Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery†
Marco
Herrera‡
 a,
Jeonghwan
Kim‡
a,
Jeonghwan
Kim‡
 a,
Yulia
Eygeris
a,
Yulia
Eygeris
 a,
Antony
Jozic
a and
Gaurav
Sahay
a,
Antony
Jozic
a and
Gaurav
Sahay
 *abc
*abc
aDepartment of Pharmaceutical Sciences, College of Pharmacy, Robertson Life Sciences Building, Oregon State University, Portland, Oregon 97201, USA
bDepartment of Biomedical Engineering, Robertson Life Sciences Building, Oregon Health & Science University, Portland, Oregon 97201, USA. E-mail: sahay@ohsu.edu
cDepartment of Ophthalmology, Casey Eye Institute, Oregon Health & Science University, Portland, Oregon 97239, USA
First published on 2nd February 2021
Abstract
Lipid-based nanoparticles (LNPs) for the delivery of mRNA have jumped to the forefront of non-viral gene delivery. Despite this exciting development, poor endosomal escape after LNP cell entry remains an unsolved, rate-limiting bottleneck. Here we report the use of a galectin 8-GFP (Gal8-GFP) cell reporter system to visualize the endosomal escape capabilities of LNP-encapsulated mRNA. LNPs substituted with phytosterols in place of cholesterol exhibited various levels of Gal8 recruitment in the Gal8-GFP reporter system. In live-cell imaging, LNPs containing β-sitosterol (LNP-Sito) showed a 10-fold increase in detectable endosomal perturbation events when compared to the standard cholesterol LNPs (LNP-Chol), suggesting the superior capability of LNP-Sito to escape from endosomal entrapment. Trafficking studies of these LNPs showed strong localization with late endosomes. This highly sensitive and robust Gal8-GFP reporter system can be a valuable tool to elucidate intricacies of LNP trafficking and ephemeral endosomal escape events, enabling advancements in gene delivery.
Introduction
The fields of nanotechnology and gene delivery continue to co-evolve for the treatment of human diseases.1–3 Within this nanoverse, extensive efforts towards RNA based therapeutics have made this nanomedicine the leading genetic tool for gene replacement, overexpression, repair and/or knockdown.2,4 One of the leading non-viral vectors for successful delivery of such nucleic acids is lipid-based nanoparticles (LNPs) (Fig. 1A).5,6 The LNP landscape continues to be developed clinically as it shows its advantageous properties for the treatment of multiple diseases including gene knockdown therapy for inherited hepatic disorders,6,7 cancer,8 and more recently, rapid vaccine development for the ongoing SARS-CoV-2 pandemic.9,10The established mechanism of LNP-assisted intracellular gene delivery is via the evolutionarily conserved endosomal trafficking pathway.11 Although this cellular network has been extensively studied, there still are a lot of mechanistic minutiae that need to be uncovered to better understand and improve the targeting of this pathway for intracellular delivery. The process starts with interactions of LNPs with the plasma membrane they come in contact with, resulting in uptake. Once they are engulfed into the cell by endocytosis, these LNP-containing vesicles get taken along a continually evolving voyage that routes them to the early endosomes which act as the sorting hub. If not routed to the extracellular space via exocytosis or other organelles, the LNPs are transported to the multi-vesicular, late endolysosomal compartments. Throughout this complicated cellular odyssey, endosomally trapped LNPs are being subjected to a gradual drop in pH. Near the end of the late endosomal maturation stage, the LNPs undergo two major fates: recycling clearance by exocytosis from the cell12 or enzymatic degradation in lysosomes.
To exert therapeutic effects, exogenously delivered genes must be retrieved from the endolysosomal pathways to the cytosol;11,13 however, a minute fraction of particles (<2%) escape into the cytoplasm following cell uptake.14 This underscores the importance of improving LNP endosomal escape to enhance potency. Despite great interest and efforts to breach the barriers faced, the exact mechanism of the endosomal escape of nanocarriers is limitedly understood. As more research focuses on understanding endosomal escape, the spotlight keeps expanding on the signaling players involved not only in the luminal endosomal system, but also in lysosomes and their closely-acting autophagosomes.15,16 Nonetheless, investigating the endosomal escape of nanocarriers is extremely difficult because it is an ephemeral, low frequency event eliciting rapid disappearance; for example, escape of siRNA delivered by LNPs occurs within 5–15 min of endocytosis.17 Current indirect visualization methods fail to give a clear picture of what is happening to the endosomes during these elusive endosomal disruption events.14,18 Therefore, improved visualization methods to illuminate the endosomal disruption events following cell uptake are highly desired.
Galectins belong to a family of evolutionarily conserved β-galactoside-binding lectins found primarily diffused in the cytosolic compartment of cells,19,20 and their biological functions include diverse pathways relating to cell homeostasis, cellular turnover, and immunity.19–24 One of their key biological functions is to sense damaged endosomes and accumulate specifically to the glycosylated leaflet of endosomal membranes.25 Out of several members of galectins, galectin 8 (Gal8) has emerged as a key sensor of damaged endosomes due to its abundant and rapid cytosolic redistribution to these sites.23,25 Recently, it has been exploited as a molecular indicator of the endosomal escape induced by non-viral vectors to crack the endosomal entrapment puzzle.17 Mechanistically, rapid recruitment of Gal8 to damaged endosomes can be used to picture the intricacies responsible for the ephemeral endosomal escape events.26,27 Here, we employed a recently reported screening method based on a Gal8-GFP reporter fusion (Gal8-GFP) for the direct visualization of endosomal disruption events following cellular uptake of LNP-encapsulated mRNA into cells.
Utilizing this Gal8-GFP based direct fluorescence-guided assay, we report differences in endosomal escape capabilities elicited by varying sterol composition of LNPs used for the delivery of mRNA. Aligned with our previous findings into sterol substitutions in LNP formulation and their effects on intracellular delivery of mRNA,28 this work further reveals the influence of sterols as it relates to the incidence of endosomal escape events by showing stark effects on this galectin-based sensor system. Especially, this Gal8-GFP sensor allows for efficiently rapid, and robust distinction of endosomal membrane integrity as an indicator of cytosolic availability of mRNA. We further elucidate the morphology of these LNPs, and correlate our findings to the bioactivity of this released cargo. Finally, we report colocalization patterns with late endosomal vesicles at the time of endomembrane disruption and attempt to connect the dots in this intricate machinery in hopes of advancing endosomal escape elucidation for the delivery of gene therapies. Our findings amplify the utility of Gal8-GFP sensors in the field of nanomedicine by showcasing clear, abundant, and easily detectable Gal8 recruitment for the quantification of endosomal escape events, which were not previously reported for LNP-encapsulated mRNA delivery.
Experimental
Materials
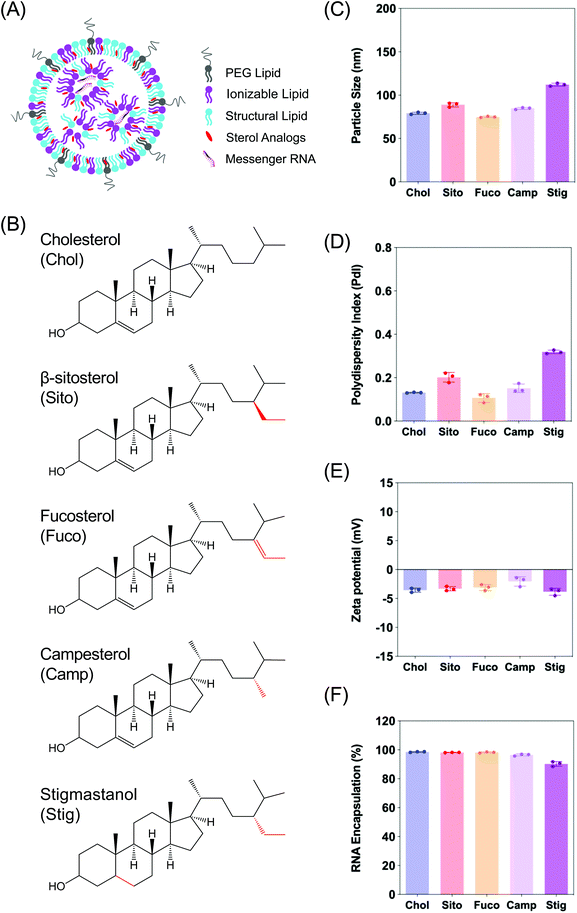
Firefly luciferase (Fluc) mRNA fully substituted with 5-methoxyuridine was purchased from Trilink Biotechnologies (L-7202). (6Z,9Z,28Z,31Z)-Heptatriacont-6,9,28,31-tetraene-19-yl 4-(dimethylamino)butanoate (DLin-MC3-DMA) was custom synthesized by Biofine International, Inc (BC, Canada). 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) was purchased from Avanti Polar Lipids, Inc (Alabaster, AL). 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2K), cholesterol, β-sitosterol, and fucosterol were obtained from Sigma Aldrich (St Louis, MO). Campesterol and stigmastanol were purchased from Cayman Chemicals (Ann Arbor, Michigan), and Matreya LLC (State College, PA), respectively. PB-GFP-Gal8 was a kind gift from Dr Jordan Green (Addgene plasmid # 127191).LNP formulation and characterization
Lipid nanoparticles (LNPs) were prepared by microfluidic mixing using a previously described method.29 In short, lipid–ethanol solutions containing DLin-MC3-DMA, a sterol, DSPC, and DMG-PEG2K, at molar ratios of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, were combined with 50 mM citrate buffer containing mRNA using a microfluidic mixer at a 1
1.5, were combined with 50 mM citrate buffer containing mRNA using a microfluidic mixer at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio. LNPs were dialyzed twice using 3 L of phosphate buffered saline (PBS, pH 7.4) and concentrated with 10 kDa Amicon® Ultra centrifuge filters (Millipore, Burlington, MA). Size distribution and zeta potential of LNPs were determined via dynamic light scattering using a Zetasizer Nano ZSP (Malvern Instruments, UK). mRNA encapsulation efficiency was determined using Quant-iT RiboGreen RNA reagent (Invitrogen, Carlsbad, CA).
3 ratio. LNPs were dialyzed twice using 3 L of phosphate buffered saline (PBS, pH 7.4) and concentrated with 10 kDa Amicon® Ultra centrifuge filters (Millipore, Burlington, MA). Size distribution and zeta potential of LNPs were determined via dynamic light scattering using a Zetasizer Nano ZSP (Malvern Instruments, UK). mRNA encapsulation efficiency was determined using Quant-iT RiboGreen RNA reagent (Invitrogen, Carlsbad, CA).
Cryo-TEM image acquisition and processing
Cryo-TEM acquisition was performed at 300 kV using FEI Titan Krios (Thermo Fisher, Hillsboro, OR) equipped with Falcon III and K3 cameras with DED. 3–5 μl of the sample was dispensed on a plasma cleaned grid (Quantifoil, R1.2/1.3 300 or 400 Cu mesh) in the FEI Vitrobot chamber at 95% relative humidity and allowed to rest for 30 s. Then, the grid was blotted for 3 s with filter paper and plunged into liquid ethane cooled by liquid nitrogen. The frozen grids were then checked for visible defects and assembled into cassettes, and the acquired images were analyzed using ImageJ. The images were processed using low-pass filter.Development of the Gal8-GFP reporter cell line and culture
All Human Embryonic Kidney 293T/17 cells (CRL-11268; ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (Hyclone Laboratories Inc., Logan, UT) and 1× penicillin/streptomycin (Thermo Fisher, Federal Way, WA). All cultures were grown in 37 °C incubators supplemented with 5% CO2 and were cultured according to suppliers’ instructions. For stable transfections, Addgene plasmid #127191 coding for Piggybac Gal8-GFP as a 3′ terminus fusion protein was co transfected with Super Piggybac Transposase expression vector from System Biosciences (Palo Alto, CA) into 293T/17 cells. GFP positive cells were obtained following fluorescence activated cell sorting using BD FACSAria™ Fusion equipped with a 488 nm laser and BD FACS Diva v8.0.1 software. Cells were sorted two separate times (day 1 post transfection and day 8 post transfection), selecting for bright green fluorescence indicative of stable genomic integrations at AATT sites. All transfections and cell uptake studies were performed in passage 10–25 of the reporter cells after successful stable transfection were established.In vitro Fluc mRNA transfection assay
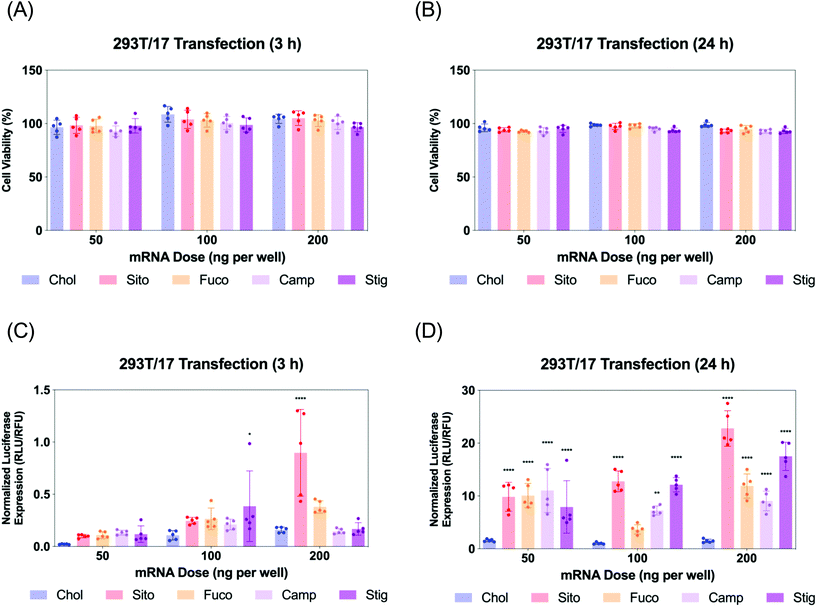
293T/17 cells were plated at 4000 cells per well in white-wall, clear-bottom, 96 well plates. After overnight incubation, cells were treated with LNPs encapsulating Fluc mRNA at various doses. mRNA transfection results were collected at 3 h or 24 h after treatment using ONE-Glo™ + Tox luciferase reporter and cell viability kit (Promega, Madison, WI) with a multimode microplate reader. Luminescent signals (luciferase expression, RLU) were divided by fluorescent signals (cell viability, RFU) for normalization of the data according to the previous study.29Visualization of Gal8 recruitment in 293T/17 Gal8-GFP cells with treatment of various LNPs
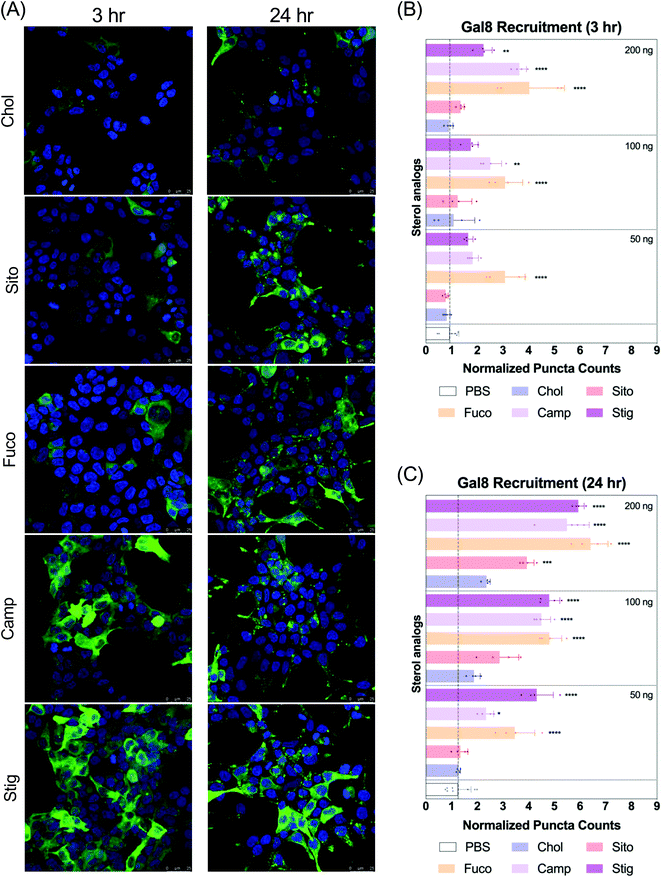
Ibidi 8-well chamber slides (Fitchburg, WI) were coated with poly-D lysine (Thermo Fisher, Waltham, MA) and rinsed with PBS before seeding the 293T/17 Gal8-GFP reporter cells at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per well in complete media and allowed to adhere and incubate overnight. The next day, different LNPs were added dropwise onto the media at the concentration of 50 ng, 100 ng and 200 ng mRNA per well and incubated for 3 h and 24 h at which point the media was aspirated, cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After fixing, cells were washed with PBS three times and DAPI (ThermoFisher, Walthman, WA) was added at 1
000 per well in complete media and allowed to adhere and incubate overnight. The next day, different LNPs were added dropwise onto the media at the concentration of 50 ng, 100 ng and 200 ng mRNA per well and incubated for 3 h and 24 h at which point the media was aspirated, cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After fixing, cells were washed with PBS three times and DAPI (ThermoFisher, Walthman, WA) was added at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS for nuclear staining. Cells were washed three times and wells were aspirated and mounted with coverslips using ProLong Diamond Antifade mountant (Thermo Fisher, Waltham, WA). Cells were imaged for GFP-positive puncta using a confocal Leica DMi8 microscope (Leica Microsystems) with oil immersion objective at 40× and 60× to report maximum intensity projections. Sample size was 4 per group.
1000 in PBS for nuclear staining. Cells were washed three times and wells were aspirated and mounted with coverslips using ProLong Diamond Antifade mountant (Thermo Fisher, Waltham, WA). Cells were imaged for GFP-positive puncta using a confocal Leica DMi8 microscope (Leica Microsystems) with oil immersion objective at 40× and 60× to report maximum intensity projections. Sample size was 4 per group.
For live-cell imaging, the reporter cells were seeded in the same way described above. Upon the LNP treatment, the cells were imaged for 16 h with 30 min intervals for time-lapse live-cell imaging using Yokogawa CSU-X1 on Zeiss Axio Observer spinning disk microscope. The images were processed and reported as the maximum intensity projections in the manufacturer software (Zen, Zeiss), followed by additional processing in ImageJ for movie export.
Puncta counting
Gal8-GFP puncta were quantified using ImageJ. Confocal images stacks were preprocessed using maximum intensity projections and Gaussian smoothing with radius of 1. Then, the puncta were counted using Find Maxima with prominence >30. The puncta counts were normalized by the nuclei counts in the analysis, which were counted using standard segmentation procedures (automatic thresholding and watershed). For live-cell imaging, the puncta were counted in the analogous manner except the counts were normalized by the ratio (initial number of sample cells)/(initial number of control cells).Immunofluorescence (ICC)
At 3 h and 24 h timepoints after LNP-Chol and LNP-Sito transfections, cells were rinsed with PBS and fixed in 4% paraformaldehyde at room temperature for 10 min. Following fixation, cells were washed three times and blocking was performed for 2 h at room temperature in PBS supplemented with 5% donkey serum and 0.2% Triton X-100 (Fisher Scientific, Waltham, MA). Cells were then rinsed with PBS and primary antibody dilutions for Rab7 (Cell Signaling, #9367) and EEA1 (Cell Signaling, #2411) were prepared at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS supplemented with 1% bovine serum albumin (BSA) and 0.2% Triton X-100 (Fisher Scientific, Waltham, MA). Primary antibody was incubated overnight at 4 °C with gentle rocking. The next day, the cells were washed with PBS three times and incubated with Alexa Fluor Plus 647-conjugated donkey anti-rabbit secondary antibody (Thermo Fisher, A32795) at 1
100 in PBS supplemented with 1% bovine serum albumin (BSA) and 0.2% Triton X-100 (Fisher Scientific, Waltham, MA). Primary antibody was incubated overnight at 4 °C with gentle rocking. The next day, the cells were washed with PBS three times and incubated with Alexa Fluor Plus 647-conjugated donkey anti-rabbit secondary antibody (Thermo Fisher, A32795) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 in PBS supplemented with 1% BSA and 0.2% Triton X-100 for 2 h at room temperature. Following incubation, cells were washed with PBS, nuclei stained with DAPI and coverslipped using ProLong Diamond Antifade mountant (Thermo Fisher, Waltham, MA). Cells were imaged using a Zeiss LSM 880 with Fast Airyscan employing a 63× oil immersed objective for maximum intensity projections reported. Sample size was 4 representative images per group. Image processing and export were done in the manufacturer's software (Zen, Zeiss).
2000 in PBS supplemented with 1% BSA and 0.2% Triton X-100 for 2 h at room temperature. Following incubation, cells were washed with PBS, nuclei stained with DAPI and coverslipped using ProLong Diamond Antifade mountant (Thermo Fisher, Waltham, MA). Cells were imaged using a Zeiss LSM 880 with Fast Airyscan employing a 63× oil immersed objective for maximum intensity projections reported. Sample size was 4 representative images per group. Image processing and export were done in the manufacturer's software (Zen, Zeiss).
Results
Characterization of LNPs consisting of various sterol analogs
In order to examine the effects of sterols in LNP trafficking in the endosomal pathway, we prepared a series of LNPs containing various sterol analogs. Cholesterol (Chol) was replaced with four different naturally occurring sterol analogs that have variations in the steroid backbone and the C24 side-chains. More specifically, β-sitosterol (Sito) and fucosterol (Fuco) have an ethyl group and an ethylidene group at C24-position, respectively. Campesterol (Camp) has a C24 methyl group and stigmastanol (Stig) is the reduced product of β-sitosterol that has a single bond at the Δ-5-position instead of a double bond (Fig. 1B). These phytosterols are known to alter cholesterol metabolism and intracellular trafficking when incorporated in liposomes, indicative of their bioactivities.30 The micellar formulations of Sito and Camp were reported to alter cholesterol influx and secretion in cells.31,32 More importantly, the chemical diversity of the phytosterols can have effects on lipid packing when incorporated in LNPs owing to different stereochemistry, affecting the physicochemical properties of the nanoparticles. For example, Fuco has a more rigid side-chain at the C24 position as compared to Sito due to a double bond, and Stig has a highly flexible steroid ring because of reduction of the Δ-5 double bond. For these reasons, we hypothesized that substitution of sterol analogs in place of Chol in LNPs will change the nanoparticle characteristics, altering the endosomal trafficking and the incidence of endosomal escape. While the sterol components were varied, DLin-MC3-DMA, DSPC, and DMG-PEG2K were used as the ionizable lipid, the structural lipid, and the PEG-lipid, respectively (Fig. 1A). LNPs containing Chol served as the baseline formulation for comparisons throughout the study.We first evaluated the physicochemical properties of a series of LNPs containing five sterol analogs (Chol, Sito, Fuco, Camp, and Stig) by dynamic light scattering (Fig. 1B). Particle sizes of LNPs containing Chol, Sito, Fuco, and Camp were similar (<100 nm) whereas LNP-Stig showed a relatively larger size (>110 nm) (Fig. 1C). Similarly, LNP-Chol, -Sito, -Fuco, and -Camp were narrowly-distributed (PdI < 0.2) whereas the size distribution of LNP-Stig were relatively wide (PdI > 0.3) (Fig. 1D). In spite of size changes in LNP-Stig, all LNPs tested showed slightly negative zeta-potentials (Fig. 1E). The results of mRNA encapsulation assay showed that LNP-Stig had a relatively lower encapsulation efficiency (90%) while other LNPs (-Chol, -Sito, -Fuco, and -Camp) had more than 95% encapsulation efficiency (Fig. 1F).
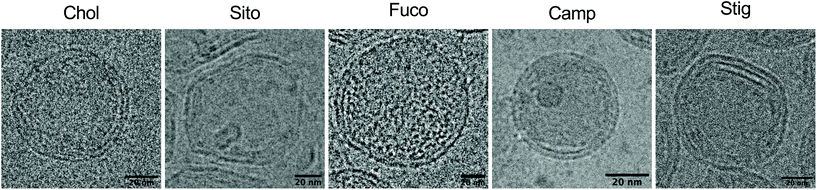
Next, we investigated whether the substitution led to morphological changes of LNPs using cryogenic transmission electron microscopy (cryo-TEM) (Fig. 2). Cryo-TEM analysis of LNP-Chol and -Camp showed spherical shapes whereas LNP-Sito and -Stig exhibited multi-faceted architecture (Fig. 2). LNP-Fuco displayed a distinctive morphology containing electron-dense pockets within the particle, which might be due to the hydrophobicity conflicts between LNP components (Fig. 2). These results concur with our previous reports that replacement of cholesterol with plant-based derivatives leads to morphological changes.28,33
Cellular entry of LNPs containing various sterol analogs
Having observed the changing physicochemical properties of the LNPs with varying sterols, we tested their cell uptake, cytotoxicity, and mRNA transfection levels in 293T/17 cells. To measure the cell entry of LNPs encapsulating mRNA, we used Cy5-labelled mRNA as a cargo. Cy5 intensity in the 293T/17 cells treated with various LNPs for 3 h was measured using flow cytometry with a viability marker staining. We found that more than 98% single viable cells were Cy5-positive and average intensities (MFI) of Cy5 within the cells was unaltered by various sterol substitutions in LNP formulations, indicating that endocytosis of various LNPs was similar regardless of sterol analogs used (Fig. S1†).Gal8 visualization indicating LNP-mediated endosomal disruption
Next, we evaluated Gal8 recruitment upon LNP treatment, suggestive of the incidence of endosomal escape events. This redistribution of Gal8 in response to endosome damage sensing is visualized as GFP puncta in the Gal8-GFP reporter cells. Therefore, we counted the GFP puncta for cells treated with our panel of LNPs to attempt to visualize differences of the elusive endosomal escape events elicited by these nanoparticles.We looked at Gal8 recruitment in cells at 3 and 24 h timepoints with LNPs. Representative images of each LNP treatment are shown in Fig. 3A, Fig. S2 and S3.† All LNPs led to some level of recruitment of Gal8-GFP in the reporter system. Increasing mRNA dose produced more counts of Gal8 puncta and the overall puncta counts were higher, and more readily detectable in the 24 h treatment compared to 3 h (Fig. 3B and C). After 3 h incubation, LNP-Fuco, -Camp, and -Stig produced greater number of Gal8 puncta than LNP-Chol (Fig. 3B). Especially, LNP-Fuco generated significantly more Gal8 puncta when compared to LNP-Chol in all mRNA doses tested (p < 0.0001, Fig. 3B). LNP-Sito led to similar puncta counts as compared to LNP-Chol in 3 h treatment (Fig. 3B). After 24 h treatment, LNP-Fuco, -Camp, and -Stig produced significantly higher degrees of easy to detect Gal8 recruitment in all mRNA doses tested when compared to LNP-Chol (Fig. 3C). LNP-Sito showed increased Gal8 recruitment in the higher doses of 100 ng per well and 200 ng per well whereas LNP-Chol induced mild Gal8 recruitment only at 200 ng per well (Fig. 3C). Despite the weaker recruitment in 3 h results, both timepoints demonstrated the potential of this Gal8-GFP system to showcase a large amount of easily detectable puncta for the elucidation of rare endosomal escape events.
mRNA transfection of LNPs containing sterol analogs
Next, we evaluated Fluc mRNA transfection by various LNPs in 293T/17 cells. We assayed cell viability and luciferase expression at two different timepoints: 3 and 24 h for LNP treatment (Fig. 4). The cell viability was not compromised by any treatment of various LNPs at any of the doses tested (Fig. 4A and B). Luciferase assay results of the 3 h treatment showed that all the other LNPs produced comparable luciferase expression to LNP-Chol at the doses of 50 and 100 ng per well (Fig. 4C). At 200 ng per well, the highest dose employed, LNP-Sito and -Fuco resulted in greater luciferase expression than LNP-Chol, while LNP-Camp and -Stig produced similar expression to LNP-Chol (Fig. 4C). Notably, LNP-Sito produced a 5-fold increase in mRNA transfection as compared to LNP-Chol in the 3 h treatment with a dose of 200 ng per well, suggesting its potency to induce endosomal escape. Influence of sterol substitutions on mRNA transfection became more pronounced in the 24 h treatment. Similarly to the results from 3 h treatment, all other LNPs (-Sito, -Fuco, -Camp, and -Stig) outperformed in terms of mRNA transfection when compared with LNP-Chol (Fig. 4D) in the 24 h data collected. In particular, LNP-Sito and -Stig significantly enhanced mRNA transfection in all doses (p < 0.0001) while LNP-Fuco and -Camp did so in the 50 ng per well and 200 ng per well conditions as compared to LNP-Chol. It is worth noting that mRNA transfection by LNP-Stig was capricious although its physicochemical properties (size, zeta-potential, mRNA encapsulation) were similar between LNP preparations. In several independent experiments, it produced variable results in in vitro Fluc mRNA transfections (Fig. 4 and S4†). We hypothesized it could be due to batch-to-batch variation of the Stig product, although we observed consistently reproducible transfection data after acquisition and testing of a new batch. In addition, it was verified that LNP treatment did not affect cell viability of the Gal8-GFP reporter cells (Fig. S5†). Taken together, varying luciferase expression is likely due to the different rates of endosomal escape given that the cellular uptake of various LNPs was similar. Although the orders of mRNA expression and Gal8 puncta counts among various LNPs were not exactly matched, sterol substitutions resulted in greater outputs than LNP-Chol in both assays.Live imaging of LNP-driven endosome damage inside Gal8-GFP reporter cells
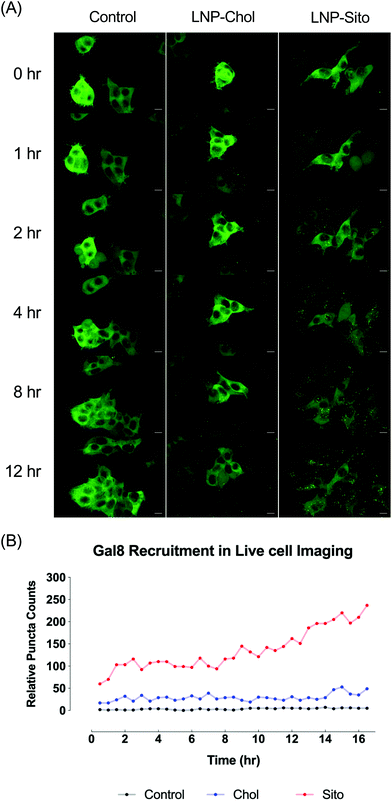
Differential Gal8 recruitment with sterol substitutions in LNPs led us to focus on the LNP-Chol and our well-characterized lead formulation, LNP-Sito. In conjunction with the moderate increase by LNP-Sito in Gal8 puncta counts, our previous findings demonstrating that Sito substitution (referred as eLNP in the previous reports) alters the morphology,33 diffusivity in endosomes, endosomal trafficking28,34 of LNPs and enhances mRNA transfection in vitro28,33 and in vivo,35 reinforced the decision to pick these two LNPs for further testing. To exclude any possible artifacts from fixation, we performed time-lapse live-cell imaging using spinning disk confocal microscopy. Representative images and time-lapse movies of control, LNP-Chol and LNP-Sito are provided in Fig. 5A and ESI videos (ESI Movie 1–3†). Both LNP-Chol and -Sito produced noticeable Gal8 recruitment while control group showed negligible levels of such (Fig. 5A). Notably, LNP-Sito strongly induced Gal8 recruitment, resulting in 10-fold higher number of puncta than LNP-Chol throughout the imaging experiments (Fig. 5A and B). Within 30 min after the LNP treatment, the Gal8 reporter cells rapidly reacted to the LNPs and produced GFP puncta (Fig. 5B). The normalized puncta count of LNP-Sito outnumbered that of LNP-Chol during 16 h live-cell imaging (Fig. 5B). Additionally, Gal8 recruitment induced by LNP-Sito showed an increasing trend as time progressed whereas that of LNP-Chol remained relatively constant, congruent with previous baseline-like results (Fig. 5B). This striking difference in the GFP puncta population suggests the highly efficient endosomal escape of LNP-Sito, which corresponds to the mRNA expression results (Fig. 4C and D).Subcellular colocalization analysis of Gal8 recruitment
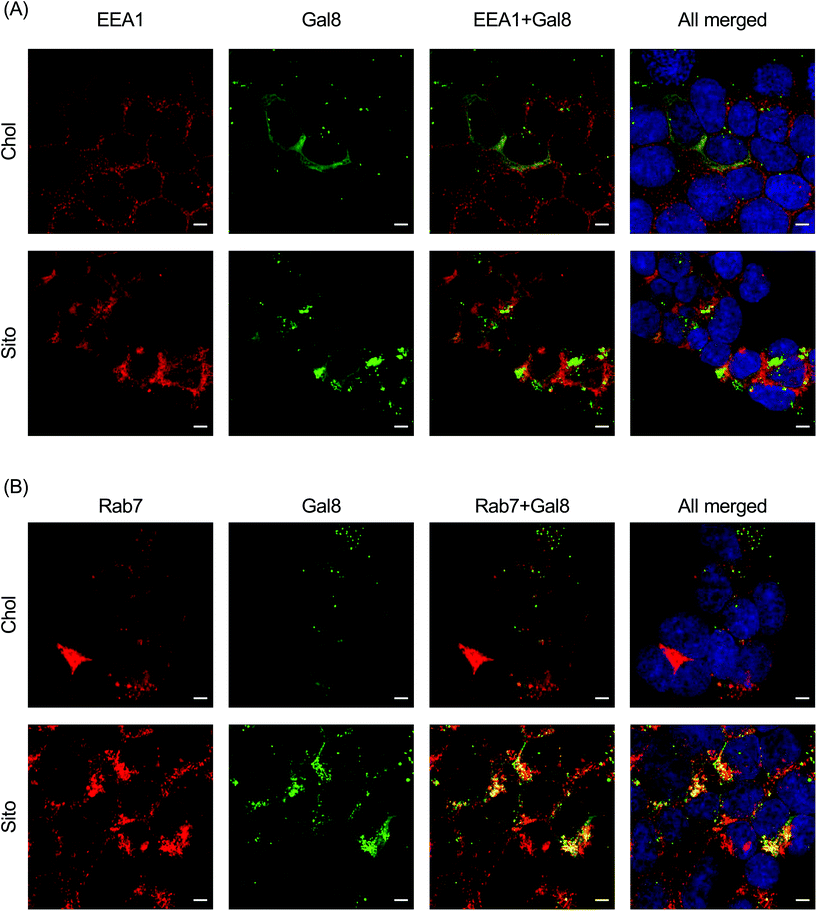
To interrogate which stage of the endosomal pathway is associated with Gal8 recruitment, we conducted immunofluorescence with endosomal markers: EEA1 (early endosome) and Rab7 (late endosome) (Fig. 6). We also quantified the colocalization of Gal8 puncta and the endosome markers by calculating the Pearson's correlation coefficient (PCC) and Mander's correlation coefficient (MCC) (Fig. S5†). In overlaid image analysis, GFP puncta representing Gal8 recruitment were not well colocalized with the EEA1 staining, which was confirmed in the correlation coefficients (Fig. 6A and Fig. S6A†). Both PCC and MCC analyses indicate that Gal8 puncta had no positive-correlation with the EEA1 staining (e.g., approximately 0 for LNP-Chol and 0.05 for LNP-Sito in MCC), suggesting that Gal8 recruitment, with associated endosome disruption, is not happening in the early endosomes (Fig. S6A†). On the other hand, many of the Gal8 puncta overlapped with the Rab7 staining as evident both from overlaid images (Fig. 6B) and metric analysis (e.g., approximately 0.2 for LNP-Chol and 0.6 for LNP-Sito in MCC) (Fig. S6B†). Particularly, LNP-Sito produced higher PCC and MCC than LNP-Chol, which can be attributed to stronger recruitment of Gal8 by LNP-Sito (Fig. S6†). Overall, better colocalization of Gal8 puncta with Rab7 late endosomal marker compared to EEA1 for both LNP-Chol and -Sito suggests that LNPs disrupted the late endosomes to enter the cytosol.Discussion
LNPs are sophisticated nanocarriers made up of multiple elements: an ionizable lipid, helper lipids (namely cholesterol and phospholipid), and a PEG–lipid (Fig. 1A). The ionizable lipid has gained a lot of attention because of its primary roles in RNA complexation, cellular uptake, and release of the genetic cargo, resulting in a number of studies to discover novel ionizable lipids.36–40 In contrast, the effects on bioactivity of cholesterol are less explored despite representing the second largest content in the clinically approved LNP formulation for siRNA delivery.36,41 Cholesterol in the LNPs modulates fluidity and permeability of lipid membrane by enhancing lipid packing.28 It is conceivable that chemical modifications to cholesterol influence these effects. For instance, esterified cholesterol variants enhanced LNP-mediated gene delivery whereas oxidized counterparts did not show any change.42 Our group also recently demonstrated that substitution of cholesterol with phytosterols in the LNPs influences nanocarrier morphology, internal structures, and transfection efficiency, forecasting changing numbers of disrupted endosomes inside the cell.28,33 Despite encouraging findings, the molecular web connecting LNP morphology to endosomal escape events and subsequent localization of nucleic acids in the cytosol remains murky, especially in the context of direct detection of endosomal destabilization events.12,43Galectins, a class of proteins that have binding affinity for β-galactoside residues, are programmed for recruitment to the sites of damaged endosomes in varied physiological states.44 For example, Gal3 can be recruited to ruptured endosomes induced by calcium phosphate precipitation and subsequently transported to autophagosomes,44,45 while Gal8 can be recruited to damaged endosomes or in other cases to phagosomes induced by bacterial invasion.23,44 Moreover, Gal9 can occasionally be recruited to the vicinity of peroxisomes.25 It is important to note that galectins bind to distinctive domains of endosomal membranes; Gal8 and Gal9 localize to the inner leaflet of damaged endosomes whereas Gal3 accumulates in the outer region of damaged endosomes.44,46 Furthermore, the rate of recruitment is different depending on galectin subtypes; upon endosomal disruption induced by Coxiella burnetii, Gal3, Gal8, and Gal9 are rapidly recruited to the endosomal rupture whereas Gal1 is accumulated relatively later.44,47 Tracking of various galectins can allow for the close scrutiny of endosomal escape triggered by nucleic acid-carrying nanoparticles.17,26,27 For example, cell lines expressing Gal8 fluorescent fusion protein were utilized to picture the endosomal escape of LNPs encapsulating siRNA,17 polymeric nanoparticles carrying siRNA,26 and ribonucleoproteins (RNPs).27
We have employed a Gal8-GFP reporter cell system to visualize endosomal destabilization induced by mRNA-loaded LNPs and found that sterol substitution in LNPs changes not only Gal8 recruitment, but also translation of delivered mRNA (Fig. 3A and 4D). These findings suggest that sterol substitution in LNPs causes endosomal escape at varying levels, affecting the mRNA transfection efficiency. Accompanying the clear difference in Gal8 puncta populations between the LNPs with substituted sterol analogs, the level of Gal8 recruitment correlated well with the mRNA transfection efficiency of all LNPs tested. Further investigation may provide valuable clues regarding the LNP-incorporated cholesterol sensing in the endosomal pathway, the cytosol and the complex other machineries involved along this escape and subsequent translation of cargo.
Live-cell imaging of LNP-Chol and -Sito evinced a visually robust temporal variation of endosome disruption depending on sterol type incorporated in LNPs. LNP-Sito, (also known as eLNP) showed highly notable levels of Gal8 puncta during 16 h live-cell imaging (Fig. 5). Given the extended retention of LNP-Sito compared to LNP-Chol inside endocytic vesicles,28 one could speculate this could lead LNP-Sito to potentially effect more efficient endosomal escape, explaining the greater number of Gal8 puncta observed. The high retention of LNP-Sito could have boosted the endosome escape by accumulating in the endosomes at the later timeframes (after 13 h, Fig. 5B). This reasoning is reinforced by the evidence showing intracellular delivery of nucleic acids is enhanced by extended retention of LNPs at the late endosomes and lysosomes in Niemann-Pick disease, type C1 (NPC1) knockout cells.12 Therefore, if NPC1 and 2 handle Sito differently from Chol, LNP-Sito could get extra time in the late endosomes, leading to more endosomal destabilization and subsequent escape. It is important to note that background puncta may be the result of Gal8 trafficking to the vicinity of peroxisomes25 as well as phagophore-associated endosome ruptures44 although we expect these effects to be negligible based on the results of media-treated controls (Fig. 5A and ESI Movie 1†).
Colocalization with endosome markers revealed the spatial distribution of Gal8 recruitment in the context of the endosomal pathway. Our results show that endosomal escape of LNPs is not colocalized well with the early endosomes (Fig. 6A and S6A†), but preferentially occurs in late endosomes (Fig. 6B and S6B†). This is corroborated in a previous report by Wittrup et al. showing that Gal8 recruitment mediated by siRNA-loaded LNPs does not occur at EEA1-positive endosomes, but at Rab5- or Rab7-positive endosomes.17 In agreement with how reported Gal8 sensing to damaged endosomes leads to the development of autophagosomes, which fuse with lysosomes for degradation or exocytosis,11 colocalization of Gal8 puncta and lysotracker in the study of Kilchrist et al. indicates a portion of damaged endosomes being transported to lysosomes.26 Another study by our group also highlights the importance of late endosome development for successful mRNA delivery, as supported by the observation that LNP-mediated mRNA transfection was suppressed in Rab7-knockout cells.34 Gal8 is also involved in signaling of mTOR (mechanistic target of rapamycin) which is recruited to late endosomes and lysosomes for modulating cell proliferation events, such as protein synthesis and autophagy, when cells are supplied with nutrients or starved.48 Our group previously showed that mTORC1 (mTOR complex 1) inhibitors decrease the translation of delivered mRNA due to diminished protein synthesis, while constitutive activation of mTORC1 by genetic deletion of tuberous sclerosis complex 2 (TSC2), which is a negative regulator of mTORC1, increases the translation of delivered mRNA.34 Conversely, Gal8 recruitment to the damaged endosomes inactivates mTORC1, inhibiting the cell proliferation events and increasing autophagy.48 These conflicts and the observed mRNA translation reported herein may be explained by perhaps quantitative easing induced by a large number of mRNA molecules escaping into the cytosol in spite of reduced mTORC1 signaling due to the endosome disruption and Gal8 redistribution. Although speculative, these hypotheses underline the highly complex molecular dynamics that are at play before, during and immediately after endosomal escape of nanoparticles. Taken together, our results support the notion that the primary site of LNP-mediated endosomal escape is ranged from late endosomes to lysosomes, which can be further enhanced by sterol substitution.
Conclusions
In sum, we confirmed that the cholesterol component in LNPs has pronounced effects on morphology, internal structure organization and mRNA transfection efficiency. The massive endosomal escape produced with C24 alkyl derivatives of cholesterol within LNPs can lead to identification of vesicles that were previously hard to pinpoint due to rarity of the escape events. Late endosomal biogenesis and trafficking emerge, once again, as the site of interest for elucidation of LNP endosomal escape. We observed stronger colocalization of LNP escape events with the Rab7 late endosomal marker than with the early marker, EEA1. Future studies should concentrate around other endosomal stage markers in conjunction with knockout cell lines of other involved players to dissect the spatiotemporal intricacies of endosomal trafficking and successful escape into the cytoplasm and how these relate to other closely interacting systems such as autophagy. Further characterization of the molecular interactions of LNP components with the endolysosomal membrane will allow us to crack some of the puzzling findings reported for LNPs containing C24 sterol derivatives. Shedding light on the structure–activity relationships of these sterol components with their interacting moieties will allow for the rational engineering of next generation LNPs. Other signaling moieties such as other galectins could be of interest especially considering the complex regulatory roles they effect in the endolysosomal system and cell physiology as a whole. Additionally, expanding our understanding to in vivo models is of greatest importance to empower nanomaterials for gene therapy. In conclusion, we advocate for the concerted probing of these known unknowns so that we can contribute to the breakthrough of this biological labyrinth.Conflicts of interest
G. S. is a co-inventor in patent application US20200129445A1 that details LNP formulations containing cholesterol derivatives.Acknowledgements
This project was supported through funding from the National Heart Lung and Blood Institute (N.H.L.B.I) 5R01HL146736 (G. S.), National Eye Institute 1R21EY031066 (G. S.), and Cystic Fibrosis Foundation SAHAY19XX0 (G. S.). A portion of this research was supported by NIH grant U24GM129547 and performed at the Pacific Northwest Cryo-EM Center (PNCC) at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. We thank Dr Stefanie Kaech Petrie, Dr Brian Jenkins, and Dr Crystal Chaw in the Advanced Light Microscopy Core in OHSU for the help for live-cell imaging. We thank the flow cytometry core in OHSU for the help for cell sorting.References
- U. Sahin, K. Karikó and Ö. Türeci, Nat. Rev. Drug Discovery, 2014, 13, 759–780 CrossRef CAS.
- J. C. Kaczmarek, P. S. Kowalski and D. G. Anderson, Genome Med., 2017, 9, 60 CrossRef.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS.
- S. F. Dowdy, Nat. Biotechnol., 2017, 35, 222–229 CrossRef CAS.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS.
- D. Adams, A. Gonzalez-Duarte, W. D. O'Riordan, C.-C. Yang, M. Ueda, A. V. Kristen, I. Tournev, H. H. Schmidt, T. Coelho, J. L. Berk, K.-P. Lin, G. Vita, S. Attarian, V. Planté-Bordeneuve, M. M. Mezei, J. M. Campistol, J. Buades, T. H. Brannagan, B. J. Kim, J. Oh, Y. Parman, Y. Sekijima, P. N. Hawkins, S. D. Solomon, M. Polydefkis, P. J. Dyck, P. J. Gandhi, S. Goyal, J. Chen, A. L. Strahs, S. V. Nochur, M. T. Sweetser, P. P. Garg, A. K. Vaishnaw, J. A. Gollob and O. B. Suhr, N. Engl. J. Med., 2018, 379, 11–21 CrossRef CAS.
- M. Balwani, E. Sardh, P. Ventura, P. A. Peiró, D. C. Rees, U. Stölzel, D. M. Bissell, H. L. Bonkovsky, J. Windyga, K. E. Anderson, C. Parker, S. M. Silver, S. B. Keel, J.-D. Wang, P. E. Stein, P. Harper, D. Vassiliou, B. Wang, J. Phillips, A. Ivanova, J. G. Langendonk, R. Kauppinen, E. Minder, Y. Horie, C. Penz, J. Chen, S. Liu, J. J. Ko, M. T. Sweetser, P. Garg, A. Vaishnaw, J. B. Kim, A. R. Simon and L. Gouya, N. Engl. J. Med., 2020, 382, 2289–2301 CrossRef CAS.
- H. He, L. Liu, E. E. Morin, M. Liu and A. Schwendeman, Acc. Chem. Res., 2019, 52, 2445–2461 CrossRef CAS.
- E. E. Walsh, R. W. Frenck, A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P.-Y. Shi, Ö. Türeci, K. R. Tompkins, K. E. Lyke, V. Raabe, P. R. Dormitzer, K. U. Jansen, U. Şahin and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2439–2450 CrossRef CAS.
- K. S. Corbett, B. Flynn, K. E. Foulds, J. R. Francica, S. Boyoglu-Barnum, A. P. Werner, B. Flach, S. O'Connell, K. W. Bock, M. Minai, B. M. Nagata, H. Andersen, D. R. Martinez, A. T. Noe, N. Douek, M. M. Donaldson, N. N. Nji, G. S. Alvarado, D. K. Edwards, D. R. Flebbe, E. Lamb, N. A. Doria-Rose, B. C. Lin, M. K. Louder, S. O'Dell, S. D. Schmidt, E. Phung, L. A. Chang, C. Yap, J.-P. M. Todd, L. Pessaint, A. Van Ry, S. Browne, J. Greenhouse, T. Putman-Taylor, A. Strasbaugh, T.-A. Campbell, A. Cook, A. Dodson, K. Steingrebe, W. Shi, Y. Zhang, O. M. Abiona, L. Wang, A. Pegu, E. S. Yang, K. Leung, T. Zhou, I.-T. Teng, A. Widge, I. Gordon, L. Novik, R. A. Gillespie, R. J. Loomis, J. I. Moliva, G. Stewart-Jones, S. Himansu, W.-P. Kong, M. C. Nason, K. M. Morabito, T. J. Ruckwardt, J. E. Ledgerwood, M. R. Gaudinski, P. D. Kwong, J. R. Mascola, A. Carfi, M. G. Lewis, R. S. Baric, A. McDermott, I. N. Moore, N. J. Sullivan, M. Roederer, R. A. Seder and B. S. Graham, N. Engl. J. Med., 2020, 383, 1544–1555 CrossRef CAS.
- S. Patel, J. Kim, M. Herrera, A. Mukherjee, A. V. Kabanov and G. Sahay, Adv. Drug Delivery Rev., 2019, 144, 90–111 CrossRef CAS.
- G. Sahay, W. Querbes, C. Alabi, A. Eltoukhy, S. Sarkar, C. Zurenko, E. Karagiannis, K. Love, D. Chen, R. Zoncu, Y. Buganim, A. Schroeder, R. Langer and D. G. Anderson, Nat. Biotechnol., 2013, 31, 653–658 CrossRef CAS.
- J. Kim, A. Narayana, S. Patel and G. Sahay, Theranostics, 2019, 9, 3191–3212 Search PubMed.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico, U. Schubert, K. Manygoats, S. Seifert, C. Andree, M. Stöter, H. Epstein-Barash, L. Zhang, V. Koteliansky, K. Fitzgerald, E. Fava, M. Bickle, Y. Kalaidzidis, A. Akinc, M. Maier and M. Zerial, Nat. Biotechnol., 2013, 31, 638–646 CrossRef CAS.
- A. Ballabio, EMBO Mol. Med., 2016, 8, 73–76 CrossRef CAS.
- S. Nakamura and T. Yoshimori, J. Cell Sci., 2017, 130, 1209–1216 CrossRef CAS.
- A. Wittrup, A. Ai, X. Liu, P. Hamar, R. Trifonova, K. Charisse, M. Manoharan, T. Kirchhausen and J. Lieberman, Nat. Biotechnol., 2015, 33, 870–876 CrossRef CAS.
- L. M. P. Vermeulen, T. Brans, S. K. Samal, P. Dubruel, J. Demeester, S. C. De Smedt, K. Remaut and K. Braeckmans, ACS Nano, 2018, 12, 2332–2345 CrossRef CAS.
- C. M. Arthur, M. D. Baruffi, R. D. Cummings and S. R. Stowell, Methods Mol. Biol., 2015, 1207, 1–35 CrossRef CAS.
- G. A. Rabinovich and M. A. Toscano, Nat. Rev. Immunol., 2009, 9, 338–352 CrossRef CAS.
- S. R. Stowell, C. M. Arthur, M. Dias-Baruffi, L. C. Rodrigues, J.-P. Gourdine, J. Heimburg-Molinaro, T. Ju, R. J. Molinaro, C. Rivera-Marrero, B. Xia, D. F. Smith and R. D. Cummings, Nat. Med., 2010, 16, 295–301 CrossRef CAS.
- C.-M. Tsai, C.-H. Guan, H.-W. Hsieh, T.-L. Hsu, Z. Tu, K.-J. Wu, C.-H. Lin and K.-I. Lin, J. Immunol., 2011, 187, 1643–1652 CrossRef CAS.
- T. L. M. Thurston, M. P. Wandel, N. von Muhlinen, Á. Foeglein and F. Randow, Nature, 2012, 482, 414–418 CrossRef CAS.
- L. Johannes, R. Jacob and H. Leffler, J. Cell Sci., 2018, 131, jcs208884 CrossRef.
- H. Du Rietz, H. Hedlund, S. Wilhelmson, P. Nordenfelt and A. Wittrup, Nat. Commun., 2020, 11, 1809 CrossRef CAS.
- K. V. Kilchrist, S. C. Dimobi, M. A. Jackson, B. C. Evans, T. A. Werfel, E. A. Dailing, S. K. Bedingfield, I. B. Kelly and C. L. Duvall, ACS Nano, 2019, 13, 1136–1152 CAS.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders, J. Karlsson, M. Lim and J. J. Green, Sci. Adv., 2019, 5, eaay3255 CrossRef CAS.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith, S. Hou, A. A. Esposito, T. Ketova, K. Welsher, J. L. Joyal, Ö. Almarsson and G. Sahay, Nat. Commun., 2020, 11, 983 CrossRef CAS.
- J. Kim, A. Jozic and G. Sahay, Cell. Mol. Bioeng., 2020, 1–12 Search PubMed.
- Y. Sato, K. Nishikawa, K. Aikawa, K. Mimura, K. Murakami-Murofushi, H. Arai and K. Inoue, Biochim. Biophys. Acta, Lipids Lipid Metab., 1995, 1257, 38–46 CrossRef.
- F. J. Field, E. Born and S. N. Mathur, J. Lipid Res., 1997, 38, 348–360 CrossRef CAS.
- R. Brauner, C. Johannes, F. Ploessl, F. Bracher and R. L. Lorenz, J. Nutr., 2012, 142, 981–989 CrossRef CAS.
- Y. Eygeris, S. Patel, A. Jozic and G. Sahay, Nano Lett., 2020, 20, 4543–4549 CrossRef CAS.
- S. Patel, N. Ashwanikumar, E. Robinson, A. DuRoss, C. Sun, K. E. Murphy-Benenato, C. Mihai, Ö. Almarsson and G. Sahay, Nano Lett., 2017, 17, 5711–5718 CrossRef CAS.
- J. Kim, A. Mukherjee, D. Nelson, A. Jozic and G. Sahay, bioRxiv, 2020 DOI:10.1101/2020.07.24.205583.
- M. Jayaraman, S. M. Ansell, B. L. Mui, Y. K. Tam, J. Chen, X. Du, D. Butler, L. Eltepu, S. Matsuda, J. K. Narayanannair, K. G. Rajeev, I. M. Hafez, A. Akinc, M. A. Maier, M. A. Tracy, P. R. Cullis, T. D. Madden, M. Manoharan and M. J. Hope, Angew. Chem., Int. Ed., 2012, 51, 8529–8533 CrossRef CAS.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. McFadyen, J. Chan, Ö. Almarsson, M. G. Stanton and K. E. Benenato, Mol. Ther., 2018, 26, 1509–1519 CrossRef CAS.
- K. J. Hassett, K. E. Benenato, E. Jacquinet, A. Lee, A. Woods, O. Yuzhakov, S. Himansu, J. Deterling, B. M. Geilich, T. Ketova, C. Mihai, A. Lynn, I. McFadyen, M. J. Moore, J. J. Senn, M. G. Stanton, Ö. Almarsson, G. Ciaramella and L. A. Brito, Mol. Ther.–Nucleic Acids, 2019, 15, 1–11 CrossRef CAS.
- Y. Dong, K. T. Love, J. R. Dorkin, S. Sirirungruang, Y. Zhang, D. Chen, R. L. Bogorad, H. Yin, Y. Chen, A. J. Vegas, C. A. Alabi, G. Sahay, K. T. Olejnik, W. Wang, A. Schroeder, A. K. R. Lytton-Jean, D. J. Siegwart, A. Akinc, C. Barnes, S. A. Barros, M. Carioto, K. Fitzgerald, J. Hettinger, V. Kumar, T. I. Novobrantseva, J. Qin, W. Querbes, V. Koteliansky, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3955–3960 CrossRef CAS.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS.
- J. A. Kulkarni, D. Witzigmann, S. Chen, P. R. Cullis and R. van der Meel, Acc. Chem. Res., 2019, 52, 2435–2444 CrossRef CAS.
- K. Paunovska, C. J. Gil, M. P. Lokugamage, C. D. Sago, M. Sato, G. N. Lando, M. Gamboa Castro, A. V. Bryksin and J. E. Dahlman, ACS Nano, 2018, 12, 8341–8349 CrossRef CAS.
- C. K. Payne, S. A. Jones, C. Chen and X. Zhuang, Traffic Cph. Den., 2007, 8, 389–401 CrossRef CAS.
- S. Bänfer and R. Jacob, Biomolecules, 2020, 10, 1232 CrossRef.
- X. Chen, B. Khambu, H. Zhang, W. Gao, M. Li, X. Chen, T. Yoshimori and X.-M. Yin, J. Biol. Chem., 2014, 289, 11162–11174 CrossRef CAS.
- M.-H. Hong, W.-H. Lin, I.-C. Weng, Y.-H. Hung, H.-L. Chen, H.-Y. Chen, P. Chen, C.-H. Lin, W. Y. Yang and F.-T. Liu, Glycobiology, 2019, 30, 49–57 Search PubMed.
- M. E. Mansilla Pareja, A. Bongiovanni, F. Lafont and M. I. Colombo, Front. Cell. Infect. Microbiol., 2017, 7, 112 CrossRef.
- J. Jia, Y. P. Abudu, A. Claude-Taupin, Y. Gu, S. Kumar, S. W. Choi, R. Peters, M. H. Mudd, L. Allers, M. Salemi, B. Phinney, T. Johansen and V. Deretic, Mol. Cell., 2018, 70, 120–135 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Cryo-TEM analysis and cellular uptake of LNPs, representative images of Gal8-GFP reporter cells treated with LNPs at various doses, colocalization analyses of Gal8 puncta with endosomal markers, time-lapse movies recording live-cell imaging of Gal8-GFP reporter cells treated with serum-free media, LNP-Chol, or LNP-Sito. See DOI: 10.1039/d0bm01947j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |