Open Access Article
Open Access ArticleEPR imaging of sinapyl alcohol and its application to the study of plant cell wall lignification†
Clémence
Simon‡
 a,
Cédric
Lion‡
a,
Cédric
Lion‡
 *a,
Hania
Ahouari
*a,
Hania
Ahouari
 b,
Hervé
Vezin
b,
Hervé
Vezin
 b,
Simon
Hawkins
a and
Christophe
Biot
b,
Simon
Hawkins
a and
Christophe
Biot
 *a
*a
aUniv. Lille, CNRS, UMR 8576 – UGSF – Unité de Glycobiologie Structurale et Fonctionnelle, 59000 Lille, France. E-mail: cedric.lion@univ-lille.fr; christophe.biot@univ-lille.fr
bUniv. Lille, CNRS, UMR 8516 – LASIRE – Laboratoire de Spectroscopie pour les Interactions, la Réactivité et l’Environnement, 59000 Lille, France
First published on 8th December 2020
Abstract
In bioimaging, bioorthogonal chemistry is most often used to visualize chemical reporters by fluorescence in their native environment. Herein, we show that TEMPO-based probes can be ligated to monolignol reporters by Diels–Alder chemistry in plant cell walls, paving the way for the study of lignification by EPR spectroscopy and imaging.
Over the past two decades, the chemical reporter strategy has allowed scientists to trace the behaviour of biomolecules in living systems according to a 2-step methodology,1–3 the first of which involves the metabolic incorporation of a reporter in the organism, tissue or cells under scrutiny. This reporter is a synthetic analogue of the biomolecule being studied bearing a small innocuous chemical tag. The second step consists of covalently binding a molecular probe to the reporter through bioorthogonal chemistry, thus allowing the detection of the metabolically tagged biomolecule directly in its natural environment. The probe used is often a fluorophore for visualising biomolecule distribution by fluorescence microscopy.
This bioorthogonal approach has been widely used in bacteria and animal cells,4 and has also been applied to plant models although to a lesser extent. Among its many applications, the chemical reporter strategy has notably been successfully employed to study lignification dynamics in plant cell walls.5–8 In addition to its vital roles for plant development and defence, lignin is a major component (together with cellulose and hemicelluloses) of lignocellulosic biomass, an economically important and renewable agro-resource utilized in many industrial processes.9 It is a polyphenolic polymer synthesized in the apoplastic cell wall domain from phenylpropanoid metabolites called monolignols. The oxidative polymerisation process is initiated by conversion of these monomers into delocalized phenoxy radicals by two classes of enzymes (laccases and/or peroxidases), followed by their spontaneous cross-coupling with radicals formed on the phenolic termini of growing lignin chains. In angiosperms, lignin is composed of three main monolignols, namely coniferyl, sinapyl and p-coumaryl alcohols. After polymerisation they are called guaiacyl (G), syringyl (S) and hydroxyphenyl (H) units, respectively.10 Although several studies have improved our knowledge about lignification, many aspects about its spatiotemporal regulation remain unknown.11,12 Of the different methodologies available to study lignification, the chemical reporter strategy seems to be highly appropriate.13 Our group and others have successfully demonstrated the metabolic incorporation of analogues of all of the three main monolignols into lignin and their detection in vivo by fluorescence after bioorthogonal click reactions.5–8 The use of such single or multiple labelling approaches has allowed a better understanding of the distribution of monolignols in the plant cell wall. However, this strategy alone cannot provide answers to certain questions. For example, it is difficult to obtain information about the physicochemical environment of the polymerised monolignol analogue from the fluorescence signal. Similarly, it is not possible to quantify the relative proportions of monolignol reporters incorporated into lignin.
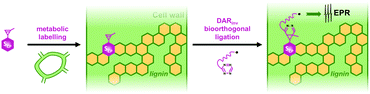
We present herein a novel approach for obtaining new insights about lignification dynamics directly in the plant cell wall. This approach combines the bioorthogonal chemical reporter strategy with Electron Paramagnetic Resonance (EPR) spectroscopy via the in situ bioconjugation of spin-labels (Fig. 1). We synthesized three paramagnetic tags based on the 2,2,6,6-tetramethyl-1-piperidinyloxyl radical (TEMPO) bearing a 1,2,4,5-tetrazine moiety, thereby enabling them to readily react with a cyclopropenyl-tagged analogue of a monolignol previously polymerized into the plant cell wall. The paramagnetic signal of the incorporated monolignol reporter in lignin was successfully measured by EPR spectroscopy, and allowed mapping of the incorporated tagged monolignols by EPR imaging as a proof of concept.
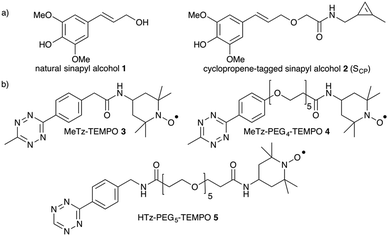
Our choice of chemical reporter was directed by biological relevance and chemical compatibility with the envisaged probes. Although copper-catalyzed alkyne–azide cycloaddition (CuAAC) has previously been used in solution for DNA labelling,14 in our hands alkyne-tagged TEMPO did not lead to successful conjugation onto azide-functionalized monolignols using Cu(I) as a catalyst in plant samples. Similarly, the classic CuSO4/sodium ascorbate catalytic system usually used in aqueous applications did not provide satisfying results either, possibly due to reduction of nitroxides to hydroxylamines in situ. As a result, we turned to Diels–Alder with inverse electronic demand (DARinv), as we deemed this reaction less prone to interfere with spin-labels. Furthermore, in our previous fluorescence imaging studies7,8 tetrazine–alkene DARinv proved to be superior to alkyne–azide chemistry in terms of specific to aspecific ratio (possibly as a result of faster kinetics), thereby facilitating subsequent data processing. Moreover, its implementation is simpler as it does not require the use of copper or polytriazole ligand. We thus opted for an analogue of sinapyl alcohol 1 bearing a methylcyclopropenyl tag that we recently developed and validated by fluorescence imaging.8 This reporter SCP2 (Fig. 2a) was synthesized in 7 steps from sinapic acid in a 12% global yield. DARinv has also been recently used by the Wittmann group for spin labelling of proteins in solution with photocaged nitroxides, as has the Suzuki–Miyaura bioorthogonal reaction.15,16
We then turned our attention to the design of suitable paramagnetic probes to detect SCP incorporation by electron paramagnetic resonance spectroscopy in situ in order to generate spin maps of plant cross-sections by EPR imaging. Because the use of nitroxide radicals is well established in biological applications, we selected TEMPO as the paramagnetic moiety. TEMPO probes are stabilized by delocalization of the radical over the N–O bond as well as by steric protection imparted by the methyl substituents, and have been shown to persist long enough in living models to enable EPR imaging.17 In order to maximize our chances and to investigate the effect of tetrazine reactivity and mobility on the detected signal, we synthesized three tetrazine-functionalized TEMPO derivatives (Fig. 2b). Firstly, a methyltetrazine (MeTz) moiety was covalently linked to 4-amino-TEMPO by reaction with an activated NHS-ester generating 3 in an 83% yield (see ESI†).
The small size of this molecular probe facilitates its mobility within the cell wall matrix allowing it to access the SCP reporters incorporated into the lignin polymer. In addition, its short and rigid spacer arm minimizes potential interactions with nearby mono- or oligolignol radicals in the cell wall, which could lead to spin-trapping and decreased lifetime of the nitroxide spin-label. Secondly, we synthesized a more flexible probe by introducing an oligo-PEG arm, that had the added benefit of improved water solubility. MeTz–PEG4–TEMPO 4 was synthesized with an 89% yield (see ESI†) and allowed us to evaluate the impact of the spacer on the efficiency of the labelling step. Finally, we also decided to synthesize a H-tetrazine (HTz) probe. Although the chemical stability of H-tetrazines is substantially lower compared to methyltetrazines, their lifetime corresponds to an acceptable range for a number of applications in chemical biology. In addition, they exhibit the fastest kinetic rates ever observed for bioorthogonal reactions and we considered that they might make suitable probes.18 HTz–PEG5–TEMPO 5 was prepared in a similar fashion to the other probes with a 99% yield (see ESI†).
After validating the efficiency of the DARinv conjugation to SCP2 in solution, we confirmed the paramagnetic behaviour of TEMPO derivatives 3, 4 and 5 by continuous wave (CW) EPR spectroscopy experiments (Fig. S1 and S2, ESI†).
As expected, the spectra obtained for the probes in aqueous solution are composed of three equidistant lines characteristic of isotropic nitroxide radical (hyperfine coupling constant α = 17 G). The chemical stability of 3, 4 and 5 in water at 4 °C was verified several times over a period of 6 weeks.
We then tested the synthesized probes in flax plants (Linum usitatissimum L.) in order to investigate whether actively lignifying tissues could be detected by EPR spectroscopy. Ex vivo metabolic incorporation of SCP (10 μM) into living flax stem cross-sections was carried out for 20 h in the light to allow its polymerization into cell wall lignin, according to a protocol previously developed in our laboratory.8 The efficiency of SCP reporter incorporation into cell walls was first confirmed by fluorescence imaging after subsequent DARinv ligation of Cy5–tetrazine (Fig. S3, ESI†). We then attempted the direct bioorthogonal ligation of the spin-label probes. EPR spectra were acquired at room temperature by depositing the labelled stem cross sections vertically on EPR-compatible quartz tubes. Initial results showed low signal to noise ratio (Fig. S4, ESI†) possibly resulting from degradation of the TEMPO radical by plant active redox enzymes and reactive oxygen species/other compounds and/or from unreacted excess free probe trapped within plant cells. We therefore decided to see whether the inclusion of a paraformaldehyde fixation step after metabolic incorporation could improve the specific to aspecific ratio. Our results (Fig. S4, ESI†) clearly indicated that signal to noise ratio was greatly improved and we therefore included this step in all further experiments. For each EPR experiment, each spectrum represents the average spectrum acquired from nine technical replicates and all measurements were performed three times on three biological replicates each time. In order to confirm that the observed EPR signal is specific to SCP labelling, we also acquired spectra from negative control samples (plant stem sections submitted to bioorthogonal labelling conditions with 3, 4 or 5, but without prior metabolic incorporation of 2). Our results (Fig. 3 and Fig. S7, ESI†) show that the EPR spectra obtained from plant samples metabolically labelled with 2 exhibit a conspicuously broadened three-band signal characteristic of anisotropic interactions. This signal can thus be attributed to slow motion paramagnetic centers immobilized within the lignocellulosic network of the cell wall easily distinguished from the narrow isotropic signal corresponding to free-probes (Fig. S2, ESI†).
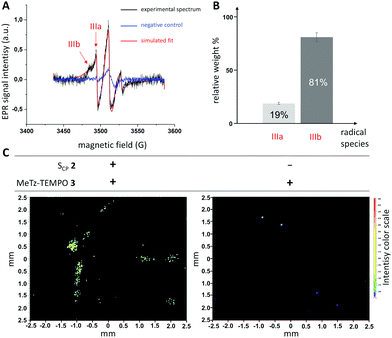 | ||
| Fig. 3 (A) CW-EPR spectra recorded at room temperature on flax cross sections using MeTz–TEMPO 3 as the spin-probe. Experiments were conducted on samples previously incubated with reporter 2 (black spectrum) or without (blue spectrum, negative control). The EPR spectra were fitted with the EasySpin toolbox (red trace) to calculate the contribution of each species. (B) relative weights of radical species IIIa and IIIb in the sample incubated with 2. (C) Ex vivo EPR imaging of flax cross-sections in the presence (left image) or absence (right image, negative control) of tagged-SCP. The signals were acquired with a field-of-view of 5 mm and gradient strength of 175 G cm−1 (see ESI†). | ||
In contrast to the results obtained with 3 and 4, the broad signal observed for 5 (Fig. S7C, ESI†) also consistently appeared in negative control spectra and in the same proportions. This result suggests that probe 5 covalently binds to the sample in a non-specific manner in unexpectedly high proportions. This could be due to degradation of the less stable H-tetrazine in this complex biological environment thereby generating intermediates that react with endogenous biomolecules.19 In this context, the presence of a 6-methyl substituent on the 1,2,4,5-tetrazine moiety seems mandatory to ensure biorthogonality. We therefore did not use 5 in the following experiments. In contrast, methyltetrazine derivatives 3 and 4 provided much more satisfying results as the characteristic broadened signal was almost completely absent in negative controls (Fig. 3 and Fig. S7A, ESI†). It can therefore be assumed that these spectra are specific of the DARinv ligation of 3 and 4 to methylcyclopropene reporter side-chains of SCP monomers polymerized into cell wall lignin, which is consistent with previously reported results using methyltetrazines for fluorescence studies.
Since spectral lines are often broadened by dynamic and/or static effects, simulations by computational methods that take orientational disorder into account are generally resorted to when interpreting EPR data. In order to get access to mobility parameters and relative quantification of paramagnetic species with different motion freedom within the same sample (i.e., specific vs. nonspecific), the room temperature CW-EPR spectra were fitted using Easyspin (see ESI†).20
Spectra obtained from plant samples tagged with 3 show the presence of two contributing species IIIA and IIIB with relative weights of 19% and 81%, respectively (Table 1 and Fig. 3B). Whereas parameters determined for the minor IIIA correspond to background residual free probe in solution (fast tumbling, Azz = 17 G), the spectral parameters of IIIB clearly indicate an immobilized TEMPO moiety with slow-motion, which can be attributed to 3 covalently bound to SCP-units. This species corresponds to specific labeling of de novo lignin within the sample (i.e., lignin biosynthesized during the metabolic incorporation experiment) as it is virtually absent from negative controls and is the major form observed (81% relative weight in the signal). Indeed, the formation of the rigid dihydropyridazine linkage together with the conformationally restricted nature of the spacer (only one sp3 carbon atom) explains its lower mobility which is clearly shown by the increased hyperfine coupling constant (Azz = 37 G) and correlation time (τc = 182 ns).
| Species | 2Azz (G) | τ c (s−1) | Relative weight ± SD | Concentration (μM per cross-section) ± SD |
|---|---|---|---|---|
| IIIA | 34 | 1.73 × 10−9 | 0.19 ± 5% | 0.36 ± 5% |
| IIIB | 74 | 1.82 × 10−7 | 0.81 ± 5% | 1.54 ± 5% |
| IVA | 34 | 3.23 × 10−9 | 0.22 ± 5% | 0.75 ± 5% |
| IVB | 75 | 4.67 × 10−8 | 0.51 ± 8% | 1.74 ± 8% |
| IVC | 101 | 8.73 × 10−8 | 0.23 ± 8% | 0.79 ± 8% |
These results represent the first time that metabolic incorporation of a biomolecule and its detection by EPR spectroscopy after bioorthogonal click labelling have been combined in a plant, and more widely in a eukaryotic model. In vivo bioorthogonal spin labeling has however, also been very recently applied to proteins in bacteria using CuAAC ligation.21
Although the presence of residual unbound probe in the sample is usually considered a hindrance in data processing, we were able to use it to our advantage to quantify the amount of labelled SCP within the sample. For this, a calibration curve was produced in the 1 to 200 μM range (Fig. S5, ESI†), which then allowed us to determine the concentration of IIIA in the measured samples. Given the relative weights calculated for each species, this technique allows us to quantify the amount of immobilized probe IIIB specifically linked to SCP monomers polymerized into lignin. Although 4 also gave a specific signal by EPR and led to similar results (Fig. S7A, ESI†), a significant difference arose (Table 1). The simulations calculated from the acquired spectra clearly show that there are two major immobilized contributors to the spectrum IVB and IVC, in addition to the residual signal for free probe IVA (Table 1 and Fig. S7B, ESI†). Whereas IVA exhibits a mobility very similar to the previously observed IIIA (Azz = 17 G, τc = 2 and 3 ns, respectively), the two other contributors IVB and IVC to the EPR signal are even more immobilized (τc = 46 and 87 ns, respectively). It could be hypothesized that the length of the spacer arm and its flexibility allows probe 4 to explore a larger environmental pocket around the tagged lignin site thereby enabling it to enter into denser and/or smaller cell wall spaces that are less accessible to 3. We then went on to use the nitroxide radical footprint of 3 specifically labeled to lignin in the plant cell wall to perform EPR imaging (Fig. 3C). The EPR images were obtained on two sets of 9 flax stem cross-sections with (left), or without (right) prior metabolic incorporation of 2. Careful examination indicated that the high signals observed in the left-hand image are associated with plant samples and most likely result from the detection of labelled lignin in flax xylem tissue. Differences in signal intensity observed between samples can be related to the fact that the flax cross-sections are orientated differently in the tube (Fig. S8, ESI†). It is unlikely that these differences reflect variation in SCP incorporation as DARinv fluorophore ligation (Fig. S3, ESI†) gave highly reproducible results in different sections in agreement with our previous studies in this species.7,8,13 In contrast, a much lower overall signal was observed in samples that had not metabolically incorporated 2. Interestingly, a closer look at the observed signal indicated that the marking spots were between 5 and 10 μm in size suggesting that there are privileged zones in which the probe is located. Since these dimensions are comparable to those of the cell walls in the lignifying region of the young xylem (Fig. S3, ESI†) it is tempting to speculate that these zones demonstrate the imaging of de novo lignification at the cellular level. However, further refinement of this approach and continued experimentation are clearly necessary to confirm this hypothesis. Although EPR imaging has previously been used in plants, to our knowledge, this is the first example of its application to plant cell wall polymers,22–24 and the only one treating with the labelling of exogenous reporters. Our work was done at RT as a proof of concept and it should be noted that imaging samples at cryogenic temperature would allow an improvement of the signal to noise ratio of the image in future studies.
In conclusion, we have demonstrated that the bioorthogonal chemical reporter strategy can be combined with EPR spectroscopy in plant tissues to detect the incorporation of a cyclopropenyl-tagged monolignol into the lignin polymer. We show that the production of de novo lignin can be detected by another method in addition to fluorescence. Simulation of the recorded EPR spectrum provides mobility parameters about the paramagnetic probe specifically tagged to lignin and should allow us to obtain spatial information about its polymerization dynamics in future studies. The spin labeling strategy presented here could also be transposed to other major plant cell wall polymers like pectin or hemicelluloses of importance in the context of lignocellulosic biomass valorization.
Conflicts of interest
There are no conflicts to declare.References
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- C. G. Parker and M. R. Pratt, Cell, 2020, 180, 605–632 CrossRef CAS PubMed.
- Kenry and B. Liu, TRECHEM, 2019, 1, 763–778 CAS.
- N. Bukowski, J. L. Pandey, L. Doyle, T. L. Richard, C. T. Anderson and Y. Zhu, Bioconjugate Chem., 2014, 25, 2189–2196 CrossRef CAS PubMed.
- Y. Tobimatsu, D. Van de Wouwer, E. Allen, R. Kumpf, B. Vanholme, W. Boerjan and J. Ralph, Chem. Commun., 2014, 50, 12262–12265 RSC.
- C. Lion, C. Simon, B. Huss, A.-S. Blervacq, L. Tirot, D. Toybou, C. Spriet, C. Slomianny, Y. Guerardel, S. Hawkins and C. Biot, Cell Chem. Biol., 2017, 24, 326–338 CrossRef CAS PubMed.
- C. Simon, C. Lion, C. Spriet, F. Baldacci-Cresp, S. Hawkins and C. Biot, Angew. Chem., Int. Ed., 2018, 57, 16665–16671 CrossRef CAS PubMed.
- Z. Chen and C. Wan, Renewable Sustainable Energy Rev., 2017, 73, 610–621 CrossRef CAS.
- R. Vanholme, B. Demedts, K. Morreel, J. Ralph and W. Boerjan, Plant Physiol., 2010, 153, 895–905 CrossRef CAS PubMed.
- Y. Tobimatsu and M. Schuetz, Curr. Opin. Biotechnol, 2019, 56, 75–81 CrossRef CAS PubMed.
- R. A. Dixon and J. Barros, Open Biol., 2019, 9, 190215 CrossRef PubMed.
- C. Simon, C. Lion, C. Biot, N. Gierlinger and S. Hawkins, Annual Plant Reviews online, American Cancer Society, 2018, pp. 1–32 Search PubMed.
- P. Ding, D. Wunnicke, H.-J. Steinhoff and F. Seela, Chemistry, 2010, 16, 14385–14396 CrossRef CAS PubMed.
- A. Kugele, B. Silkenath, J. Langer, V. Wittmann and M. Drescher, ChemBioChem, 2019, 20, 2479–2484 CrossRef CAS PubMed.
- A. Kugele, T. S. Braun, P. Widder, L. Williams, M. J. Schmidt, D. Summerer and M. Drescher, Chem. Commun., 2019, 55, 1923–1926 RSC.
- G. Yan, L. Peng, S. Jian, L. Li and S. E. Bottle, Chin. Sci. Bull., 2008, 53, 3777–3789 CAS.
- B. L. Oliveira, Z. Guo and G. J. L. Bernardes, Chem. Soc. Rev., 2017, 46, 4895–4950 RSC.
- M. R. Karver, R. Weissleder and S. A. Hilderbrand, Bioconjugate Chem., 2011, 22, 2263–2270 CrossRef CAS PubMed.
- S. Stoll and A. Schweiger, J. Magn. Reson., 2006, 178, 42–55 CrossRef CAS PubMed.
- P. Widder, J. Schuck, D. Summerer and M. Drescher, Phys. Chem. Chem. Phys., 2020, 22, 4875–4879 RSC.
- K. Nakagawa and B. Epel, Spectrochim. Acta, Part A, 2014, 131, 342–346 CrossRef CAS PubMed.
- K. Nakagawa and H. Hara, Free Radical Res., 2015, 49, 1–6 CrossRef CAS PubMed.
- J. L. Berliner and H. Fujii, Science, 1985, 227, 517–519 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc05218c |
| ‡ Co-first authors with equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |