Open Access Article
Open Access ArticleTMT-based quantitative proteome profiles reveal the memory function of a whole heart decellularized matrix for neural stem cell trans-differentiation into the cardiac lineage†
Changyong
Wang‡
,
Xiaoning
Yang‡
,
Xiao
Zhang‡
,
Baijun
Liu‡
,
Wei
Liu
,
Yuan
Shen
,
Zhongbao
Gao
,
Qi
Yin
,
Chunlan
Wang
and
Jin
Zhou
 *
*
Beijing Institute of Basic Medical Sciences, 27 Taiping Rd, Beijing 100850, PR China. E-mail: sisun819@yahoo.com
First published on 22nd March 2021
Abstract
Whole organ or tissue decellularized matrices are a promising scaffold for tissue engineering because they maintain the specific memory of the original organ or tissue. A whole organ or tissue decellularized matrix contains extracellular matrix (ECM) components, and exhibits ultrastructural and mechanical properties, which could significantly regulate the fate of stem cells. To better understand the memory function of whole organ decellularized matrices, we constructed a heart decellularized matrix and seeded cross-embryonic layer stem cells – neural stem cells (NSCs) to repopulate the matrix, engineering cardiac tissue, in which a large number of NSCs differentiated into the neural lineage, but besides that, NSCs showed an obvious tendency of trans-differentiating into cardiac lineage cells. The results demonstrated that the whole heart decellularized microenvironment possesses memory function. To reveal the underlying mechanism, TMT-based quantitative proteomics analysis was used to identify the differently expressed proteins in the whole heart decellularized matrix compared with a brain decellularized matrix. 937 of the proteins changed over 1.5 fold, with 573 of the proteins downregulated and 374 of the proteins upregulated, among which integrin ligands in the ECM serve as key signals in regulating NSC fate. The findings here provide a novel insight into the memory function of tissue-specific microenvironments and pave the way for the therapeutic application of personalized tissues.
1. Introduction
Whole organ or tissue decellularization is a significant advancement that provides a promising alternative as a traditional scaffold for tissue reconstruction and therapeutic strategies.1,2 Various methods that derive scaffolds from decellularized organs and tissues have been developed and optimized, including the lungs, liver and heart. After cell removal from an organ or a tissue, it preserves the specific microenvironment, including ECM biochemical components, 3D architecture, microvasculature structure, mechanical properties and bioactivity.2,3 The intact scaffolds exhibit a memory function of the specific organ or tissue and provide a suitable substrate for the attachment and differentiation of reseeded cells.Whole heart decellularized scaffolds have received much attention in the tissue engineering field. It has been reported that a whole heart decellularized matrix can influence stem cell behavior and fate decision.4–6 In a previous study, cardiac or endothelial cells seeded into a whole heart decellularized matrix could differentiate into the cardiac lineage.7 Besides, pluripotent stem cells, human induced pluripotent stem cell (hiPSC)-derived cardiovascular progenitors, human embryonic stem cells (hESCs) and human mesendodermal cells (hMECs) derived from hESCs were all differentiated into the cardiac lineage in situ in a decellularized heart.8,9 These studies demonstrate that the whole heart decellularized matrix could maintain the specific memory of the original organ and guide cell differentiation without inducible factors.
Whole-organ decellularized scaffolds provide a local micro-environment for stem cells, which is also called the stem cell niche. The engineering of stem cell niches is rapidly expanding and they are widely known to regulate stem cell self-renewal, proliferation and differentiation.10 Interestingly, recent studies have indicated that stem cell niches are capable of directing stem cell types through influencing the complex interactions between stem cells and scaffolds. The vacated ovarian germline stem cell niche is a stable structure capable of inducing cell division of foreign surrounding somatic stem cells to generate ovarian follicle cells.11 Also, bone marrow stem cells can transdifferentiate into neurons in the brain and neural stem cells into hematopoietic cells.12 All of these studies suggest that stem cell niches have the capacity to modulate cell trans-differentiation.
Previous studies about the heart-specific microenvironment are mostly focused on cardiac stem cells and pluripotent stem cells, which possess the innate capacity to differentiate into cardiomyocytes. We wondered whether a whole heart decellularized matrix has a memory function sufficient to direct adult stem cells originating from other lineages to differentiate into the president cardiovascular lineage. To address this question, we reseeded cross-embryonic layer NSCs in a whole heart decellularized scaffold. It was found that both neural specific markers and cardiac specific markers were detected at the mRNA and protein level at day 14 post recellularization. ECM proteins between decellularized heart and decellularized brain were comparatively analyzed through TMT labelling quantitative proteomics. We found that ECM proteins of the decellularized heart regulate stem cells through activating integrin-focal adhesion. Our study illuminates that a tissue specific microenvironment can provide potential in determining stem cell fate.
