Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An anisotropic nanocomposite hydrogel guides aligned orientation and enhances tenogenesis of human tendon stem/progenitor cells†
Yichi
Xu‡
ab,
Heyong
Yin‡
cd,
Jin
Chu
 c,
David
Eglin
c,
David
Eglin
 a,
Tiziano
Serra
a,
Tiziano
Serra
 *a and
Denitsa
Docheva
*a and
Denitsa
Docheva
 *c
*c
aAO Research Institute Davos, Clavadelerstrasse 8, Davos Platz, Switzerland. E-mail: tiziano.serra@aofoundation.org
bDepartment of Plastic and Reconstructive Surgery, The First Medical Center, Chinese P.L.A. General Hospital, Beijing, China
cExperimental Trauma Surgery, Department of Trauma Surgery, University Regensburg Medical Centre, Regensburg, Germany. E-mail: denitsa.docheva@ukr.de
dDepartment of Orthopedics, Beijing Friendship Hospital, Capital Medical University, Beijing, China
First published on 12th February 2021
Abstract
The uniform and aligned arrangement of tendon cells is a marker of tendon tissue morphology and the embodiment of its biological anisotropy. However, most of the hydrogels used for tendon tissue engineering do not present anisotropic structures. In this work, a magnetically-responsive nanocomposite hydrogel composed of collagen type I (COL I) and aligned iron oxide nanoparticles (IOPs) was investigated for potential application in tendon tissue engineering. COL I with a mixture of remotely aligned IOPs (A/IOPs) and human tendon stem/progenitor cells (COL I-A/IOPs-hTSPCs) was prepared and the alignment of IOPs was induced under a remote magnetic field. Following the gelation of COL I, a stable and anisotropic nanocomposite COL I-A/IOPs hydrogel was formed. In addition, hTSPCs embedded in COL I with random IOPs (COL I-R/IOPs-hTSPCs) and in pure COL I (COL I-hTSPCs) were used as control groups. Cell viability, proliferation, morphology, cell row formation, and alignment of IOPs and hTSPCs were evaluated over time. In addition, a comprehensive gene expression profile of 48 different genes, including tendon-related genes and lineage/cross-linking genes, was obtained by implementing designer quantitative RT-PCR plates. The hTSPCs morphology followed the orientation of the anisotropic COL I-A/IOPs hydrogel with increased row formation in comparison to pristine COL I and COL-R/IOPs. Moreover, higher proliferation rate and significant upregulation of tendon gene markers were measured in comparison to hTSPCs cultivated in the COL I-R/IOPs and COL I. Thus, we suggest that providing the cells with aligned focal contact points, namely the aligned IOPs, is sufficient to provoke an immense effect on the formation of aligned cell rows. Taken together, we report a novel strategy for directing stem cell behavior without the use of exogenous growth factors or pre-aligned COL I fibers, and propose that anisotropic nanocomposite hydrogels hold great potential for tendon tissue engineering applications.
Tendon injuries prevail in orthopaedics and trauma surgery, accounting for up to 45% of all musculoskeletal injuries. However, the intrinsic repair potential of tendon tissue is very limited.1 Tendon injuries tend to become chronic conditions, and it is rare that ruptured tendon can restore its strength and function to the initial physiological state.2 Recently, methods for tendon injury repair, including conservative treatments such as anti-inflammatory drugs, physical therapy, active movement, and surgical options, e.g. suturing, tendon autograft or allograft transplantation, have been intensively applied. However, these treatments do not achieve satisfactory long-term clinical outcome and so alternative therapeutic methods need to be developed.3 Stem cell-based tendon tissue engineering technology is one of the most attractive and widely explored strategies that has shown great potential for boosting tendon repair.4 Cell source is a key element in tendon tissue engineering. Tendon stem/progenitor cells (TSPCs), first identified by Bi Y et al. in 2007, are considered to be a very promising candidate for tendon repair.5 In addition, the choice of cell-biomaterial carrier is another key factor for the success of tendon tissue engineering therapies.6 Biomaterials provide a specific microenvironment that can influence stem cell stemness and fate.7 Hydrogels are widely studied in tendon tissue engineering, since they can provide a three-dimensional (3D) environment for cell growth, proliferation and matrix remodeling in vitro, and can be implanted in vivo as cell delivery carriers.8 COL I is the most abundant collagen matrix in native healthy tendon tissue. It contains a GFOGER sequence that is an integrin receptor recognition site and thereby, mediates cell attachment and survival and shows excellent biocompatibility for biological applications.2,9
