Open Access Article
Open Access ArticleSquaric acid as a new chemoselective moiety for mass spectrometry-based metabolomics analysis of amines†
Weifeng
Lin
 a,
Zhen
Yang
a,
Amanpreet
Kaur
a,
Zhen
Yang
a,
Amanpreet
Kaur
 a,
Annika
Block
a,
Miroslav
Vujasinovic
b,
J.-Matthias
Löhr
bc and
Daniel
Globisch
a,
Annika
Block
a,
Miroslav
Vujasinovic
b,
J.-Matthias
Löhr
bc and
Daniel
Globisch
 *a
*a
aDepartment of Chemistry–BMC, Science for Life Laboratory, Uppsala University, Box 599, Uppsala SE-75124, Sweden. E-mail: Daniel.globisch@scilifelab.uu.se
bDepartment for Digestive Diseases, Karolinska University Hospital, Stockholm, Sweden
cDepartment of Clinical Science, Intervention and Technology (CLINTEC), Karolinska Institute, Stockholm, Sweden
First published on 12th August 2021
Abstract
The investigation of microbiome-derived metabolites is important to understand metabolic interactions with their human host. New methodologies for mass spectrometric discovery of undetected metabolites with unknown bioactivity are required. Herein, we introduce squaric acid as a new chemoselective moiety for amine metabolite analysis in human fecal samples.
The gut is one of the most important organs of human metabolism. It harbours complex interconnected communities of bacteria, fungi, viruses, and yeast, which is collectively referred to as gut microbiota.1 Within the past decade, an increasing number of studies has demonstrated the complexity of the gut microbiota and their impact on human physiology. The microbiome can also be considered as an additional organ or even a “second brain” as the gut-brain axis has been identified.2,3 These microbial communities are highly metabolically active and exchange metabolites with other microbes and their host.4 While metagenomic analyses rapidly elucidate details on microbial species and community changes, our understanding of the metabolism in the gut is still limited. Mass spectrometric-based metabolomics is the method-of-choice for investigation of this metabolic relationship and the elucidation of affected pathways. However, the majority of these metabolites are still unknown due to the complexity of metabolic processes in this interspecies communication and limited analytical methods.5,6 The detailed elucidation of the metabolic exchange between the human host and its gut microbiota has a tremendous potential for the discovery of small molecule biomarkers of diseases and bioactive compounds.7
Amine-containing metabolites have important roles in biological processes and this class of metabolites contains the largest percentage of gut microbiota-derived metabolites.8 Especially, amino acids, short peptides as well as neurotransmitters are crucial metabolites for human physiology. However, it is estimated that many metabolites still need to be discovered in human fecal samples and their chemical structures elucidated.7 The identification of these metabolite classes in fecal samples is very important to provide greater insight into gut microbiota metabolism associated with human health as this sample type contains a high abundance of gut microbiota-derived metabolites. Chemical modification is a useful technique to enrich metabolites and to enhance their detection.9–11 However, the available chemical derivatization methods have to overcome matrix interferences in mass spectrometric analysis. For examples, N-hydroxysuccinimide (NHS) is the most commonly used moiety for labelling amine-containing metabolites but it rapidly hydrolyzes under neutral conditions. Other moieties have also been reported for investigation of amine metabolites such as methyl acetimidate and dansyl chloride.9
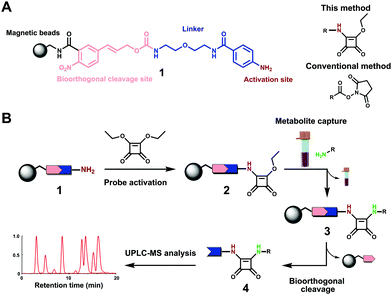
We have recently introduced a new methodology using chemoselective probes immobilized to magnetic beads for mass spectrometric analysis of human samples.12–14 To gain access to additional amine metabolites, we sought to activate our chemoselective probe with a new and more stable moiety for chemoselective amine analysis. 3,4-Diamino analogues of squaric acid (squaramides) are utilized for selective amine conjugation in diverse research fields such as materials science, medicinal chemistry and sensors.15 In addition, squaric acids are used for bioconjugation of macromolecules, specific cell labelling and bioisosteric replacement due to a high selectivity for reaction with amines under slightly basic conditions. Additionally, Tietze et al. reported the efficient use of squaric acid diesters for sequential conjugation of two amines with different structures in 1991.16 However, this chemoselective functional group has so far not been reported for metabolomics-based investigations.
Due to these beneficial properties of the squaric acid moiety, we have activated our recently developed chemoselective probe with this moiety for analysis of amine-containing metabolites in human samples. In contrast to previous reports, we have shortened the scaffold synthesis for immobilization to amine activated beads 1 (Fig. 1A). This chemical biology tool is immobilized to magnetic beads, which allows for separation of the captured metabolites from the sample matrix. Straightforward amidation of the squaric acid ester with magnetic beads-immobilized probe 1 is performed (Fig. 1B). Activated probe 2 can be incubated with any metabolite extract. The analytes for analysis with ultra-performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS) were released from the magnetic beads under mild bioorthogonal conditions using Pd(0) catalysis (Scheme S1, ESI†).14
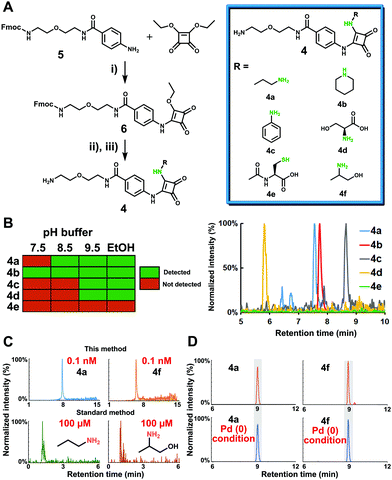
Before applying this new methodology to human samples, we first tested the properties of the probe activation, conjugation chemistry and stability of the released metabolite conjugates on synthesized simplified probe 6 (Fig. 2A and Scheme S2, ESI†). The conditions for synthesis of 6 from intermediate 5 with squaric acid diethylester were optimized for activation of immobilized probe 1. Monosquaramide 6 was then tested for reactivity and chemoselectivity with five representative metabolites to cover diverse amine functionalities present in metabolites. The reaction of 6 with 1-aminopropane (primary amine), piperidine (secondary amine), aniline (aromatic amine), L-serine (amino acid), and N-acetyl-L-cysteine (thiol) was monitored under different reaction conditions such as pH 7.5, pH 8.5, pH 9.5 as well as 1% trimethylamine in ethanol (Fig. 2B and Fig. S1, ESI†). The results were analyzed via UPLC–MS analysis and confirmed that basic conditions or addition of an organic base leads to higher reactivity.17 Importantly, this moiety only reacted with amine-containing molecules and not with the thiol nucleophile N-acetyl-L-cysteine demonstrating the chemoselectivity for amines under the tested conditions. Based on these successful results, we synthesized metabolite conjugates 4a–e to determine the limit of detection (LOD). The lowest LOD value was determined to be 80 pM for the conjugate of 1-aminopropane 4a in positive MS ionization mode, which corresponds to 400 amol. As our method is based on magnetic beads separation from the sample matrix prior to cleavage and UPLC–MS analysis, this high mass spectrometric sensitivity determined by the LOD measurements for metabolite conjugates is similar to captured metabolites from human samples due to reconstitution to the same solution. The LOD for unconjugated 1-aminopropane was determined to be 100 μM, which demonstrates a mass spectrometric sensitivity increase by a factor of 1.25 million for 4a. The LOD for the other tested metabolites ranged from 100 pM to 1.0 μM (Fig. 2C and Table 1, Table S1, ESI†).
| Concentration | Signal/Noise | ||||
|---|---|---|---|---|---|
| 4a | 4b | 4c | 4d | 4f | |
| 100 μM | 1285.81 | 1181.49 | 123.09 | 43.39 | 886.35 |
| 10 μM | 1056.63 | 1101.21 | 42.98 | 37.25 | 586.38 |
| 1.0 μM | 629.76 | 812.36 | 14.41 | 10.39 | 283.95 |
| 100 nM | 102.18 | 182.47 | 4.43 | 1.02 | 67.36 |
| 10 nM | 34.84 | 60.43 | ND | ND | 21.89 |
| 1.0 nM | 23.71 | 46.83 | ND | ND | 13.75 |
| 100 pM | 11.85 | 20.78 | ND | ND | 6.78 |
| 80 pM | 3.68 | ND | ND | ND | 1.28 |
| 50 pM | ND | ND | ND | ND | ND |
Next, we determined the stability of squaramides of the general structure 4 under the cleavage conditions for the bioorthogonal moiety p-nitrocinnamyloxycarbonyl (Noc) that is the key part of the probe design. The stability under cleavage conditions using Pd(OAc)2, PPh3 and dimethylbarbituric acid was analyzed to eliminate the possibility of undesired decomposition of the captured metabolites in human samples. All conjugates were found unaltered even after 16 hours of treatment confirming the stability of 4 throughout our entire sample preparation procedure (Fig. 2D and Fig. S2, ESI†).
Upon method optimization, we treated human fecal samples collected from three different pancreatic cancer patients with probe 2. The analysis of this sample type is crucial as only a small percentage of metabolites present has been discovered and a high number of metabolites are derived from microbiome metabolism.8 The new method in this study can be applied for the analysis of known and the discovery of yet unknown metabolites. The mass spectrometric and metabolomics-based discovery of biomarkers or disease modulators in human fecal samples has so far been limited partly due to the lack of advanced analytical tools.
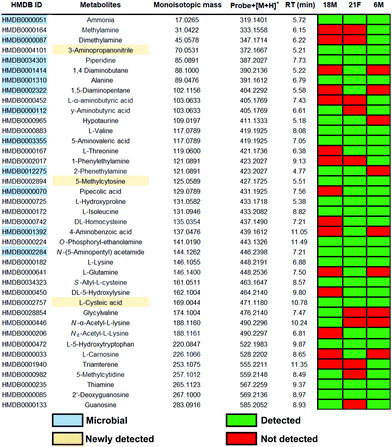
The treatment of fecal samples with 2 was performed according to our previously reported procedures with minor modification.13 In brief, metabolites from fecal samples were extracted using a specialized lysing matrix-based homogenizer and the samples were reconstituted in ethanol. Probe 2 was next incubated with metabolite extracts to form stable squaramides with amine metabolites as well as a control sample with unconjugated magnetic beads (Fig. 3).13 After separation from the sample matrix, we treated the captured metabolites 3 with the bioorthogonal cleavage conditions to release the metabolite conjugates for subsequent UPLC–MS analysis. In parallel, unmodified beads were also incubated and treated with identical cleavage conditions as control for the bioinformatic analysis. The data was processed in R using the XCMS metabolomics framework. The data analysis identified more than 3000 significantly altered features after removal of features following these criteria: (i) m/z values smaller than the ammonium conjugate (<319.1401 Da), (ii) less abundant in the metabolite extract compared to the control sample, (iii) not significantly altered features (p-values > 0.05), (iv) features that eluted before 1.5 min. The filtering process was followed by the comparison of the m/z values from the candidate features with the Human Metabolome Database (HMDB) to determine their tentative chemical structures.18
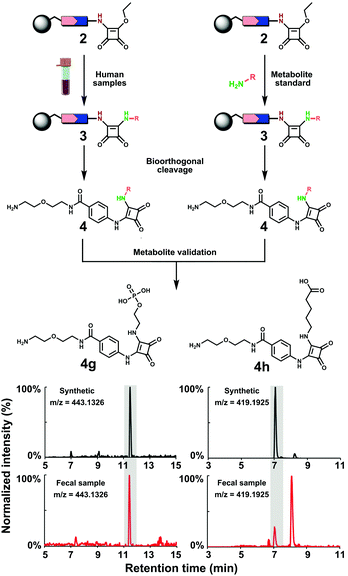 | ||
| Fig. 3 Metabolite validation and library construction. Workflow for co-injection experiments with conjugated O-phosphoryl-ethanolamine (4g) and 5-aminovaleric acid (4h) as representative examples. | ||
This analysis resulted in 165 metabolites that were annotated with a chemical structure with a 10 ppm accuracy (Fig. S3–S6, ESI†). This number exceeds conventional metabolomics investigations as well as other amine derivatization methods.19,20 For determination of the metabolite structure at the highest confidence level, a synthetic reference metabolite is required to distinguish between regioisomers.21 This level of structure validation is the current bottleneck of metabolomics research. We have constructed a metabolite conjugate library directly from conjugated probe 2 and treatment of a series of amine metabolites from our in-house metabolite library (Fig. 3). Combinatorial treatment of standard metabolites at low quantity (less than 0.1 mg) yielded a metabolite conjugate library of 92 metabolites of the general structure 4 (Table S2, ESI†). This library includes diverse classes of amine metabolites: microbial metabolites, small aliphatic amines, amino acids and their modifications as well as nucleoside metabolites.
Using our constructed in-house conjugated metabolite library, we have confirmed 39 metabolites at the highest confidence level by UPLC–MS validation experiments (Fig. 4). Most of these validated metabolites have been reported to be linked to development of diseases (Table S3, ESI†). For example, the neurotransmitter γ-aminobutyric acid (GABA) has been associated with development of Alzheimer's disease.22 Additionally, it has recently been identified that GABA is produced by human Intestinal Bacteroides spp. to counter acid stress.23 A representative example for the efficient metabolite validation procedure is the conjugate of O-phosphoryl-ethanolamine (4g). O-Phosphoryl-ethanolamine is a part of sphingomyelin and exists in all living species ranging from bacteria to humans.24 Furthermore, we have validated the three amines 3-aminopropanonitrile, 5-methylcytosine and L-cysteic acid that are newly detected in fecal samples, which we believe stems from the enhanced mass spectrometric sensitivity of our method.
Furthermore, we have detected 12 microbiota-derived metabolites in these fecal samples that can be considered a direct readout for gut microbiota metabolism and interaction with the human host. For example, 5-aminovaleric acid (4h) has been associated with colorectal cancer.25 Other microbial metabolites of importance are the volatile metabolite ammonium, the two diamines 1,4-diaminobutane and 1,5-diaminopentane, as well as 4-aminobenzoic acid. The latter compound cannot be produced by humans due to the lack of the enzymes. Additionally, our method allows to successfully map metabolic pathways from the microbiome in these fecal samples e.g. the valine, leucine and isoleucine pathway. Within this pathway, threonine is an essential amino acid that is produced by microbes through the conversion of glycine from serine.26 Isoleucine is one of the essential branched-chain amino acids (BCAA) that is produced through bacterial metabolism in the human body.27 Parallel analysis of these microbial metabolites utilizing a single method has a high potential to monitor microbial bioactivity in human samples.
Conclusions
This study represents the first application of squaric acid for chemoselective analysis of amine metabolites in metabolomics-based studies. The high stability of the formed conjugates of squaramide with metabolites and the enhanced ionization properties of these conjugates led to successful capture and analysis of amine-containing metabolites in human fecal samples. The highly improved LOD at pM concentrations yielded three previously undetected metabolites in this sample type as well as a series of microbiota-derived metabolites. Furthermore, we have designed a reliable and straightforward method to synthesize a 93 amine-containing metabolite library for efficient validation of metabolites with coverage of different metabolite classes. This method can now be utilized for monitoring amine metabolites in any biological sample.Ethical statement
Patient fecal samples were obtained in accordance with the World Medical Association Declaration of Helsinki and all patients gave written informed consent. Approval for the study was obtained from the ethical committee at Karolinska Institute Hospital (Ethical approval number: Dnr 2017/290-31). Fecal samples were collected using routine clinical collection protocols and all patient codes have been removed in this publication. All samples were stored at −80 °C.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for funding by the Swedish Research Council (VR 2016-04423/VR 2020-04707), the Swedish Cancer Foundation (19 0347 Pj), and a generous start-up grant from the Science for Life Laboratory to D.G.References
- T. S. B. Schmidt, J. Raes and P. Bork, Cell, 2018, 172, 1198–1215 CrossRef CAS PubMed.
- R. Mehrian-Shai, J. K. V. Reichardt, C. C. Harris and A. Toren, Trends Cancer, 2019, 5, 200–207 CrossRef CAS PubMed.
- E. Sherwin, S. R. Bordenstein, J. L. Quinn, T. G. Dinan and J. F. Cryan, Science, 2019, 366 Search PubMed.
- N. Koppel, V. Maini Rekdal and E. P. Balskus, Science, 2017, 356 Search PubMed.
- R. R. da Silva, P. C. Dorrestein and R. A. Quinn, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12549–12550 CrossRef CAS PubMed.
- Y. Qiao, H. Hayashi and S. Chong Teo, Chem. – Asian J., 2020, 15, 2117–2128 CrossRef CAS.
- A. Milshteyn, D. A. Colosimo and S. F. Brady, Cell Host Microbe, 2018, 23, 725–736 CrossRef CAS PubMed.
- G. V. Sridharan, K. Choi, C. Klemashevich, C. Wu, D. Prabakaran, L. B. Pan, S. Steinmeyer, C. Mueller, M. Yousofshahi, R. C. Alaniz, K. Lee and A. Jayaraman, Nat. Commun., 2014, 5, 5492 CrossRef CAS.
- Y. Iwasaki, Y. Nakano, K. Mochizuki, M. Nomoto, Y. Takahashi, R. Ito, K. Saito and H. Nakazawa, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2011, 879, 1159–1165 CrossRef CAS PubMed.
- C. C. Hughes, Nat. Prod. Rep., 2021 10.1039/d0np00034e.
- E. E. Carlson and B. F. Cravatt, J. Am. Chem. Soc., 2007, 129, 15780–15782 CrossRef CAS PubMed.
- W. Lin, L. P. Conway, A. Block, G. Sommi, M. Vujasinovic, J. M. Lohr and D. Globisch, Analyst, 2020, 145, 3822–3831 RSC.
- L. P. Conway, N. Garg, W. Lin, M. Vujasinovic, J. M. Lohr and D. Globisch, Chem. Commun., 2019, 55, 9080–9083 RSC.
- N. Garg, L. P. Conway, C. Ballet, M. S. P. Correia, F. K. S. Olsson, M. Vujasinovic, J. M. Lohr and D. Globisch, Angew. Chem., Int. Ed., 2018, 57, 13805–13809 CrossRef CAS PubMed.
- F. R. Wurm and H. A. Klok, Chem. Soc. Rev., 2013, 42, 8220–8236 RSC.
- L. F. Tietze, C. Schröter, S. Gabius, U. Brinck, A. Goerlach-Graw and H. J. Gabius, Bioconjugate Chem., 1991, 2, 148–153 CrossRef CAS.
- M. Ximenis, E. Bustelo, A. G. Algarra, M. Vega, C. Rotger, M. G. Basallote and A. Costa, J. Org. Chem., 2017, 82, 2160–2170 CrossRef CAS.
- D. S. Wishart, Y. D. Feunang, A. Marcu, A. C. Guo, K. Liang, R. Vázquez-Fresno, T. Sajed, D. Johnson, C. Li, N. Karu, Z. Sayeeda, E. Lo, N. Assempour, M. Berjanskii, S. Singhal, D. Arndt, Y. Liang, H. Badran, J. Grant, A. Serra-Cayuela, Y. Liu, R. Mandal, V. Neveu, A. Pon, C. Knox, M. Wilson, C. Manach and A. Scalbert, Nucleic Acids Res., 2018, 46, D608–D617 CrossRef CAS PubMed.
- K. Guo, C. Ji and L. Li, Anal. Chem., 2007, 79, 8631–8638 CrossRef CAS PubMed.
- A. Lkhagva, C. C. Shen, Y. S. Leung and H. C. Tai, J. Chromatogr. A, 2020, 1610, 460536 CrossRef CAS PubMed.
- C. Ballet, M. S. P. Correia, L. P. Conway, T. L. Locher, L. C. Lehmann, N. Garg, M. Vujasinovic, S. Deindl, J. M. Lohr and D. Globisch, Chem. Sci., 2018, 9, 6233–6239 RSC.
- A. N. Fonteh, R. J. Harrington, A. Tsai, P. Liao and M. G. Harrington, Amino Acids, 2007, 32, 213–224 CrossRef CAS PubMed.
- N. Otaru, K. Ye, D. Mujezinovic, L. Berchtold, F. Constancias, F. A. Cornejo, A. Krzystek, T. de Wouters, C. Braegger, C. Lacroix and B. Pugin, Front. Microbiol., 2021, 12, 656895 CrossRef PubMed.
- D. Schiroli, L. Ronda and A. Peracchi, FEBS J., 2015, 282, 183–199 CrossRef CAS PubMed.
- D. G. Brown, S. Rao, T. L. Weir, J. O'Malia, M. Bazan, R. J. Brown and E. P. Ryan, Cancer Metab., 2016, 4, 11 CrossRef PubMed.
- Z. X. Cheng, C. Guo, Z. G. Chen, T. C. Yang, J. Y. Zhang, J. Wang, J. X. Zhu, D. Li, T. T. Zhang, H. Li, B. Peng and X. X. Peng, Nat. Commun., 2019, 10, 3325 CrossRef.
- C. Nie, T. He, W. Zhang, G. Zhang and X. Ma, Int. J. Mol. Sci., 2018, 19 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cb00132a |
| This journal is © The Royal Society of Chemistry 2021 |