2. Materials and methods
2.1. Preparation of the whole heart decellularized matrix
Animal organs were harvested in accordance with approved animal protocols and institutional guidelines. Sprague-Dawley rats (260–280 g) were used in this study. The following is the preparation process of the 3D heart decellularized matrix. Rat donors were pre-treated with barbital sodium, exposed to general anesthesia, and then injected with a dose of heparin (2.0 U g−1 of body weight) into the post cava. The cannulated heart was removed from the body and kept frozen at −80 °C. Before decellularization, the rat hearts were thawed at room temperature and connected to a perfusion pump (Leifu). The perfusion velocity was kept at 3 mL min−1. The hearts were firstly perfused with distilled water for 15 min, and then 1% sodium dodecyl sulfate (SDS, Invitrogen) was perfused for 12 h, followed by 1% Triton-X100 (Sigma) for 0.5 h, and finally phosphate-buffered saline (PBS) for at least 72 h to remove the residual Triton-X100 and SDS. The decellularized heart was perfused with high-glucose Dulbecco's Modified Eagle's Medium (H-DMEM, Gibco) overnight before cell seeding. The decellularized hearts were characterized by immunohistochemistry and histochemical staining.The preparation process of the 2D heart decellularized matrix is mainly divided into two steps: (1) Preparation of a heart decellularized matrix hydrogel: cut the 3D extracellular scaffold into pieces, pre-freeze it in a refrigerator at −70 °C, and then freeze-dry it in a vacuum freeze-dryer at −80 °C for 12 h. The lyophilized decellularized matrix material was accurately weighed and incubated with hydrochloric acid-pepsin solution at a concentration of 10 mg ml−1 with a magnetic stirrer at room temperature for 48 hours. After enzyme digestion, the extracellular scaffold particles gradually dissolved into the hydrogel state, and the hydrogel was collected by centrifuging at 4 °C for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 to remove the incompletely dissolved particles. (2) Preparation of 2D heart decellularized matrix hydrogel sheets: soak 10 × 10 mm coverslips in concentrated acid overnight and clean them with ultrapure water. Then, they were soaked in absolute ethyl alcohol for 1–2 h and air-dried. The heart decellularized matrix hydrogel was dripped onto the coverslips and dried at 37 °C for 4 h. Finally, a 2D sheet of heart decellularized matrix hydrogel was obtained.
000 rpm min−1 to remove the incompletely dissolved particles. (2) Preparation of 2D heart decellularized matrix hydrogel sheets: soak 10 × 10 mm coverslips in concentrated acid overnight and clean them with ultrapure water. Then, they were soaked in absolute ethyl alcohol for 1–2 h and air-dried. The heart decellularized matrix hydrogel was dripped onto the coverslips and dried at 37 °C for 4 h. Finally, a 2D sheet of heart decellularized matrix hydrogel was obtained.
2.2. Histology and immunofluorescence
Scaffolds were paraffin-embedded and sectioned, followed by hematoxylin and eosin (H&E) staining, Masson's Trichrome and immunofluorescence staining. H&E staining, Masson's Trichrome and Immunofluorescence staining were used to determine the presence of cellular components, collagen in the decellularized matrix, and matrix proteins collagen I, collagen III, collagen IV and fibronectin, respectively. All the procedures were carried out following standard protocols. Both rat native hearts and the decellularized matrix were immunostained with antibodies against anti-actinin and anti-Cx43. All of the secondary antibodies were from Invitrogen. Images were recorded with OpenLab 5.5.1 (Improvisation) on a Leica DMRA microscope (Leica). The recellularized constructs were subjected to paraffin sectioning and immunostained with the following antibodies at an appropriate dilution ratio: anti-Troponin T; anti-α-actinin; anti-rabbit β-3-Tubulin; anti-rabbit GFAP; anti-MAP2; anti-c-kit; anti-Cx43; anti-Nkx2.5; anti-GATA4; anti-nestin; anti-Ki67.2.3. DNA quantification
DNA quantification was used to quantify the DNA residue post-decellularization. Approximately 50 mg of native heart (n = 3) and decellularized matrix (n = 3) were quantified, each of them were subjected to DNA extraction using a DNeasy Blood and Tissue kit (DP318 TIANGEN) according to the manufacturer's instructions. The total amount of DNA was quantified using an ultra-micro ultraviolet spectrophotometer. The results were confirmed by gel electrophoresis analysis.2.4. Scanning electron microscopy (SEM)
The native hearts and decellularized matrix were sectioned into small blocks (8 mm3) and then fixed with 2.5% (vol/vol) glutaraldehyde for 2 h at room temperature, followed by three times PBS washing (5 min each). Later, they were serially dehydrated using 30, 50, 70, 90, and 100% ethanol and then freeze-dried by a lyophilizer. The samples were coated with 2 μm AuPd using a sputter coater system (sputter module 108auto, Cressington Scientific, Watford, UK). Images were captured with a Jeol6335F field emission SEM with a backscatter detector (Nova200 NanoLab).2.5. AFM for matrix stiffness measurement
Atomic Force Microscopy (AFM) measurements in the ECM require immobilization of the sample on a rigid substrate. To achieve this, we cut thin slices (10–50 μm) of decellularized tissue samples using a cryostat and then placed them on top of a positively charged glass slide. Freezing/thawing processes had little effect on the mechanical properties of the cellular scaffold. The samples were placed on an Asylum 1-D AFM (FASTSCANBIO, Bruker) and indented by a pyramid-tipped probe (Veeco, Santa Barbara, CA), ksp 60 pN nm−1. Force-indentation profiles were obtained immediately adjacent to the tissue or decellularized matrix, and each indentation profile was fit up to the point at which probe indentation into the secreted matrix stopped with a Hertz cone model.13–15In addition, instead of a pyramidal tip, a sphere attached to the end of the cantilever could be used to obtain an average value of E over a wider region of the sample. When a flat surface of an elastic body was probed with a spherical tip, the F–δ relationship was described by the Hertz contact model:
2.6. Protein extraction and TMT sample preparation
Decellularized matrices of the heart and brain were first ground by liquid nitrogen, then sonicated three times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 M Urea, 2 mM EDTA, 10 mM DTT and 1% Protease Inhibitor Cocktail III). The protein extraction and digestion of the decellularized matrix was performed according to a previous report.16,17After trypsin digestion, peptide was desalted by a Strata X C18 SPE column (Phenomenex) and vacuum-dried. Peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer's protocol for the 6-plex TMT kit. For labeling, each TMT reagent (defined as the amount of reagent required to label 100 μg of protein) was thawed and reconstituted in 24 μl ACN. The peptide mixtures were then incubated for 2 h at room temperature and pooled. The samples, labeled as 126 (heart1), 127 (heart2), 128 (heart3), 129 (brain1), 130 (brain2) and 131 (brain3), were multiplexed and vacuum dried.
2.7. Liquid chromatography tandem mass analysis
The sample was then fractionated into fractions by high pH reverse-phase HPLC using an Agilent 300Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). Briefly, peptides were firstly separated with a gradient of 2% to 60% acetonitrile in 10 mM ammonium bicarbonate (pH 10) over 80 min into 80 fractions. Then, the peptides were combined into 18 fractions and dried by vacuum centrifugation.The peptides were dissolved in 0.1% FA, directly loaded onto a reversed-phase pre-column (Acclaim PepMap 100, Thermo Scientific). Peptide separation was performed using a reversed phase analytical column (Acclaim PepMap RSLC, Thermo Scientific). The gradient consisted of an increase from 6% to 22% solvent B (0.1% FA in 98% ACN) over 24 min, 22% to 35% in 8 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl min−1 on an EASY-nLC 1000 UPLC system, and the resulting peptides were analysed by a Q ExactiveTM plus hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific).
For mass spectrum (MS) analysis, the peptides were analysed in positive ion mode. The electrospray voltage applied was 2.0 kV. Full MS spectra for intact peptides were acquired in the Orbitrap with a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Peptides were selected for MS/MS using the NCE setting at 30; ion fragments were detected in the Orbitrap at a resolution of 17
000. Peptides were selected for MS/MS using the NCE setting at 30; ion fragments were detected in the Orbitrap at a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. For the MS survey scan, the top 20 precursor ions above a threshold ion count of 104 with 30.0 s dynamic exclusion were selected. Automatic gain control (AGC) was used to prevent overfilling of the ion trap, and 5 × 104 ions were accumulated for the generation of the MS/MS spectra. For MS scans, the m/z scan range was 350 to 1800. The fixed first mass was set as 100 m/z.
500. For the MS survey scan, the top 20 precursor ions above a threshold ion count of 104 with 30.0 s dynamic exclusion were selected. Automatic gain control (AGC) was used to prevent overfilling of the ion trap, and 5 × 104 ions were accumulated for the generation of the MS/MS spectra. For MS scans, the m/z scan range was 350 to 1800. The fixed first mass was set as 100 m/z.
2.8. Bioinformatic analysis
The Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). The proteins were classified by Gene Ontology annotation based on three categories: biological process, cellular component and molecular function.The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to annotate the protein pathway. The KEGG Pathways mainly include: Genetic Information, Processing Metabolism, Environmental Information Processing, and Cellular Processes. Identified protein domain functional descriptions were annotated by the InterPro domain database in InterProScan (a sequence analysis application), based on the protein sequence alignment method. InterPro (http://www.ebi.ac.uk/interpro/) is a database that integrates diverse information about protein families, domains and functional sites, and is available to the public via web-based interfaces and services for free.
2.9. Cell culture
NSCs were isolated from the rat cortex on embryonic day 14 (E14).45 Cells were floating cultured in proliferation medium, DMEM/F12 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; Invitrogen, Carlsbad, CA) containing 20 ng mL−1 basic fibroblast growth factor (Invitrogen), 20 ng mL−1 epidermal growth factor (Invitrogen), and 2% B27 supplements (Invitrogen), and passaged every 3 days. These embryonic NSCs were able to proliferate and self-renew until passage 10.
1; Invitrogen, Carlsbad, CA) containing 20 ng mL−1 basic fibroblast growth factor (Invitrogen), 20 ng mL−1 epidermal growth factor (Invitrogen), and 2% B27 supplements (Invitrogen), and passaged every 3 days. These embryonic NSCs were able to proliferate and self-renew until passage 10.
2.10. Recellularization of a heart decellularized matrix
Approximately 3 × 107 NSCs were resuspended in 300 μL of basic differentiation media (H-DMEM + 20% FBS) and manually injected into the decellularized heart matrix five times. In order to induce a stable interaction of cells with the matrix, we didn't perfuse the recellularized heart as soon as the cells were seeded, avoiding the cells being washed out in the reseeded decellularized matrix by perfusion. In the first 2 days, the recellularized construct was maintained in the medium in two-dimensional non-perfused tissue-culture dishes. After 2 days of static culture, we mounted the recellularized constructs into a perfusion system and pulled them down to submerge them in the medium. Another 24 h later, the recellularized construct was raised up and continuously perfused with medium. The medium was changed every 48 h. We obtained the recellularized heart on 3, 7 or 14 days post-recellularization and performed a functional assessment.2.11. Reverse transcription and quantitative polymerase chain reaction
The recellularized heart matrix was perfusion cultured for 14 days, and then the total RNA was extracted using Trizol (Invitrogen). Total RNA was subjected to DNase I (Invitrogen, CA, USA) treatment to eliminate the genomic DNA carry over. DNase-treated RNA was then reverse transcribed using a high-capacity cDNA kit (Toyobo). Real-time quantitative PCR (q-PCR) was performed with Fast SYBR Green Master Mix (Toyobo).2.12. Statistical analysis
One-way ANOVA and a Student's unpaired t-test were used for statistical significance, respectively, using Origin software. Data are shown as mean ± SD. Tests with a P value less than 0.05 were considered statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001.3. Results
3.1. The whole heart decellularized scaffold possessed the tissue specific microenvironment of the original organ
Fig. 1 illustrates the overall schema for this study; we constructed, mapped, and compared the different proteins between heart or brain derived decellularized matrices to investigate functional molecular and signaling pathways underlying the stimulation of the whole heart decellularized matrix by quantitative proteomics analysis. Given the complex roles of molecular signals in dynamically regulating mRNA transcription, transducing the extracellular signals and promoting cell–matrix interactions, we hypothesized that a global shift in the molecular signals could explain the differences in determining NSC fate. | ||
| Fig. 1 A schematic illustration of the recellularization of the whole heart decellularized scaffold and procedural image of the research. | ||
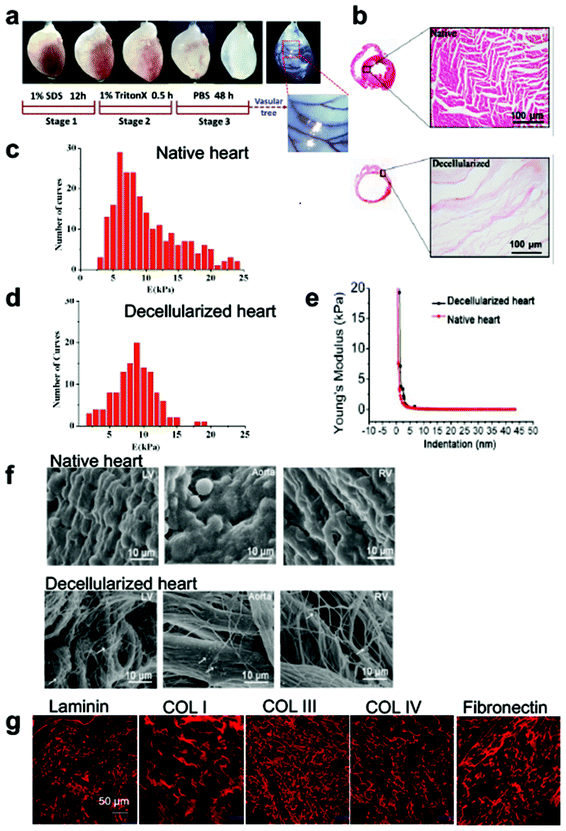
The whole heart decellularized scaffold was achieved by antegrade coronary perfusion with decellularization buffer to lyse the cellular content in rat hearts. A translucent decellularized heart was obtained by progressive discoloration until the time at which the apparent vascular tree was visualized with second and third-level branches (Fig. 2a). To evaluate the degree of decellularization, the expression level of intracellular structural proteins and quantification of DNA were assessed. Immunofluorescent staining showed that the positive expression of Cx43 and α-actinin was observed in the native heart, while intracellular proteins and DAPI positive cells were not detected in the decellularized heart (Fig. S1a†). No remaining nuclei and cellular content were observed in the whole heart decellularized scaffold (Fig. 2b), and DNA quantification analysis suggested that approximately 98% of the DNA was removed post decellularization (Fig. S1b†). The stiffness of the samples was characterized by atomic force microscopy (AFM). The heart decellularized scaffold as shown in Fig. 2c had a similar stiffness to native heart in the left ventricle region (Fig. 2d). The Young's modulus of the heart decellularized scaffold was decreased with increasing indentation depth, with the values slightly greater than that of the native heart, suggesting that the decellularization process had comparatively little effect on stiffness (Fig. 2e).
The ultrastructure of the decellularized scaffold was preserved after the removal of the precedent cells and examined by scanning electron microscope (SEM) (Fig. 2f). The tissue was intact and the structure of the ECM was connected with the cells and connective tissue in the native heart, while the cells and connective tissue were removed in the decellularized scaffold, leaving a filament like appearance. Within the region of the left ventricle, it presented a highly fibrous network structure, whereas in the decellularized aortic wall and right ventricle, the fiber orientations and compositions were partly conserved as those in the native heart. The ribbon-shaped fibers with random arrangement were considered to be attached glycosaminoglycans (white arrows), forming porous, net-like 3D microstructures. Masson's Trichrome staining on the sections of the decellularized heart showed the preservation of collagen, exhibiting a honeycomb structure (Fig. S1c†). Furthermore, typical ECM components such as collagen I, collagen III, collagen IV, laminin and fibronectin were detected by immunofluorescent staining and believed to remain in the decellularized matrix (Fig. 2g). Besides, the main ECM components exhibited a porous structure and distributed in an oriented direction as shown in the immunofluorescent staining images, confirming that both the structure and basement membrane components of the ECM were preserved from the native heart.
3.2. NSCs seeded and recellularized in the whole heart decellularized scaffold
To determine whether NSCs would reprogram into a non-neural lineage by the stimulation of a tissue specific microenvironment, we disassociated the clonal neurospheres into monodispersed cells (approximately 3 × 107) and injected them into the decellularized heart scaffold. The NSC spheres were sorted based on size and their self-renewal ability was identified by immunostaining the specific marker nestin of NSCs before recellularization (Fig. S2a†). The recellularized constructs turned from translucent to the pink color of the culture medium (Fig. S2b†). In order to characterize the vascularization of the recellularized tissue, immunofluorescence staining of vWF was conducted (Fig. S2c†). The H&E staining results showed that most of the NSCs dispersed individually on the scaffold, and adhered onto the surface of the matrix at the early stage (3 days) of perfusion culturing (Fig. 3a). The interactions between the cells and the matrix were enhanced over days, where most of the cells spread out along the surface of the matrix at day 7 (Fig. 3a). Interestingly, some cells presented a cardiac-like spindle appearance at day 14 (Fig. 3a and b), and the percentage of these cells is about 2.3% (Fig. 3b). The distribution and the putative potential of the cells were further confirmed by immunofluorescent staining to determine the presence of nestin-positive cells (Fig. 3c). The results showed that the total number of cells was not increased much over the time of culturing, with the number of nestin-positive cells decreasing after 14 days of perfusion, suggesting that the majority of NSCs were differentiated and their pluripotency was gradually decreased (Fig. 3c and Fig. S2d†).3.3. NSCs differentiate into cells of neural lineage and trans-differentiate into cells of cardiac lineage
To evaluate the differentiation of NSCs after recellularization, flow cytometry and immunofluorescence analysis were firstly used. We assessed the expression of neural lineage-specific markers, including MAP2 for the presence of neurons, and GFAP – a canonical specific marker of astrocytes. The results showed that differentiated cells originating from NSCs were stained positive for MAP2 and GFAP antibodies (Fig. 3d and Fig. S3†). Flow cytometry analysis further confirmed their presence and the percentage of MAP2 positive and GFAP positive cells was 39.2% and 18.84%, respectively (Fig. 3e). However, at day 7, some cells were stained positive for MAP2 antibodies and maintained the expression of nestin, while few cells were stained positive for nestin antibody at day 14 (Fig. 3f). In addition, the cytoskeletal structure of the cells was investigated by immunofluorescence analysis. The results showed that both the recellularized constructs at day 7 and day 14 presented a positive expression of β-3-tubulin (Fig. 3g), and the cells maintained the normal cytoskeletal structure of neural lineage cells. The results suggested that NSCs maintained their original differentiation capacity in the whole heart decellularized scaffold. To evaluate the proliferating behavior of the differentiated neurons, we analysed the cell cycle progression of the recellularized construct by immunofluorescent staining with the active cell cycle specific marker Ki67 (Fig. 3h). The results showed that few neurons were stained positive for Ki67 after both 7 and 14 days of perfusion culture, which indicated that the neurons originated from NSCs had limited proliferation capacity.It was reported that NSCs could undergo direct reprogramming under special conditions to give rise to non-neural cell types.18,19 In our case, to further demonstrate whether the stem cell niche within the decellularized heart could determine the lineage commitment of NSCs to form the resident cell types, we investigated the expression of cardiac specific markers. Firstly, we conducted qPCR analysis to evaluate the variation tendency of the mRNA expression level over the days of culturing after recellularization. The results showed that the expression level of pluripotent genes, nestin, was decreased in the recellularized construct (Fig. 4a). The expression level of transcriptional factors (GATA4 and Nkx 2–5) and specific genes of cardiac stem cells (c-kit) were significantly increased, together with a growing expression level of cardiac specific genes (α-actinin and cTNT) over the course of culturing. On the other hand, the expression of specific genes for the presence of neurons (MAP2, β-3-tubulin) and astrocytes (GFAP) was also found to be significantly increased (Fig. 4a). Immunofluorescent staining analysis was then performed and we found a negative expression of cTNT at day 7, but cTNT+ cells were observable at day 14 (Fig. 4b). In addition, α-actinin+ cells spread and became spindle-shaped at day 14 post recellularization (Fig. 4c, Fig. S4a, and S4b†). Flow cytometry analysis was performed later and further validated these findings, and the percentage of α-actinin+ cells was found to take up about 1.76% (Fig. S4c†). Besides, Cx43 (the predominant cardiac gap-junction protein) and N-cadherin (a specific marker of adherens junctions) were detected to be expressed in the recellularized constructs and specially distribute in the cell–cell contact region (Fig. 4d and Fig. S4b†), which indicated better cardiac maturation. Furthermore, we conducted immunofluorescent and immunohistochemical analysis to assess the specific gene expression of cardiac stem cells, including Nkx2–5, GATA4 and c-kit (Fig. 4e–g and Fig. S5†), and the positive expression of those markers was noticed at day 14, although negative results were presented at day 3 and day 7 post recellularization. All these results clearly confirmed that, through the support of the whole heart decellularized scaffold, a portion of NSCs was able to trans-differentiate into the cardiac lineage, while most NSCs maintained the capacity of differentiating into the neural lineage.
To better understand the memory function of the heart decellularized matrix, it is necessary to investigate the cell fate of NSCs in the native brain microenvironment. We prepared a brain decellularized matrix, characterized the structure through SEM (Fig. S6a†) and H&E staining analysis (Fig. S6b†), and determined the common ECM components, including collagen I, collagen III, collagen IV, laminin and fibronectin by immunofluorescent staining (Fig. S6c†), which proved that the brain decellularized matrix preserved the ultrastructure and chemical components. NSCs were seeded in the brain decellularized scaffold and the cardiac and neural specific markers were detected by immunofluorescent analysis. We found the positive expression of β-3-tubulin, but negative expression of cardiac specific markers, including α-actinin, cTNT, and N-cadherin (Fig. 5), which shows that the NSCs have no tendency of transdifferentiating into cardiac lineage cells in their native microenvironment.
3.4. Decellularized heart maintains the tissue specific ECM proteins to regulate NSCs
To get a mechanistic insight into the effect of the whole heart decellularized scaffold on NSC differentiation and trans-differentiation, we used TMT protein labelling and mass spectra to comparatively quantify the proteome of the whole heart decellularized matrix compared to the whole brain decellularized matrix, and we were able to identify the signaling pathways that are involved in the regulation of stem cell fate (Fig. S7a†). It was found that the relative quantities of the proteins differed largely between the decellularized heart matrix and brain matrix. Specifically, 3477 proteins were identified and 2275 proteins were quantified through the comparison between the heart matrix and brain matrix, among which 937 of them were changed over 1.5 fold, with 374 upregulated and 573 downregulated (Fig. S7b†). GO enrichment-based clustering including cellular components, molecular functions and biological processes was then performed. The up-regulated proteins were highly enriched in extracellular matrix and extracellular region in the cellular component category (Fig. S8a and S9†), where most of the differently expressed proteins were annotated as binding and were found to be significantly enriched in Q4 for the molecular function analysis (Fig. S10†); for analysis of biological processes, the proteins annotated as extracellular structure and extracellular matrix organizations were significantly enriched in Q4 (Fig. S11†). In addition, the KEGG pathway analysis of the quantitatively changed proteins showed a number of vital pathways (Fig. S8b†), among which, the signal pathways of focal adhesion (Fig. 6a) and ECM-receptor interaction (Fig. 6b) were enriched. The results suggested that the extracellular signals can be mediated by the ECM-receptor interaction and focal adhesion into the cells. | ||
| Fig. 6 Quantitative proteomic profiling of decellularized heart. Significantly enriched pathways obtained by KEGG analysis. (a) Focal adhesion pathway. (b) ECM-receptor interaction pathway. | ||
3.5. Inactivation of Integrins αvβ3 changes the lineage fate of NSCs
The results of TMT labelling quantitative analysis give us a clue that the integrin-mediated signaling pathway may be involved in NSC fate determination, regulated by the whole heart decellularized scaffold. To further validate this hypothesis, a control experiment was performed, where NSCs were incubated with/without cilengitide before recellularization. Cilengitide can selectively and potently block the communication between the αvβ3/αvβ5 integrins and tissue specific matrix proteins such as vitronectin, fibronectin, fibrinogen, tenascin, etc.,20 where tenascin is an extracellular matrix component that is involved in the regulation of neural stem cell development (Fig. 6b).21 q-PCR and immunofluorescence staining were conducted to comparatively evaluate the trans-differentiation of cilengitide treated or untreated groups after the recellularized constructs were perfusion cultured for 14 days. Remarkably, NSCs treated with cilengitide suffered impaired trans-differentiation with decreased expression of cardiac specific markers. As shown in Fig. 7a, the expressions of cardiac specific genes α-actinin, cTNT, GATA4, Nkx2–5 and MYH in the cilengitide-treated group were significantly decreased compared with that in the untreated group. In addition, the inhibited trans-differentiation was further confirmed by immunofluorescence staining, where the expression of cTNT+ cells was not observable in the cilengitide treated samples, while the cTNT+ cells presented as elongated spindle cells in the cilengitide untreated group (Fig. 7b). Therefore, we verified our hypothesis and concluded that integrin plays a critical role in the whole heart decellularized matrix, which triggered NSC trans-differentiation.4. Discussion
Whole organ decellularized matrices are a promising scaffold used for tissue engineering because they provide specific cues for the reseeded cells that are thought to better mimic the microenvironment of residential tissue.6–8 The stem cell niche of a whole organ decellularized matrix can promote stem cell growth and differentiation,22–24 and especially switch the intrinsic specification of stem cells.4 In this study, we reported for the first time that whole heart decellularized matrix possesses the potential to reprogram NSCs. Then, a model was proposed to elucidate the mechanism that is responsible for NSC trans-differentiation triggered by the decellularized heart (Fig. 8). Our study demonstrated that whole heart decellularized matrix maintained the “memory” of resident cells to reprogram seeded NSCs, which is in agreement with recent studies, which suggested that native ECM possessed the defining “zip codes” to reprogram stem cells to their residential lineages.1,25–27It is rational that the specific composition and ultrastructural organization of the cultured cells can be altered by the source tissue/organ, since ECM is the secretive product of the resident cell populations.1 We investigated the effect of the ECM on cell differentiation performance by evaluating the behaviors of NSCs under the stimulation of heart- and brain-derived decellularized scaffolds. At day 14 post recellularization, the expression of neural specific markers (Nestin, β-3-Tubulin, GFAP, MAP2) and cardiac specific markers (α-actinin, cTNT, Cx43 and N-cadherin) was detected in the recellularized heart construct (Fig. 3 and 4). However, only the expression of neural specific marker β-3-tubulin could be detected in the recellularized brain construct, while the expression of cardiac specific markers, such as α-actinin, cTNT and N-cadherin, were not observed (Fig. 5). In accordance with previous studies, ES and iPSC have the ability to spontaneously differentiate into cardiomyocytes, while NSCs could not differentiate into cardiomyocytes across germ layers in a native environment. In conclusion, the results suggested that the tissue specific microenvironment of the whole heart decellularized scaffold possessed the cues of the precedent cells to induce NSC trans-differentiation. To further interrogate the tissue specific ECM components of the decellularized heart, we compared the protein profiles of the heart and brain decellularized matrix by TMT labelling. The differently expressed proteins were highly enriched in the extracellular region, which induced the high enrichment of integrin related ECM–receptor interaction and focal adhesion signaling pathways in mediating the extracellular signals into the cells. These results match previous findings where ECM adhesion receptors provide signals to regulate stem cell behavior and cell lineage specification.28–30 For example, Kraehenbuehl et al. reported that cells can interact with the ECM by cell-surface receptors (integrin) and transmit signals across the cell membrane.31 The tissue specific ECM component of decellularized heart promotes cell membrane–membrane interactions, cellular adhesion, growth, and migration of NSCs.32 In our quantitative proteome profiles, the significantly up-regulated proteins in the heart ECM, Tenascin and Fibronectin, could bind integrin αvβ3. It has been reported that αvβ3 integrin binds to RGD motifs present in the ECM proteins and participates in cell proliferation and differentiation of some cell types. We speculated that the αvβ3 integrin-mediated signaling pathway transmits the chemical components, ultrastructure and mechanical properties of the heart ECM to NSCs and promotes NSC trans-differentiation. When we blocked the integrin avb3 signaling pathway with cilengitide, the trans-differentiation of NSCs was greatly inhibited. These findings provide evidence confirming that ECM proteins in the heart stimulated NSC behavior by activating integrin signaling pathways.
In the cell microenvironment, the stimulating factors mainly include chemical composition, matrix stiffness and ultrastructure properties, which could collectively affect the fate of cells. We have discussed the effect of chemical composition on the differentiation behaviors of stem cells by comparing the heart and brain ECM proteins. In addition to ECM components, physical signals such as stiffness and ultrastructural characteristics also play essential roles in modulating stem cell behaviors, either by regulating the cells’ ability to migrate into and attach to specific locations or by influencing stem cell specification.26,27,33–35 We compared the cell behaviors of NSCs on 2D and 3D heart extracellular matrices. Although they have similar biological components, there are obvious differences in ultrastructure and mechanical properties.36 Immunofluorescence experiments showed that NSCs on the 2D ECM did not show a trans-differentiation trend after 14 days of culture (Fig. S12†). This interesting phenomenon indicated that tissue specific ECM components of the heart were not sufficient to induce NSC trans-differentiation. Matrix stiffness and ultrastructural characteristics are likely to be important factors in regulating cell mechanical responses and different ECM stiffness can induce NSCs into different cell lineages.37–39 Similar findings were shown by previous researchers. Fibroblasts cultured within a 3D matrix have enhanced cell adhesion and cell morphology when compared with those cultured on 2D flat surfaces.40 Changes of substrate stiffness direct human mesenchymal stem cells along neuronal, muscle or bone lineages.41,42 In our study, decellularized heart preserved the typical ECM components, matrix stiffness and ultrastructural characteristics similar to native heart, which confirmed that it could be regarded as a better mimic of native heart in the research of regenerative medicine.43,44
5. Conclusions
In conclusion, a recellularized construct was used as a model to investigate if the microenvironment of a whole heart decellularized scaffold possessed the “zip code” of the resident cells to stimulate cross-embryonic layer differentiation of NSCs. It was found that NSCs trans-differentiate into cells of the cardiac lineage with the stimulation of the whole heart decellularized scaffold; meanwhile, they maintained their original potency to differentiate into the neural lineage. The tissue specific ECM proteins of decellularized heart can activate integrin–focal adhesion to regulate the fate of NSCs that trans-differentiate into the cardiac lineage. Our study provides significant implications for understanding the tissue-specific microenvironment of decellularized heart, and, more importantly, provides fundamental understanding for the use of interdisciplinary cardiac tissue engineering research in clinically relevant applications.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of the Chinese Academy of Military Medical Science and approved by the Animal Ethics Committee of the Chinese Academy of Military Medical Science.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Key Program of the National Key Research and Development Program of China (No. 2017YFA0106100, No. 2016YFY1101303), Key Program of National Natural Science Foundation of China (No. 31830030), and Beijing NOVA Program of China (No. 2016B615).References
- S. Sabetkish, A. M. Kajbafzadeh, N. Sabetkish, R. Khorramirouz, A. Akbarzadeh, S. L. Seyedian, P. Pasalar, S. Orangian, R. S. Beigi and Z. Aryan, J. Biomed. Mater. Res., Part A, 2015, 103, 1498–1508 CrossRef PubMed.
- J. J. Song and H. C. Ott, Trends Mol. Med., 2011, 17, 424–432 CrossRef CAS PubMed.
- J. P. Guyette, J. Charest, R. W. Mills, B. Jank, P. T. Moser, S. E. Gilpin, J. R. Gershlak, T. Okamoto, G. Gonzalez and D. J. Milan, Circ. Res., 2016, 118, 56–72 CrossRef CAS PubMed.
- D. Kuraitis, C. Hou, Y. Zhang, B. Vulesevic, T. Sofrenovic, D. Mckee, Z. Sharif, M. Ruel and E. J. Suuronen, J. Mol. Cell Cardiol., 2011, 51, 187–197 CrossRef CAS PubMed.
- J. S. Hammond, T. W. Gilbert, D. Howard, A. Zaitoun, G. Michalopoulos, K. M. Shakesheff, I. J. Beckingham and S. F. Badylak, J. Hepatol., 2011, 54, 279–287 CrossRef CAS PubMed.
- J. F. Mano, G. A. Silva, H. S. Azevedo, P. B. Malafaya, R. A. Sousa, S. S. Silva, L. F. Boesel, J. M. Oliveira, T. C. Santos and A. P. Marques, J. R. Soc., Interface, 2011, 4, 999–1030 CrossRef PubMed.
- H. C. Ott, T. S. Matthiesen, S. K. Goh, L. D. Black, S. M. Kren, T. I. Netoff and D. A. Taylor, Nat. Med., 2008, 14, 213–221 CrossRef CAS PubMed.
- S. L. J. Ng, K. Narayanan, S. Gao and A. C. A. Wan, Biomaterials, 2011, 32, 7571–7580 CrossRef CAS PubMed.
- T. Y. Lu, B. Lin, J. Kim, M. Sullivan, K. Tobita, G. Salama and L. Yang, Nat. Commun., 2013, 4, 1431–1442 CrossRef PubMed.
- T. Vazin and D. V. Schaffer, Trends Biotechnol., 2010, 28, 117–124 CrossRef CAS PubMed.
- T. Kai and A. Spradling, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4633–4638 CrossRef CAS PubMed.
- T. R. Brazelton, F. M. V. Rossi, V. I. Keshet and H. M. Blau, Science, 2000, 290, 1775–1779 CrossRef CAS PubMed.
- I. Andreu, T. Luque, A. Sancho, B. Pelacho, O. Iglesias-García, E. Melo, R. Farré, F. Prósper, M. R. Elizalde and D. Navajas, Acta Biomater., 2014, 10, 3235–3242 CrossRef CAS PubMed.
- A. J. Booth, R. Hadley, A. M. Cornett, A. A. Dreffs, S. A. Matthes, J. L. Tsui, K. Weiss, J. C. Horowitz, V. F. Fiore and T. H. Barker, Am. J. Respir. Crit. Care Med., 2012, 186, 866–876 CrossRef CAS PubMed.
- E. Melo, N. Cárdenes, E. Garreta, T. Luque, M. Rojas, D. Navajas and R. Farré, J. Mech. Behav. Biomed. Mater., 2014, 37, 186–195 CrossRef CAS PubMed.
- A. Lundby, K. Lage, B. T. Weinert, D. B. Bekkerjensen, A. Secher, T. Skovgaard, C. D. Kelstrup, A. Dmytriyev, C. Choudhary and C. Lundby, Cell Rep., 2012, 2, 419–431 CrossRef CAS PubMed.
- W. Jin and F. Wu, PLoS One, 2016, 11, e0147586 CrossRef PubMed.
- R. L. Rietze, H. Valcanis, G. F. Brooker, T. Thomas, A. K. Voss and P. F. Bartlett, Nature, 2001, 412, 736–739 CrossRef CAS PubMed.
- R. Galli, U. Borello, A. Gritti, M. G. Minasi, C. Bjornson, M. Coletta, M. Mora, M. G. De Angelis, R. Fiocco and G. Cossu, Nat. Neurosci., 2000, 3, 986–991 CrossRef CAS PubMed.
- D. A. Reardon and D. Cheresh, Genes Cancer, 2011, 2, 1159–1165 CrossRef PubMed.
- E. Garcion, A. Halilagic, A. Faissner and C. ffrench-Constant, Development, 2004, 131, 3423–3432 CrossRef CAS PubMed.
- T. W. Gilbert, T. L. Sellaro and S. F. Badylak, Biomaterials, 2006, 27, 3675–3683 CAS.
- D. A. Taylor, Curr. Opin. Biotechnol., 2009, 20, 598–605 CrossRef CAS PubMed.
- J. M. Wainwright, C. A. Czajka, U. B. Patel, D. O. Freytes, K. Tobita, T. W. Gilbert and S. F. Badylak, Tissue Eng. Part C Methods., 2010, 16, 525–532 CrossRef CAS PubMed.
- T. H. Petersen, E. A. Calle, L. Zhao, E. J. Lee, L. Gui, M. B. Raredon, K. Gavrilov, T. Yi, Z. W. Zhuang and C. Breuer, Science, 2010, 329, 538–541 CrossRef CAS PubMed.
- J. Cortiella, J. Niles, A. Cantu, A. Brettler, A. Pham, G. Vargas, S. Winston, J. Wang, S. Walls and J. E. Nichols, Tissue Eng., Part A, 2010, 16, 2565–2580 CrossRef CAS PubMed.
- T. L. Sellaro, A. Ranade, D. M. Faulk, G. P. Mccabe, K. Dorko, S. F. Badylak and S. C. Strom, Tissue Eng., Part A, 2009, 16, 1075–1082 CrossRef PubMed.
- M. R. Walker, K. K. Patel and T. S. Stappenbeck, J. Pathol., 2009, 217, 169–180 CrossRef CAS PubMed.
- K. Lee, E. A. Silva and D. J. Mooney, J. R. Soc., Interface, 2011, 8, 153–170 CrossRef CAS PubMed.
- P. Karpowicz, S. Willaimemorawek, L. Balenci, B. Deveale, T. Inoue and D. K. D. Van, J. Neurosci., 2009, 29, 3885–3896 CrossRef CAS PubMed.
- T. P. Kraehenbuehl, R. Langer and L. S. Ferreira, Nat. Methods, 2011, 8, 731–736 CrossRef CAS PubMed.
- W. Huang, L. Zhang, R. Niu and H. Liao, In Vitro Cell. Dev. Biol.: Anim., 2009, 45, 10–14 CrossRef CAS PubMed.
- J. Irianto, C. R. Pfeifer, Y. Xia and D. E. Discher, Cell, 2016, 165, 1820–1820 CrossRef CAS PubMed.
- J. C. Friedland, M. H. Lee and D. Boettiger, Science, 2009, 323, 642–644 CrossRef CAS PubMed.
- K. Saha, A. J. Keung, E. F. Irwin, Y. Li, L. Little, D. V. Schaffer and K. E. Healy, Biophys. J., 2008, 95, 4426–4438 CrossRef CAS PubMed.
- V. M. Jessica, A. DeQuach, A. Miglani, S. Lange, G. M. Keller, F. Sheikh and K. L. Christman, PLoS One, 2010, 5, 5–12 Search PubMed.
- A. D. Doyle, R. J. Petrie, M. L. Kutys and K. M. Yamada, Curr. Opin. Cell Bio., 2013, 25, 642–649 CrossRef CAS PubMed.
- K. N. An, Y. L. Sun and Z. P. Luo, Biorheology, 2004, 41, 239–246 CAS.
- J. A. Pedersen and M. A. Swartz, Ann. Biomed. Eng., 2005, 33, 1469–1490 CrossRef PubMed.
- A. D. Doyle, N. Carvajal, A. Jin, K. Matsumoto and K. M. Yamada, Nat. Commun., 2015, 6, 8720 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- S. Evenram, V. Artym and K. M. Yamada, Cell, 2006, 126, 645–647 CrossRef CAS PubMed.
- B. K. Teo, S. T. Wong, C. K. Lim, T. Y. Kung, C. H. Yap, Y. Ramagopal, L. H. Romer and E. K. Yim, ACS Nano, 2013, 7, 4785–4798 CrossRef CAS PubMed.
- N. Watanabe, T. Kato, A. Fujita, T. Ishizaki and S. Narumiya, Nat. Cell Biol., 1999, 1, 136–143 CrossRef CAS PubMed.
- Y. Chen, V. Balasubramaniyan, J. Peng, E. C. Hurlock, M. Tallquist, J. Li and Q. R. Lu, Nat. Protoc., 2007, 2, 1044–1051 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01287d |
| ‡ The first 4 authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2021 |