The anisotropy of biological tissue is the basic characteristic of its development, function and regeneration.10 The uniform and parallel row arrangement of tendon cells is the hallmark of tendon tissue and embodiment of its biological anisotropy.11 However, most of the hydrogels used for tendon tissue engineering do not present anisotropic structures or macromolecular organization to instruct embedded tendon progenitor cells. Nonetheless, hydrogels (e.g. collagen), remain an attractive carrier material due to the ease of production and handling, great biocompatibility to cells and ease of modification according to the foreseen application in tissue engineering.12 Hydrogels are highly responsive to heat and pH condition with high controllability and operability.13 Moreover, minimally invasive delivery of the material was achieved alone or functionalized with cells to the tissue or organ target site. Bae W G et al. (2015) used photolithography to artificially guide cell growth orientation on the surface of the hydrogels.14 External electric or magnetic fields can be used to form linear chains of colloidal particles.15 Tognato R et al. (2019) reported the synthesis and processing of cellularized magnetic nanocomposite hydrogels for the development of anisotropic cell-guidance 3D matrices and for the generation of bio-inspired soft-robotic systems.16 The hydrogel anisotropy can be achieved by mixing magnetic particles and applying low-intensity external magnetic field to control and regulate the alignment of the particles.17 Paramagnetic iron oxide nanoparticles (IOPs) are widely used as a magnetically-responsive element. The dipole–dipole magnetic attraction between these particles leads to self-assembly into uniform linear alignment along the remote magnetic field.18,19 Moreover, the IOPs, showing satisfactory biological compatibility, have been widely used in biomedical fields.20 Hu et al. reported the preparation of anisotropic hydrogels by self-assembling the magnetic particles in an external magnetic field, which were then solidified into gels by polymerization at high temperature (approx. 50 °C).21 However, this process is not suitable for use with living cells. Another study reported that hydrogels containing conductive carbon nanotubes (containing carbon nanotubes, CNTs) enforce the alignment of C2C12 cells on the surface of the gel.22 However, in this study, the alignment and maturation of the muscle cells may be not only due to the formation of anisotropic CNT tissue but also due to the polarization of small proteins in hydrogels caused by dielectrophoresis.23 Other limitations that need to be addressed are the complexity of the techniques for constructing hydrogel networks18 and the potential toxicity of the materials24 in order to ensure the biosafety of biological manufacturing processes and the material biocompatibility of the constructed composite anisotropic hydrogel.
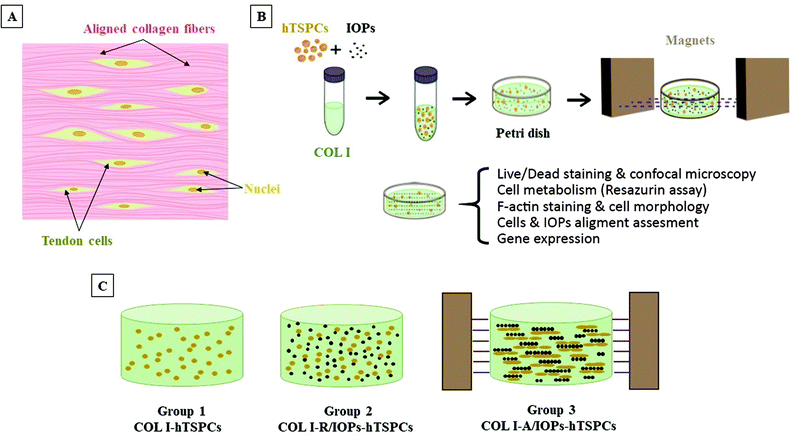
The uniform arrangement of tendon cells along the hierarchically aligned collagen fibers is the symbol of the tendon tissue and embodiment of its biological anisotropy (Fig. 1A). So far, there have been no previous studies on TSPCs morphology and differentiation into tendon cells can be stimulated by embedding them into an anisotropic COL I microenvironment. In our study, we explored an original method to functionalize COL I with IOPs and prepared a nanocomposite hydrogel with excellent biocompatibility, non-toxicity, biological anisotropy, and controllable responsiveness through low intensity magnetic field and collagen gelation (Fig. 1B). The hTSPCs were isolated from non-ruptured Achilles tendon biopsies of three young and healthy human male patients (28 ± 5 years). The isolation of the human cells was performed in accordance with the Guidelines of the Ethical Grant No. 166-08, and experiments were approved by the Ethics Committee at the Medical Faculty of the Ludwig-Maximilians-University (LMU), Munich, Germany (Kohler et al.25). Informed consents were obtained from the human participants of this study. The cells used in this study were previously isolated and characterized by Kohler et al.25 Three study groups were designed: hTSPCs embedded in hydrogels containing 0.1% (5 μm pore size)26 COL I (COL I-hTSPCs), in COL I with random IOPs (COL I-R/IOPs-hTSPCs) or in COL I with an aligned IOPs group (COL I-A/IOPs-hTSPCs) (Fig. 1C). IOPs were synthetized using a previously described protocol.20 The induction of biological anisotropy of the nanocomposite hydrogel was achieved under a remote magnetic field according to Tognato R et al. (2019):16 two pieces of neodymium magnet (NdFeB, a permanent magnet made from an alloy of neodymium, iron, and boron forming the Nd2Fe14B tetragonal crystalline structure) were set on a Teflon plate in an ice box with a distance of 5 cm, which provided a 0.02 Tesla linear and homogeneous magnetic field environment. Briefly, a mixture of COL I-IOPs-hTSPCs in a Petri dish (35 mm diameter) was placed in the middle of the two magnets. After 20 minutes of exposure to the magnetic field in the ice box, the mixture was then transferred into a 37 °C incubator and cross-linking and gelation occurred in approx. 15 minutes. Afterwards, 2 ml culture medium was added into the Petri dish and cell-loaded hydrogels were cultured for up to 7 days. Cell viability, proliferation, morphology, cell row or string formation, and alignment of IOPs and hTSPCs were evaluated. In addition, a comprehensive profiling of 48 different genes, including tendon-, lineage-, and cross-linking-related genes, was performed.
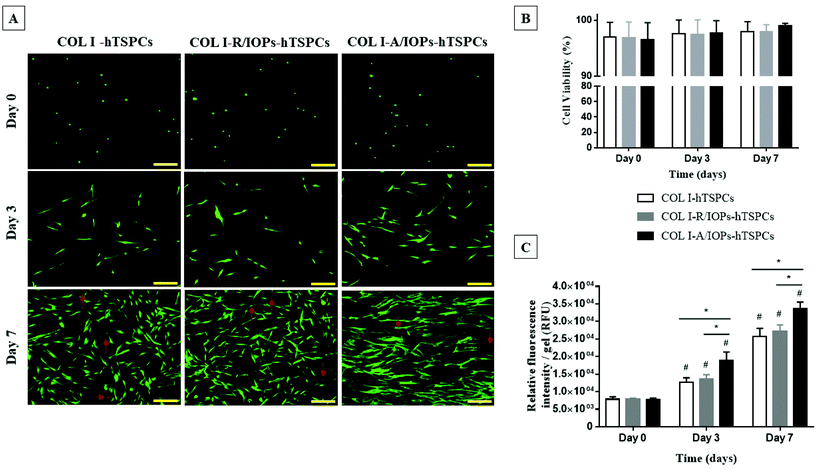
Cell viability and proliferation were evaluated by Live/Dead staining and the resazurin metabolic assay. As shown in Fig. 2A, the cells were viable and round-shaped at day 0. From day 3, the hTSPCs exhibited high cell viability and elongated cell morphology. Interestingly, at day 7, hTSPCs showed a clear and aligned orientation along the IOP strings, as well as augmented cell elongation in the COL I-A/IOPs-hTSPCs group. In contrast, hTSPCs cultured in the other two groups were randomly distributed and exhibited less stretched cell shapes. In all the groups, very few dead cells could be observed (red narrows in Fig. 2A). Quantitative analysis of cell viability was in line with these observations (Fig. 2B, p > 0.05).
The resazurin cell metabolic results showed an identical cell amount at day 0 for all groups. From day 3 to day 7, cell proliferation occurred in all groups, with a significantly higher proliferation rate in the COL I-A/IOPs-hTSPCs group compared to the other groups (p < 0.05; Fig. 2C). There was no significant difference in cell proliferation between the COL I-hTSPCs and COL I-R/IOPs-hTSPCs groups at all time points (p > 0.05; Fig. 2C).
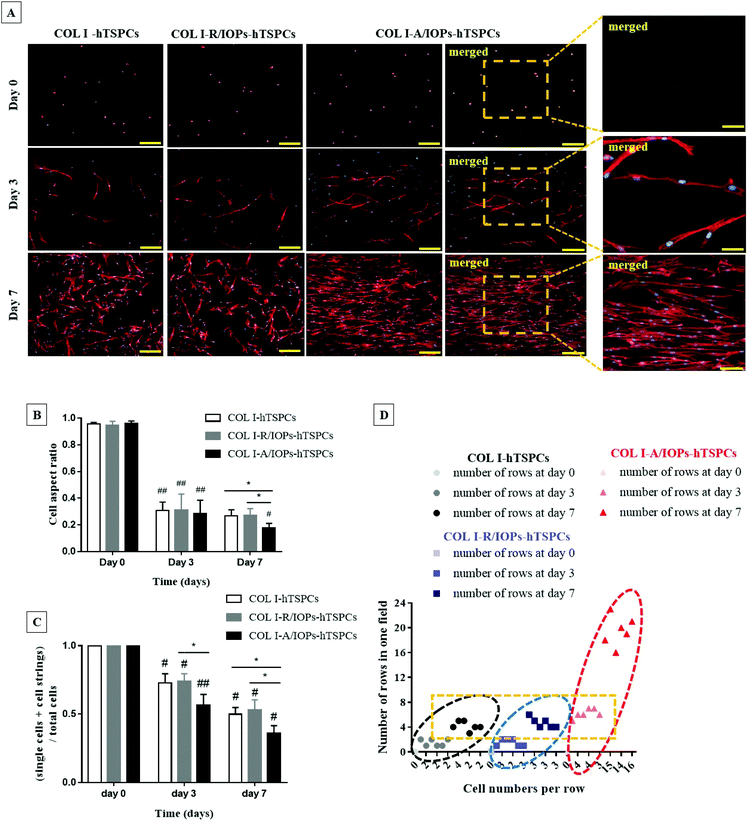
Fluorescently labeled phalloidin-based F-actin staining, counterstained with DAPI, was applied to image the cytoskeletal structure and cell shape. As shown in Fig. 3A and similar to that shown in Fig. 2A, from day 3, hTSPCs became elongated in all groups. Interestingly, they preferentially stretched and elongated in the direction of the IOPs alignment in the COL I-A/IOPs-hTSPCs group. Such preferential alignment was not observed with the other two groups. At day 7, an even greater cell alignment with the formation of parallel rows was observed in the COL I-A/IOPs-hTSPCs group, whilst hTSPCs in the other two groups were randomly distributed. Next, the qualitative analysis of cell morphology was conducted by calculating the cell aspect ratio (Fig. S1†), a ratio of the horizontal axis to the vertical axis of individual cells. As shown in Fig. 3B, at day 0, the cell aspect ratio was close to 1 in the three study groups, indicating an approximately spherical cell shape.
At day 3, the aspect ratio of each group was significantly smaller than that at day 0 (p < 0.05), with no significant differences detected amongst the groups (p > 0.05). At day 7, the hTSPC aspect ratio in the COL I-A/IOPs-hTSPCs group was significantly smaller than the other two groups (p < 0.05), indicating a more elongated cell shape. Furthermore, the formation of cell rows or chains was also analyzed in the F-actin images. The number of individual cells (n1), cell chains of 2 or more cells (n2) and total nuclei (n3) from each view was counted, then the value of (n1 + n2)/n3 was calculated to evaluate cell chain formation (cell fusion index), as per the formula; the lower the calculated value, the higher the cell chain formation, which is indicative of increased cell fusion via cell–cell contact. As shown in Fig. 3C, at day 0, no cell rows were formed in all the groups, and were single cell, n2 = 0, with initial ratio (n1 + 0)/n1 equal to 1. From day 3–7, the ratio of the COL I-A/IOPs-hTSPCs group was significantly lower than that of the other two groups (p < 0.01 and p < 0.05, respectively), whilst the difference between the COL I-hTSPCs and COL I-R/IOPs-hTSPCs groups was not statistically significant (p > 0.05). To summarize, our results demonstrated a very significant effect of the anisotropy of the COL I-A/IOPs matrix on hTSPCs cell shape and row formation.
Additionally, the number of cell rows and cell number per row were also determined. As shown in Fig. 3D, there was no cell chains generated at day 0 for all the groups, so the number of cell rows and cell number of each row were 0. From day 3 onwards, the number of cell rows and connected cells in each row in the COL I-A/IOPs-hTSPCs group was significantly higher than the other two groups. This confirmed that the uniform parallel arrangement of IOPs led to the aligned orientation of hTSPCs into significantly higher number of rows containing significantly higher number of connected cells.
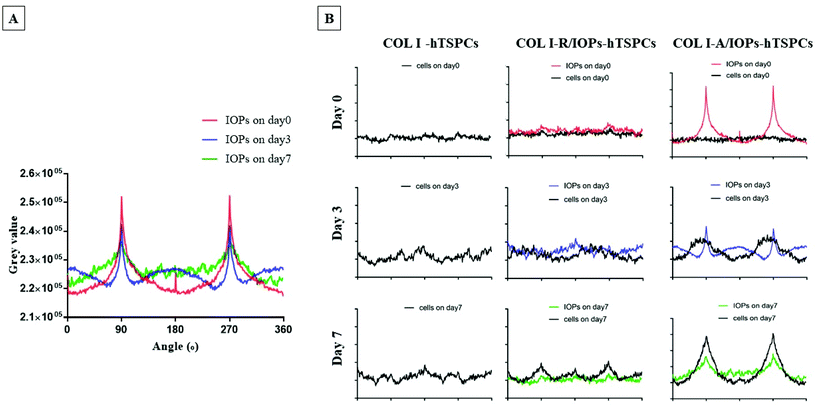
To explore how the degree of uniform alignment of IOPs changes over time, alignment kinetic curves were plotted at each time point. As shown in Fig. 4A and Fig. S2,† the alignment of IOPs was evaluated using a bright-field optical image and analyzed with the two-dimensional fast Fourier transform (2D-FFT) using ImageJ software. From day 0–7, the peaks were reduced and became inhomogeneous, which indicated that although the parallel array of IOPs still exists, the number of IOPs positioned in other directions increased over time. Nevertheless, as shown in Fig. 4B, in the COL I-hTSPCs group without the presence of IOPs, the distribution and orientation of the cells were random at the three different time points and few peaks appear at different angle degrees. In the COL I-R/IOPs-hTSPCs group, there were no aligned IOPs and cells. Here, the amplitudes of the cell curve increased slightly between day 0 and 7 as this increase was related to the natural stretching of the cells. However, during the 2D-FFT, the accumulated cell directions were random and visible not only at 90 and 270 angle degrees, but also at 0 and 180 degrees. In the COL I-A/IOPs-hTSPCs group, very aligned IOP rows were formed after exposure to a remote magnetic field. Intriguingly, from day 3, hTSPCs within the nanocomposite hydrogel were also organized in parallel rows overlapping with the IOP rows. At day 7, the hTSPCs and IOPs curves had high amplitudes at 90 and 270 angle degrees and were highly co-aligned suggesting that the aligned IOPs directed the hTSPCs alignment as well. One possibility is that hTSPCs use the IOPs, as focal points of cell to matrix contact, pulling onto them and undergoing stretching, cell–cell contact and forming a continuous cell row.
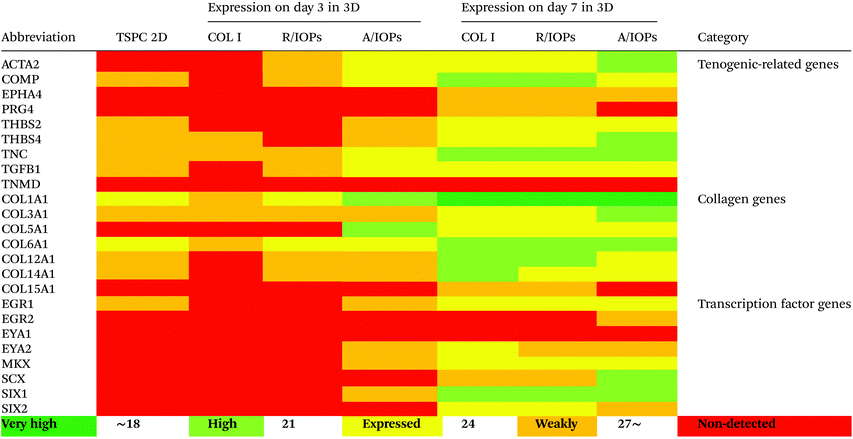
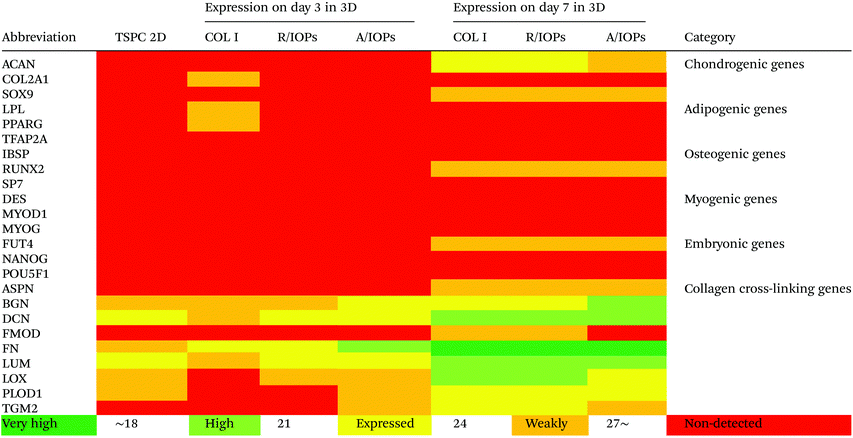
Since anisotropic materials can significantly improve the activity, proliferation and other biological behaviors of stem cells, it would be of importance to investigate if they can influence the fate of hTSPCs.26 Therefore, we conducted a comprehensive analysis of TSPCs gene expression upon cultivation in COL I-A/IOPs hydrogel, including lineage specificity (e.g., chondrogenesis, osteogenesis, lipogenesis, myogenesis, and embryonicity) and multiple tendon-related gene markers, covering virtually all potential differentiation directions of tendon stem/progenitor cells. Specifically, in order to explore the gene expression profile of hTSPCs, custom-designed quantitative RT-PCR plates (96-well format) for 48 different genes, including tenogenic, lineage and collagen cross-linking genes (listed in Tables 1 and 2, for full gene names, refer to Table S1 in the ESI†) were previously designed27 and experiments were carried out to compare the gene expression amongst the study groups. Based on delta Ct values, a gene expression heatmap was generated and is shown in Tables 1 and 2. Furthermore, as compared to hTSPCs from standard cell culture (prior 3D cultivation) and hTSPCs cultivated for 3 days in COL I hydrogels (without IOPs), we have included the corresponding heatmaps in Tables 1 and 2 based on data derived from our previous publications.27,28 Next, differences in genes showing a significant change in expression at day 7 were expressed as fold change to the COL I-hTSPC group and plotted in Fig. 5C and D. First, our gene expression results indicated that the tenogenic phenotype of hTSPC was stable during culture in all groups since the majority of genes related to other lineages (chondrogenic, adipogenic, osteogenic, myogenic and embryogenic) were neither expressed nor upregulated over time (Table 2). Expression of multiple tendon-related and collagen-cross linking genes was detected, confirming the tendon origin of the cells. Moreover, many of these genes upregulated with prolonged 3D cultivation (day 7 vs. day 3 and 2D). The hTSPCs, during expansion as well as during 3D cultivation, were supplemented with cell culture medium containing 10% Fetal Bovine Serum (FBS). FBS is known to contain different growth factors; however, a previous study29 with bone marrow-derived mesenchymal stem cells cultivated in medium supplemented with FBS versus such medium enriched with additional growth factors did not reveal that FBS itself can induce tenogenic differentiation. In addition, all study groups were treated equally with FBS, and based on the gene expression profiling in Tables 1 and 2, the effects were related to days of cultivation and to the alignment of the IOPs. Nevertheless, for clinical translation, FBS should be avoided due to immunological complications and hTSCPs should be cultivated ideally in autologous human serum.
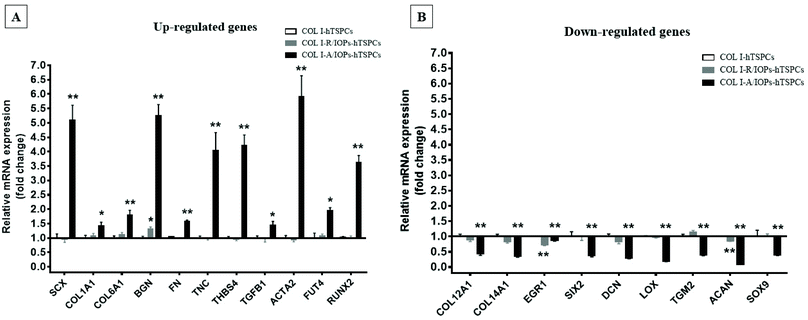 | ||
| Fig. 5 (A) Up-regulated genes in the COL I-A/IOPs-hTSPCs group at day 7, and immunofluorescence staining of COL1A1, COL6A1 and SCX is presented in the ESI Fig. 11.† (B) Down-regulated genes in the COL I-A/IOPs-hTSPCs group at day 7. GAPDH was used as the reference gene. Only genes with significant change in expression are plotted. The qPCR has been conducted with n = 3 TSPC donors, 3 gels/donor. Statistical differences between two groups (COL I-R/IOPs-hTSPCs or COL I-R/IOPs-hTSPCs vs. COL I-hTSPCs) were determined using unpaired Student's t-test p < 0.05 (*) and p < 0.01 (**). | ||
| The heatmaps for hTSPCs from standard cell culture (prior 3D cultivation) and hTSPCs cultivated for 3 days in COL I hydrogels (without IOPs) are based on data derived from previous publications by our team.27,28 |
|---|
|
| The heatmaps for hTSPCs from standard cell culture (prior 3D cultivation) and hTSPCs cultivated for 3 days in COL I hydrogels (without IOPs) are based on data derived from previous publications by our team.27,28 |
|---|
|
Tenomodulin (TNMD) is a gene that is highly expressed in tendons and ligaments30–32 and has been shown to play an important role in regulating TSPC function and COL I fibrils during tendon development and mechanical loading. Loss of TNMD expression in mice resulted in a pathological thickening of COL I fibrils.30 In our study, the expression of TNMD was not detected in either of the experimental groups (Table 1), which is consistent with our previous results that TNMD mRNA shows rapid loss during hTSPC in vitro culture.30–32 In the future, it would be of interest to test if mechanical stimulation of the nanocomposite hydrogels can lead to TNMD gene expression rescue.
Our data at day 3 showed no obvious difference in the gene expression levels between the COL I-R/IOP-hTSPCs and COL I-A/IOP-hTSPCs groups. Expression of some tendon-related genes such as ACTA2, COMP, THBS2, THBS4, TNC, and TGF-b1 and some collagen cross-linking genes such as BGN, FN, PLOD and TGM2 were slightly higher in the COL I-A/IOP-hTSPCs group. Most strikingly, at day 7, the majority of tenogenic genes including the well-known master transcription factor SCX that promotes fibroblast proliferation and collagen synthesis in tendon and tendon-related matrix genes COL1A1, COL6A1, BGN, FN, TNC, and THBS4 were significantly up-regulated in the COL I-A/IOP-hTSPCs group (p < 0.05; Fig. 5), indicating a clear positive effect of the COL I-A/IOP microenvironment. COL 1 is the most abundant matrix protein in tendon tissues. COL 6 is also presented in the tendon matrix and plays an important role in regulating the formation of COL I fibers.2 BGN plays a critical role during connective tissue development and has been reported to be a critical component of the TSPC niche.5 TNC is expressed in tendon matrix and it contributes to tendon repair.2 THBS4 is abundant in tendon and regulates ECM synthesis, composition, and organization.33 In addition, some other genes, like TGF-b1 that induces the expression of the transcription factor SCX during embryonic development,34 ACTA2 and RUNX2 (the master osteoblast transcription factor)35 were also found to be significantly up-regulated in the COL I-A/IOP-hTSPCs group (p < 0.05). Specifically, for RUNX2, a follow up validation would be necessary to clarify if the detected upregulation results in higher RUNX2 protein levels. In addition, several tendon-related genes (COL12A1, COL14A1, EGR1, SIX2, DCN, LOX, TGM2) and cartilage-related genes (ACAN, SOX9) were down-regulated in the COL I-A/IOP-hTSPCs group (p < 0.05). Pilot validation analysis at the protein level was carried out via immunohistochemical staining on hydrogel cryosections with anti-COL 1, COL 6 and SCX antibodies, showing the detection of all three antigens with a tendency of higher levels in the COL I-A/IOPs-hTSPCs group (Fig. S3†). To decipher the potential mechanism, additional protein quantitative analyses, such as western blotting or ELISA, are planned for subsequent studies. Interestingly, the COL 1 staining of gels loaded with A/IOPs (with or without cells) indicated no alignment of the COL I fibrils in the 0.1% hydrogels, thus further strengthening the major effect on cell behavior that was triggered by the aligned nanoparticles in the composite hydrogel.
The results of gene profiling indicated that at day 3 of culture, although the cells in the COL I-A/IOPs-hTSPCs group have initiated aligned arrangement, no significant effect on the gene expression levels was detected. At day 7, hTSPCs have formed a large number of aligned rows in the COL I-A/IOPs-hTSPCs group, and a significant up-regulation in tendon-related genes was revealed. These results suggest that in order to detect an effect at the mRNA level, a certain time is required for the transcriptional machinery to operate. Moreover, it is possible that there is a threshold in the number of rows above which an upregulation of tendon-specific gene markers (e.g. scleraxis) can be detected. Thus, our model may be very useful to uncover instructive cues influencing molecular feedback during the formation of cell rows in tendon tissue morphogenesis in vitro. Another concept that can be drawn from our model is that it can be used to investigate how specific molecular factors are sensitive to cell alignment and how this is translated “outside-in” within the cell.
Magnetotherapy for tendon regeneration36 is a rapidly increasing field based on the use of magnetically responsive biomaterials as well as the direct application of magnetic fields to tendon cells, thus generating physical forces that can modulate their activity and fate. For potential clinical translation of the nanocomposite hydrogels, it is important to carry out clinically relevant preclinical studies. In the future, we plan to investigate the performance of this anisotropic hydrogel combined with hTSPCs in established models for Achilles tendon injury in rodents.37–39 A possible area for the application of anisotropic COL I-A/IOPs hydrogel combined with cells, growth factors or drugs are partial tendon defects or conservatively treated tendon injuries via minimally invasive percutaneous injections.
Conclusion
In conclusion, we have demonstrated that an anisotropic matrix composed of COL I and aligned IOPs guided hTSPCs behavior with aligned cellular orientation and enhanced tenogenic differentiation. Thus, our study provides a novel strategy for directing stem cell behavior without the use of exogenous growth factors and indicates that the anisotropic nanocomposite hydrogel has great potential for tendon tissue engineering application.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by four scientific and clinic departments from Switzerland, Germany and China: AO foundation, Switzerland; Institute of Orthopedics, Chinese P. L. A. General Hospital, Beijing, China; Experimental Trauma Surgery, Department of Trauma Surgery, University Regensburg Medical Centre, Regensburg, Germany; and Department of Orthopedics, Beijing Friendship Hospital, Capital Medical University, Beijing, China. D. D. acknowledges the financial support of of the EU H2020-WIDESPREAD-05-2017-Twinning Grant “Achilles: Overcoming specific weakness in tendon biology to design advanced regenerative therapies” Proposal Nr. 810850. Y. X. is grateful for the financial support from the China Scholarship Council [CSC, no. 201703170097] and the Sino Swiss Science and Technology Cooperation [SSSTC, no. EG08-122016]. The authors thank Dr Gabriele Giancane for providing the IOPs and Dr Girish Pattappa for English proof-reading.Notes and references
- R. Costa-Almeida, I. Calejo and M. E. Gomes, Int. J. Mol. Sci., 2019, 20, 3002 CrossRef CAS.
- D. Docheva, S. A. Muller, M. Majewski and C. H. Evans, Adv. Drug Delivery Rev., 2015, 84, 222–239 CrossRef CAS.
- J. G. Snedeker and J. Foolen, Acta Biomater., 2017, 63, 18–36 CrossRef.
- P. P. Lui, Stem Cells Cloning: Adv. Appl., 2015, 8, 163–174 Search PubMed.
- Y. Bi, D. Ehirchiou, T. M. Kilts, C. A. Inkson, M. C. Embree, W. Sonoyama, L. Li, A. I. Leet, B. M. Seo, L. Zhang, S. Shi and M. F. Young, Nat. Med., 2007, 13, 1219–1227 CrossRef CAS.
- X. Zhang, Y. C. Lin, Y. F. Rui, H. L. Xu, H. Chen, C. Wang and G. J. Teng, Stem Cells Int., 2016, 2016, 4076578 Search PubMed.
- S. Naahidi, M. Jafari, M. Logan, Y. Wang, Y. Yuan, H. Bae, B. Dixon and P. Chen, Biotechnol. Adv., 2017, 35, 530–544 CrossRef CAS.
- N. Juncosa-Melvin, G. P. Boivin, M. T. Galloway, C. Gooch, J. R. West, A. M. Sklenka and D. L. Butler, Tissue Eng., 2005, 11, 448–457 CrossRef CAS.
- M. Schneider, P. Angele, T. A. H. Jarvinen and D. Docheva, Adv. Drug Delivery Rev., 2018, 129, 352–375 CrossRef CAS.
- N. S. Kalson, Y. Lu, S. H. Taylor, T. Starborg, D. F. Holmes and K. E. Kadler, eLife, 2015, 4, e05958 CrossRef.
- T. Mammoto and D. E. Ingber, Development, 2010, 137, 1407–1420 CrossRef CAS.
- Z. Yan, H. Yin, M. Nerlich, C. G. Pfeifer and D. Docheva, J. Exp. Orthop., 2018, 5, 1 CrossRef.
- A. S. Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Nat. Mater., 2016, 15, 413–418 CrossRef.
- W. G. Bae, J. Kim, Y. H. Choung, Y. Chung, K. Y. Suh, C. Pang, J. H. Chung and H. E. Jeong, Biomaterials, 2015, 69, 158–164 CrossRef CAS.
- B. Bharti and O. D. Velev, Langmuir, 2015, 31, 7897–7908 CrossRef CAS.
- R. Tognato, A. R. Armiento, V. Bonfrate, R. Levato, J. Malda, M. Alini, D. Eglin, G. Giancane and T. Serra, Adv. Funct. Mater., 2019, 29, 1804647 CrossRef.
- A. I. Goncalves, M. T. Rodrigues and M. E. Gomes, Acta Biomater., 2017, 63, 110–122 CrossRef CAS.
- B. Bharti, A. L. Fameau, M. Rubinstein and O. D. Velev, Nat. Mater., 2015, 14, 1104–1109 CrossRef CAS.
- V. Bonfrate, D. Manno, A. Serra, L. Salvatore, A. Sannino, A. Buccolieri, T. Serra and G. Giancane, J. Colloid Interface Sci., 2017, 501, 185–191 CrossRef CAS.
- Y. Bao, J. A. Sherwood and Z. Sun, J. Mater. Chem. C, 2018, 6, 1280–1290 RSC.
- W. Hu, G. Z. Lum, M. Mastrangeli and M. Sitti, Nature, 2018, 554, 81–85 CrossRef CAS.
- J. Ramon-Azcon, S. Ahadian, M. Estili, X. B. Liang, S. Ostrovidov, H. Kaji, H. Shiku, M. Ramalingam, K. Nakajima, Y. Sakka, A. Khademhosseini and T. Matsue, Adv. Mater., 2013, 25, 4028–4034 CrossRef CAS.
- N. Abd Rahman, F. Ibrahim and B. Yafouz, Sensors (Basel), 2017, 17, 449 CrossRef.
- S. Lanone, P. Andujar, A. Kermanizadeh and J. Boczkowski, Adv. Drug Deliv. Rev., 2013, 65, 2063–2069 CrossRef CAS.
- J. Kohler, C. Popov, B. Klotz, P. Alberton, W. C. Prall, F. Haasters, S. Muller-Deubert, R. Ebert, L. Klein-Hitpass, F. Jakob, M. Schieker and D. Docheva, Aging Cell, 2013, 12, 988–999 CrossRef CAS.
- S. Ramanujan, A. Pluen, T. D. McKee, E. B. Brown, Y. Boucher and R. K. Jain, Biophys. J., 2002, 83, 1650–1660 CrossRef CAS.
- H. Yin, Z. Yan, R. J. Bauer, J. Peng, M. Schieker, M. Nerlich and D. Docheva, Biomed. Mater., 2018, 13, 034107 CrossRef.
- H. Yin, F. Strunz, Z. Yan, J. Lu, C. Brochhausen, S. Kiderlen, H. Clausen-Schaumann, X. Wang, M. E. Gomes, V. Alt and D. Docheva, Biomaterials, 2020, 236, 119802 CrossRef CAS.
- M. Bottagisio, S. Lopa, V. Granata, G. Talo, C. Bazzocchi, M. Moretti and A. B. Lovati, Differentiation, 2017, 95, 44–53 CrossRef CAS.
- S. Dex, P. Alberton, L. Willkomm, T. Sollradl, S. Bago, S. Milz, M. Shakibaei, A. Ignatius, W. Bloch, H. Clausen-Schaumann, C. Shukunami, M. Schieker and D. Docheva, EBioMedicine, 2017, 20, 240–254 CrossRef.
- S. Dex, D. Lin, C. Shukunami and D. Docheva, Gene, 2016, 587, 1–17 CrossRef CAS.
- D. Docheva, E. B. Hunziker, R. Fassler and O. Brandau, Mol. Cell. Biol., 2005, 25, 699–705 CrossRef CAS.
- O. Stenina-Adognravi and E. F. Plow, Matrix Biol., 2019, 75–76, 300–313 CrossRef CAS.
- C. L. Mendias, J. P. Gumucio, M. E. Davis, C. W. Bromley, C. S. Davis and S. V. Brooks, Muscle Nerve, 2012, 45, 55–59 CrossRef CAS.
- S. Vimalraj, B. Arumugam, P. J. Miranda and N. Selvamurugan, Int. J. Biol. Macromol., 2015, 78, 202–208 CrossRef CAS.
- T. Pesqueira, R. Costa-Almeida and M. E. Gomes, J. Cell Physiol., 2018, 233, 6395–6405 CrossRef CAS.
- D. Lin, P. Alberton, M. D. Caceres, E. Volkmer, M. Schieker and D. Docheva, Cell Death Dis., 2017, 8, e3116 CrossRef CAS.
- C. F. Hsieh, P. Alberton, E. Loffredo-Verde, E. Volkmer, M. Pietschmann, P. Muller, M. Schieker and D. Docheva, Nanomedicine, 2016, 11, 1153–1167 CrossRef CAS.
- C. F. Hsieh, P. Alberton, E. Loffredo-Verde, E. Volkmer, M. Pietschmann, P. E. Muller, M. Schieker and D. Docheva, Eur. Cells Mater., 2016, 32, 228–240 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01127d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |