 which could not meet the requirement of real sample detection. Numerous CRISPR/cas-based biosensors improve the sensitivity to aM, or even zM by target amplification.20–22 However, these methods are often time-consuming, and could produce false positive amplification due to the effect of impurities in a complex sample. Therefore, developing a sensitive and specific CRISPR/cas biosensor without resorting to target amplification could be valuable for rapid and accurate miRNA detection.
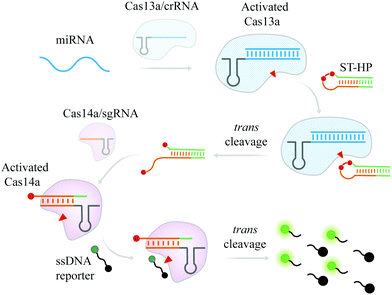
In this study, a cascade CRISPR/cas for miRNA detection, termed as casCRISPR, was developed. This strategy makes use of the trans-cleavage activity of two CRISPR/cas systems, Cas13a and Cas14a, and can achieve a detection limit at fM level without target amplification. Although the detection sensitivity needs to be further improved, the high-fidelity CRISPR/cas system can endow the method with single-base specificity. As an important biomarker, miRNA has attracted a lot of attention in the biomedical field.23–27 A potential breast cancer biomarker, miR-17, was selected to evaluate the detection ability of casCRISPR. The principle of casCRISPR is illustrated in Scheme 1.
When target miRNA is introduced, the trans-cleavage activity of Cas13a/crR is activated to collaterally cleave the phosphodiester bond next to rU of ST-HP (locked-trigger for Cas14a/sgRNA with two uracil ribonucleotides in the loop) and make ST-HP transform the stable hairpin structure into a duplex structure with a 5′-toehold, which can initiate the interaction with Cas14a/sgR through strand displacement. After that, the trans-cleavage activity of Cas14a that is activated by cleaved ST-HP can efficiently hydrolyze the fluorophore (FAM) and the quencher (BHQ1) labelled poly-T DNA reporter (FQTR) (sequence in Table S1, ESI†). As a result, the fluorophore of the FQTR gets rid of the quenching effect and releases fluorescence signals.
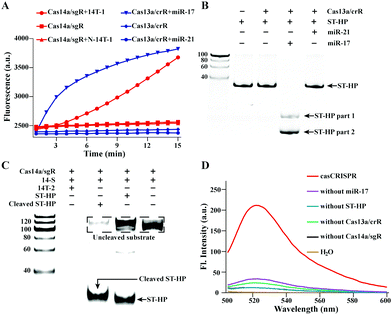
LbuCas13a used in this study was characterized in our previous work.28 The purified Cas14a was analysed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. S1, ESI†). Then, the trans-cleavage activity of Cas13a/crR and Cas14a/sgR was respectively tested by monitoring the miR-17 and ssDNA target (14T-1) caused fluorescence release of FQ5U (200 nM) and the FQTR (500 nM) (Fig. 1A). All these results confirmed that we have successfully obtained efficient Cas13a and Cas14a. Next, it is necessary to explore whether Cas13a can collaterally cleave hairpin structured DNA oligonucleotides ST-HP with unpaired rU. The urea-denaturation-PAGE result is shown in Fig. 1B. It can be clearly seen that miR-17 activated Cas13a/crR cleaved ST-HP into two fragments with lower molecular weight. Relatively, non-target miR-21 did not cause cleavage. The result confirmed that only target miR-17 can activate Cas13a/crR to cleave ST-HP. Next, the cleaved ST-HP activated trans-cleavage ability of Cas14a/sgR was also verified by PAGE. A 100 nt ssDNA (14-S) was used as the non-target substrate of Cas14a instead of the FQTR. As shown in Fig. 1C, Cas13a/crR-cleaved ST-HP can effectively activate Cas14a/sgR to hydrolyze 14-S, while the intact ST-HP could not cause the cleavage of 14-S.
Further, the casCRISPR was confirmed by fluorescence spectra analysis. FQTR was served as the reporter. The result in Fig. 1D shows that only when miR-17, Cas13a/crR, Cas14a/sgR and ST-HP existed simultaneously, there was an obvious fluorescence signal.
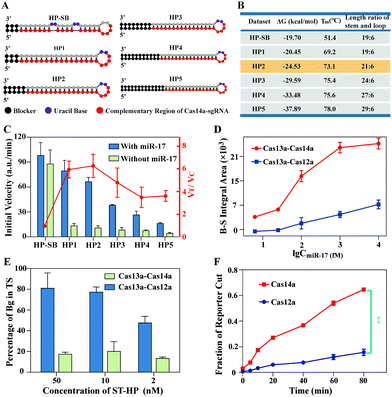
The ST-HP as a bridge between Cas13a and Cas14a plays two roles: one is to serve as the substrate of Cas13a trans-cleavage, and the other is to trigger the trans-cleavage activity of Cas14a. Thus, the properties of ST-HP are the most critical factor for the performance of casCRISPR. A series of ST-HP with different locations of rU and length ratios of stem to loop were designed. The structure and the corresponding key parameters of the ST-HP are respectively shown in Fig. 2A and B. And, the detection results of the casCRISPR with different ST-HP are shown in Fig. 2C. As shown, the bubbles in the stem increase the chances of sgRNA invading, thereby leading to obvious growth of the background signal. Thus, the rU was decided to be set in the loop of ST-HP. Next, to ensure that the cleaved ST-HP could be recognized by Cas14a/sgR efficiently, the 4 nt in the loop can serve as a toehold to efficiently initiate the subsequent strand displacement reaction after the HP is cleaved. Meanwhile, a blocker at the end of the stem is necessary to prevent the DNA breathing effect caused undesirable interference of 14a-sgR. From the analysis results of casCRISPR with HP1–HP5, it can be seen that the HP2 with a length ratio of stem-loop of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 and 5 base pairs blocker can obtain a maximum ratio of VT to VC. Therefore, the HP2 was applied in the following experiments.
6 and 5 base pairs blocker can obtain a maximum ratio of VT to VC. Therefore, the HP2 was applied in the following experiments.
Since Cas12a also can be severed as ssDNA-activated DNase, another version of casCRISPR coupled Cas13a with Cas12a (casCRISPR v2) was constructed and compared with Cas13a–Cas14a (casCRISPR v1). The IA (integral area) of the fluorescence intensity curve (Fig. 2D) demonstrated that casCRISPR v1 can detect miR-17 as low as ∼6.25 fM, while casCRISPR v2 can detect ∼100 fM miR-17. As shown in Fig. 2E, casCRISPR v2 has higher background leakage, which may be caused by the nonspecific binding and unwinding abilities of Cas12a to dsDNA.29 Moreover, Cas14a possesses higher cleavage efficiency than Cas12a when ssDNA is used as the activator (Fig. 2F). Overall, Cas13a–Cas14a can achieve a lower detection limit and a higher signal to background ratio than Cas13a–Cas12a.
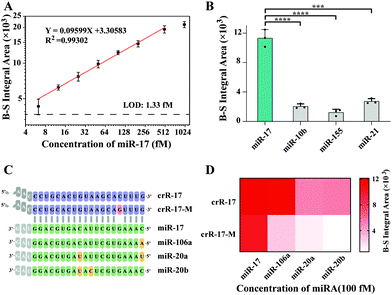
Under the optimal experimental conditions (Fig. S2, ESI†), the sensitivity of casCRISPR was tested by detecting various concentrations of miR-17. The IA of the fluorescence intensity curve was proportional to the miR-17 concentration range from 6.25 to 500 fM. The limit of detection (LOD) was calculated to be 1.33 fM. The correlation equation was obtained as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IA = 3.30583 + 0.09599 × log
IA = 3.30583 + 0.09599 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CmiR-17(fM) (R2 = 0.99302) (Fig. 3A). When the miRNA concentration exceeds 500 fM, the background-subtracted (B-S) integral area has no obvious growth, which may be due to the depletion of the cleaved ST-HP. To better understand the signal amplification efficiency of casCRISPR, a stoichiometric titration experiment was performed and the result shows that target ssDNA (14T-2) activated Cas14a/sgR catalyzed trans-cleavage of ssDNA with a rate of ∼1504 turnover per second, and the catalytic efficiency (Kcat/Km) was 1.397 × 109 s−1 M−1 (Fig. S3, ESI†).
CmiR-17(fM) (R2 = 0.99302) (Fig. 3A). When the miRNA concentration exceeds 500 fM, the background-subtracted (B-S) integral area has no obvious growth, which may be due to the depletion of the cleaved ST-HP. To better understand the signal amplification efficiency of casCRISPR, a stoichiometric titration experiment was performed and the result shows that target ssDNA (14T-2) activated Cas14a/sgR catalyzed trans-cleavage of ssDNA with a rate of ∼1504 turnover per second, and the catalytic efficiency (Kcat/Km) was 1.397 × 109 s−1 M−1 (Fig. S3, ESI†).
To evaluate the selectivity of casCRISPR, several different miRNA, including miR-17, miR-10b, miR-155 and miR-21, were respectively added into casCRISPR system with the same concentration of 50 pM. The real-time fluorescence monitoring results are shown in Fig. 3B. It can be seen that only miR-17 can result in an obvious fluorescence increase compared to other miRNA.
Furthermore, we explored the discrimination ability of casCRISPR for highly homologous miRNA family members (miR-17, 106a, 20a and 20b sequences are shown in Fig. 3C). The detection results are shown in Fig. 3D. It can be seen that casCRISPR with perfectly matched crRNA (crR-17) can distinguish miR-17 from miR-20a and miR-20b, which have two base differences to miR-17, while miR-106a only possesses a single base difference at the 5′ end compared to miR-17 and also displayed significant fluorescence signals. Accordingly, an extra mismatch base was introduced into the position 16 of the spacer of the crRNA for Cas13a (crR-17-M, the sequence shown in Fig. 3C) to improve the specificity. As shown in Fig. 3D, casCRISPR with mismatch bearing crR-17-M possesses stronger discrimination ability. This result also verified that casCRISPR can provide single-base resolution by flexibly designing the crR sequence. We also evaluated the performance of casCRISPR with crR-17-M for miR-17 detection when miR-17 was co-existing with its homologous family members, which have only one or two base differences compared to miR-17. The detection results are shown in Fig. S4 (ESI†). It can be seen that there is no significant difference in the fluorescence signal among only miR-17 and other complex miRNA samples. The result indicates that casCRISPR has high stability and can resist the interference of mismatched miRNA.
We evaluated the application ability of casCRISPR by detecting miRNA in serum samples. At first, miR-17 with different concentrations were spiked into 20 times diluted normal human umbilical cord serum and tested by casCRISPR. The results in Fig. S5A and B (ESI†) show that the IA was proportional to the miR-17 concentration from 20 to 100 fM. The LOD was calculated to be 22.3 fM. When the miRNA concentration exceeds 100 fM, the growth of the B-S integral area slows down. It can be speculated that some serum components could affect the trans-cleavage activity of the Cas enzyme. The correlation equation was obtained as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IA = 0.13776 + 0.1122 × log
IA = 0.13776 + 0.1122 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CmiR-17(fM) (R2 = 0.98811). The recovery (%) was defined as the ratio of the mean tested concentration (Cte) by casCRISPR and the spiked concentration of miR-17 (Csp). It can be confirmed that casCRISPR possesses a good reproducibility for miRNA detection in the simulated serum sample. Subsequently, three serum extract samples from breast adenocarcinoma patients were tested, and one sample from a healthy person was served as control. As shown in Fig S5C (ESI†), the relative expression of miR-17 in all of the three patient samples displayed varying degrees of increase compared to the healthy person's sample. And the results are consistent with the qRT-PCR analysis results (Fig. S6 and Table S3, ESI†).
CmiR-17(fM) (R2 = 0.98811). The recovery (%) was defined as the ratio of the mean tested concentration (Cte) by casCRISPR and the spiked concentration of miR-17 (Csp). It can be confirmed that casCRISPR possesses a good reproducibility for miRNA detection in the simulated serum sample. Subsequently, three serum extract samples from breast adenocarcinoma patients were tested, and one sample from a healthy person was served as control. As shown in Fig S5C (ESI†), the relative expression of miR-17 in all of the three patient samples displayed varying degrees of increase compared to the healthy person's sample. And the results are consistent with the qRT-PCR analysis results (Fig. S6 and Table S3, ESI†).
In addition, the miR-17 expression level in HepG2 (liver hepatocellular carcinoma), MDA-MB-231 (human breast adenocarcinoma cell), and LO2 (human normal liver cells) was detected by casCRISPR. Fig. S5D (ESI†) shows that the detection result of casCRISPR is consistent with the qRT-PCR result that the miR-17 expression in HepG2 was much higher than in LO2 and MDA-MB-231. We compared the features and performance of casCRISPR with qRT-PCR (Table S4, ESI†). casCRISPR can shorten the reaction time by avoiding complex reverse transcription and target amplification processes. And, the isothermal reaction process makes casCRISPR more suitable for point-of-care detection. More importantly, casCRISPR can provide higher specificity over qRT-PCR due to the high-fidelity recognition ability of Cas13a/crRNA.
In summary, this is the first time a cascade CRISPR/cas system, termed casCRISPR, has been developed to realize high-sensitive and specific miRNA detection without target amplification. Under optimized conditions, the homogenous casCRISPR fluorescence detection system achieved a detection limit of 1.33 fM for miR-17, and dramatically distinguished miR-17 from its high-homologous family members. The casCRISPR also can be employed to detect miRNA in complex cell extracts and serum samples. Furthermore, another version of casCRISPR, Cas13a–Cas12a, was constructed and systemically compared with Cas13a–Cas14a. With newer and functionally improved Cas protein being discovered or created, more versions of casCRISPR with different Cas/sgRNA combinations could be constructed to play greater roles in biosensing and molecular diagnosis.
We give special thanks to Professor Yanli Wang for the generous gift of the LbuCas13a expression plasmid. This work was supported by the National Natural Science Foundation of China (NSFC) (81630046); the Natural Science Foundation of Guangdong Province, China (2020A1515010525); the China Postdoctoral Science Foundation (2018M643113); and the Science and Technology Program of Guangzhou (2019050001).
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709 CrossRef CAS.
- L. A. Marraffini and E. J. Sontheimer, Science, 2008, 322, 1843 CrossRef CAS.
- J. Qiao, W. Sun, S. Lin, R. Jin, L. Ma and Y. Liu, Chem. Commun., 2019, 55, 4707–4710 RSC.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262–1278 CrossRef CAS.
- X. Zhu, M. M. Lv, J. W. Liu, R. Q. Yu and J. H. Jiang, Chem. Commun., 2019, 55, 6511–6514 RSC.
- W. Chang, W. Liu, H. Shen, S. Chen, P. Liao and Y. Liu, Anal. Chim. Acta, 2020, 1112, 46–53 CrossRef CAS.
- J. Wang, T. Bing, N. Zhang, X. Liu and D. Shangguan, Chem. Commun., 2020, 56, 10038–10041 RSC.
- Y. Li, S. Li, J. Wang and G. Liu, Trends Biotechnol., 2019, 37, 730–743 CrossRef CAS.
- R. Zhou, Y. Li, T. Dong, Y. Tang and F. Li, Chem. Commun., 2020, 56, 3536–3538 RSC.
- Y. Chen, S. Yang, S. Peng, W. Li, F. Wu, Q. Yao, F. Wang, X. Weng and X. Zhou, Chem. Sci., 2019, 10, 2975–2979 RSC.
- M. Huang, X. Zhou, H. Wang and D. Xing, Anal. Chem., 2018, 90, 2193–2200 CrossRef CAS.
- K. Zhang, R. Deng, X. Teng, Y. Li, Y. Sun, X. Ren and J. Li, J. Am. Chem. Soc., 2018, 140, 11293–11301 CrossRef CAS.
- G. Dugar, R. T. Leenay, S. K. Eisenbart, T. Bischler, B. U. Aul, C. L. Beisel and C. M. Sharma, Mol. Cell, 2018, 69(893–905), e897 Search PubMed.
- S. Y. Li, Q. X. Cheng, J. K. Liu, X. Q. Nie, G. P. Zhao and J. Wang, Cell Res., 2018, 28, 491–493 CrossRef CAS.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839 CrossRef CAS.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839 CrossRef CAS.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436 CrossRef CAS.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, J. Am. Chem. Soc., 2020, 142, 207–213 CrossRef CAS.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, Adv. Mater., 2019, 31, 1970365 CrossRef.
- R. Wang, X. Zhao, X. Chen, X. Qiu, G. Qing, H. Zhang, L. Zhang, X. Hu, Z. He, D. Zhong, Y. Wang and Y. Luo, Anal. Chem., 2020, 92, 2176–2185 CrossRef CAS.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X. F. Yu, Nat. Commun., 2018, 9, 5012 CrossRef.
- K. Zhang, R. Deng, Y. Li, L. Zhang and J. Li, Chem. Sci., 2016, 7, 4951–4957 RSC.
- Y. Sun and T. Li, Anal. Chem., 2018, 90, 11614–11621 CrossRef CAS.
- A. Chen, G. F. Gui, Y. Zhuo, Y. Q. Chai, Y. Xiang and R. Yuan, Anal. Chem., 2015, 87, 6328–6334 CrossRef CAS.
- K. Chen, Q. Huang, T. Fu, G. Ke, Z. Zhao, X. Zhang and W. Tan, Anal. Chem., 2020, 92, 7404–7408 CrossRef CAS.
- Q. Zhang, F. Chen, F. Xu, Y. Zhao and C. Fan, Anal. Chem., 2014, 86, 8098–8105 CrossRef CAS.
- W. Wu, T. Zheng and Y. Tian, Chem. Commun., 2020, 56, 8083–8086 RSC.
- Y. Shan, X. Zhou, R. Huang and D. Xing, Anal. Chem., 2019, 91, 5278–5285 CrossRef CAS.
- W. Jiang, J. Singh, A. Allen, Y. Li, V. Kathiresan, O. Qureshi, N. Tangprasertchai, X. Zhang, H. P. Parameshwaran, R. Rajan and P. Z. Qin, ACS Omega, 2019, 4, 17140–17147 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc06412b |
| This journal is © The Royal Society of Chemistry 2021 |