Open Access Article
Open Access ArticleVolatile atmospheric pressure chemical ionisation mass spectrometry headspace analysis of E. coli and S. aureus†
Kerry
Rosenthal
 a,
Eugenie
Hunsicker
b,
Elizabeth
Ratcliffe
c,
Martin R.
Lindley
*ad,
Joshua
Leonard
e,
Jack R.
Hitchens
e and
Matthew A.
Turner
de
a,
Eugenie
Hunsicker
b,
Elizabeth
Ratcliffe
c,
Martin R.
Lindley
*ad,
Joshua
Leonard
e,
Jack R.
Hitchens
e and
Matthew A.
Turner
de
aSchool of Sport, Exercise & Health Sciences, Loughborough University, Loughborough, UK. E-mail: m.r.lindley@lboro.ac.uk
bDepartment of Mathematical Sciences, Loughborough University, Loughborough, UK
cDepartment of Chemical Engineering, Loughborough University, Loughborough, UK
dTranslational Chemical Biology Research Group, Loughborough University, Loughborough, UK
eDepartment of Chemistry, Loughborough University, Loughborough, UK
First published on 15th November 2021
Abstract
Identifying the characteristics of bacterial species can improve treatment outcomes and mass spectrometry methods have been shown to be capable of identifying biomarkers of bacterial species. This study is the first to use volatile atmospheric pressure chemical ionisation mass spectrometry to directly and non-invasively analyse the headspace of E. coli and S. aureus bacterial cultures, enabling major biological classification at species level (Gram negative/positive respectively). Four different protocols were used to collect data, three utilising discrete 5 min samples taken between 2 and 96 h after inoculation and one method employing 24 h continuous sampling. Characteristic marker ions were found for both E. coli and S. aureus. A model to distinguish between sample types was able to correctly identify the bacteria samples after sufficient growth (24–48 h), with similar results obtained across different sampling methods. This demonstrates that this is a robust method to analyse and classify bacterial cultures accurately and within a relevant time frame, offering a promising technique for both clinical and research applications.
Introduction
The rapid identification of bacterial species is crucial to provide appropriate treatment as misuse of antibiotics is a primary cause of antimicrobial resistance.1 Additionally, it is important to identify features of antimicrobial sensitivity such as the presence of a bacterial biofilm infection, which requires higher concentrations of antibiotics to treat compared to planktonic bacteria.2 Traditional tests such as Gram-staining and microscopy, combined with biochemical tests and antibiograms for antibiotic resistance profiling,3,4 require skilled operators to perform, as these techniques often include a subjective visual judgement.5 More recent techniques include polymerase chain reaction (PCR), which can provide information on genotypic but not phenotypic antimicrobial resistance.6Mass spectrometry methods are capable of measuring biomarkers of bacterial infection. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can determine the species of a bacterial sample within a few hours with 80% accuracy,7 much faster than traditional methods. However, the ionisation method destroys the sample so it cannot be used to monitor changes. Gas chromatography-mass spectrometry (GC-MS) is the ‘gold-standard’ for analysing volatile organic compounds (VOCs) and has been commonly used to analyse VOCs from the headspace of bacterial cultures.8–16 Using gas chromatography to separate the volatiles before they reach the mass spectrometer allows for a more accurate identification of the compounds but complex samples can take an hour to analyse17 and differences in the materials used for pre-treatment have been shown to affect the VOCs detected from bacterial samples.12,13
Real-time analysis methods such as proton transfer reaction-mass spectrometry (PTR-MS) and selected ion flow tube mass spectrometry (SIFT-MS) can be used to obtain a snapshot of a sample within minutes,18 or measure the changes of a sample over time.19 This allows for monitoring of the growth of a bacterial culture or the response to antibiotics over time. These methods are both soft ionisation techniques, which makes the resulting ions easier to interpret as there is less fragmentation. Previous studies using PTR-MS headspace analysis have found 23 m/z windows as potential biomarkers over a 24 h growth period of E. coli (across a mass range of m/z 18–150)19 and 15 m/z windows with clear differences between blank broth and growing S. aureus cultures, across a mass range of m/z 20–200 from inoculation to 350 min.20
This research uses volatile atmospheric pressure chemical ionisation coupled to a single quadrupole mass spectrometer (vAPCI-MS) which facilitates the direct analysis of VOCs using a venturi jet-pump. Therefore, no sample pre-treatment is required, reducing potential sources of interlaboratory variation, thereby increasing the potential for direct comparison between studies. Additionally, no pre-treatment step allows for real-time analysis of VOCs in air samples such as exhaled breath21,22, bloodstains,23 and the headspace of food.17 The headspace of bacterial cultures can be analysed without any disruption to the sample; therefore, allowing for observations over time, similar to PTR-MS or SIFT-MS, but unlike the most commonly used method MALDI-TOF MS. Direct analysis can be used to measure changes such as bacterial growth or the reaction to antibiotics. Additionally, the system is much smaller than traditional bench-top mass spectrometers at 66 × 28 × 56 cm, requiring less laboratory space and potentially making it transportable due to the compact footprint. It also has a simplistic design and user-friendly software which does not require expertise to operate. These factors make it a promising method for analysing bacterial samples in both research and clinical contexts.
Many existing metabolomics data processing protocols to reduce noise and identify potential biomarkers are not suitable for the data structure produced by vAPCI-MS analysis, or have not been tested. Existing open-source software, such as Metaboanalyst,24 have various data processing tools available, including variable filtering and scaling methods. However, these packages were often designed for mass spectrometry methods with chromatographic separations, which provide a discrete signal separated over two dimensions, and are therefore challenging to adapt for direct analysis techniques. Developing a standardised method for vAPCI-MS data processing and analysis is crucial to ensure the results are representative of the samples and allow for comparison between studies.
The purpose of this study was to determine the capability of the vAPCI-MS to detect markers relating to the presence and characterisation of bacterial samples and establish a data processing protocol, by analysing two existing datasets collected by different researchers under slightly different conditions.
Materials and methods
Bacterial species
Two medically important bacterial species corresponding to major biological classification of bacteria based on their cell wall structure differences were obtained from Sigma-Aldrich UK. E. coli ATCC25922 is a Gram-negative clinical isolate and a control strain used for antimicrobial susceptibility testing with further applications in bioinformatics, food testing, medium testing, and quality control. S. aureus NCTC 6571 is a Gram-positive clinical isolate, first added to the NCTC collection in 1943 by Sir Alexander Fleming in the testing and development of penicillin and used as a standard in Public Health England Standards for Microbiology Investigations and control strain for antimicrobial susceptibility testing.Instrumentation
All samples were analysed using an expression single quadrupole mass spectrometer (Advion, NY, USA) with a commercially available vAPCI source (Advion, NY, USA). This instrument uses a quadrupole mass analyser with an electron multiplier detector. It has a mass accuracy of ±0.1 Da units across the 30–300 m/z range.Optimisation of mass spectrometer conditions
10 mg of six VOCs known to be produced by E. coli (acetone, acetic acid, methyl propionate, indole, 2-octanol and 2′-aminoacetophenone (Sigma-Aldrich, UK)12,19,25–28) were mixed with methanol in a 10 ml volumetric flask. This solution was further diluted in a solution of methanol and deionised water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) to obtain a final concentration of 100 μg ml−1. This solution was infused directly into the vAPCI source at a rate of 10 μl min−1 to optimise and tune the vAPCI-MS e the vAPCI-MS detection parameters. The optimum instrument conditions were found to be: capillary temperature 250 °C, capillary voltage 120 V, source voltage offset 25 V, source gas temperature 350 °C, corona discharge voltage 5 kV, transfer line temperature 100 °C.
50) to obtain a final concentration of 100 μg ml−1. This solution was infused directly into the vAPCI source at a rate of 10 μl min−1 to optimise and tune the vAPCI-MS e the vAPCI-MS detection parameters. The optimum instrument conditions were found to be: capillary temperature 250 °C, capillary voltage 120 V, source voltage offset 25 V, source gas temperature 350 °C, corona discharge voltage 5 kV, transfer line temperature 100 °C.
The instrument was operated in positive ion mode for all experiments, over a mass range of 30–300 m/z. The system was flushed by connecting an empty 20 ml capacity headspace vial pressurised with a nitrogen gas flow, between samples to limit any potential contamination.
Sample preparation and analysis
Samples were incubated at 37 °C for timepoint analysis at 2, 6, 24, 48 and 72 h post inoculation. During analysis, headspace temperature was maintained at 35–38 °C by incubating headspace vials in a thermal heating block with a modified heated transfer line (Fig. 1a). The modification attached tubing to the lid of the headspace vial to prevent vacuum formation and maintain aerobic conditions for the bacterial cultures. For each condition, triplicate headspace vials were analysed, all measured with a 5 min run time, at 0.9 scans per second. A blank agar sample was analysed with a 5 min run time, before and after each time point.
The headspace of individual cultures, one of E. coli and one of S. aureus, were analysed continuously for 24 h using the vAPCI-MS, to allow for changes over the growth period to be examined. Laboratory air was passed through a 20 ml vial containing deionised water prior to reaching the headspace vial containing the bacteria. This humidified the atmosphere to maintain the integrity of the agar for the entire 24 h experiment. A blank agar sample was analysed with a 5 min run time, before and after each 24 h sample.
The headspace of the plates was analysed by the vAPCI-MS at 2, 6, 24, 48, 72, and 96 h from initial inoculation. The sample was placed into the Petri dish VOC sampler (Fig. 1b), with the temperature kept between 32 and 38 °C. The heated transfer line was attached to the adapter in the lid and left to equilibrate for 1 min before the sample was acquired. All samples were analysed for 5 min, at 2.5 scans per second. After analysis the sample was placed back into the incubator.
Data processing and statistics
The data was binned into windows of 1 m/z width, as done previously.22 All following data analysis was performed using R 3.6.1.29 As the bacterial samples act as permeation sources and were expected to be biologically stable over the discrete sampling period, and no separation was performed prior to the headspace entering the vAPCI-MS, each scan in a single 5 min run was treated as technical replicates. Fig. S1 (ESI†) shows some examples of the total intensity count over the 5 min run time.• ≥50% of the values were non-zero.
• The signal-to-noise ratio was above three.
• The fold-change was above two.
Percentage of non-zero values, signal-to-noise ratio and fold-change have been used previously to filter variables and ensure data quality.22,30–32
Variables that met these criteria for at least one time point for a bacteria sample were selected for that plate. Variables that were selected in at least two thirds of the plates were considered in the further analysis. The variables selected by the 5 min samples in dataset 1 were used to model the difference between the sample types (as described below). The variable selection in the other samples was for comparison.
The multinom function of the nnet R package33 was used to fit the maximum likelihood multinomial log-linear model via neural networks. The neural networks allow for one model to be created with more than two outcomes, instead of concatenating the resulting probabilities of multiple models. This is relevant to multinomial regression, as each class other than the base corresponds to a separate outcome. The result of this approach is a multinomial logistic regression model, not a more complicated neural network. Prior to model creation, the dataset was mean centred and scaled, as recommended by the nnet authors.
The model was created containing only two predictor variables; m/z 118 and m/z 231, chosen as follows. For each bacteria strain, the variable with the highest median signal-to-noise ratio across time points for the dataset 1 5 min samples was selected. Visual examination of intensity plots for dataset 2 revealed the clearest differences between blank agar, E. coli and S. aureus could be detected at 24 h (ESI, Fig. S3†). Therefore, the model was trained on the data taken at 24 h post-inoculation to ensure the model highlighted the differences between the sample types after sufficient growth.
A random selection of 19 samples from dataset 2 (six S. aureus, nine E. coli, and four blank samples) was used as the training set to create the model, and the remaining samples used as the test set. The model was also tested on the blood agar samples.
To evaluate the model, first the performance measure Youden's index was used. When fit, a multiple logistic regression model assigns to each pair of predictor values a vector of class weights. We can obtain a classifier by setting a threshold for each class of a class weight over which the predictor vector is assigned to that class. Youden's index is the threshold for which the sum of sensitivity and specificity of the classifier for that class is maximized. In our setting, we get three Youden's indices, one for each of E. coli, S. aureus, and blank agar. When we consider samples all from the same class, specificity will always equal zero. Hence, only sensitivity (proportion correctly classified) is relevant and is therefore the performance measure used.
Results and discussion
Selected variables
In total, 30 different m/z windows were selected across the sample collection methods for E. coli, with three being selected across all four experiments: m/z 117, 118, and 122. Thirteen different m/z windows were selected for S. aureus, with m/z 231 selected by the three 5 min sample experiments. A table of all the selected variables across all the methods are shown in ESI, Fig. S4a† (E. coli) and S4b (S. aureus).This method successfully selected ions also found in other studies to be markers of E. coli, including indole at m/z 118 (ref. 12, 13, 19, 34 and 35) which was selected across all four methods in this study, along with ions at m/z 117 and 119, and an unidentified ion at m/z 132,19 which was found across both dataset 1 methods. An ion at m/z 109 was selected as a marker for S. aureus in the dataset 1 5 min samples which has been found previously and tentatively identified it as 4-methylphenol.28 This replication of results indicates that the method is valid and capable of measuring VOCs actually present in the samples. Further research could employ MS/MS techniques to investigate the chemical structure and hopefully find identifications for the unknown markers in this study, in particular m/z 231.
Plots over time of some of the selected variables for in dataset 1 are shown in Fig. 2. Many of the selected m/z windows for E. coli show a similar pattern, with the highest measured intensity at 6 h but remaining considerably higher than the blank agar samples for all subsequent measurements. The highest measured intensity for S. aureus was at 48 h for most of the selected m/z windows. However, m/z 231 continued to rise over time, with the final measurement at 72 h recording the highest intensity. The only method that did not select m/z 231 as a potential biomarker of S. aureus was the 24 h continuous sample, which may be due to this delayed peak.
Plots over time of the variables only selected by the 24 h continuous samples are shown in ESI,† Fig. S5a (E. coli) and S5b (S. aureus). For the E. coli samples, there were many lower masses that were detected only in the first two hours. This has been reported in similar previous research, with m/z 59 only detectable for around 100 min.19 This shows the advantage of continuous sampling, which can detect compounds which would otherwise have been missed if only discrete 5 min samples had been utilised. However, as there was only one continuous sample of each strain, further research is required to determine the consistency of the profiles, and the optimum time points for discrete sampling. Additionally, the identification and biological relevance of these biomarkers has yet to be determined but could provide insight into changes in cell metabolism over time.
Classification model
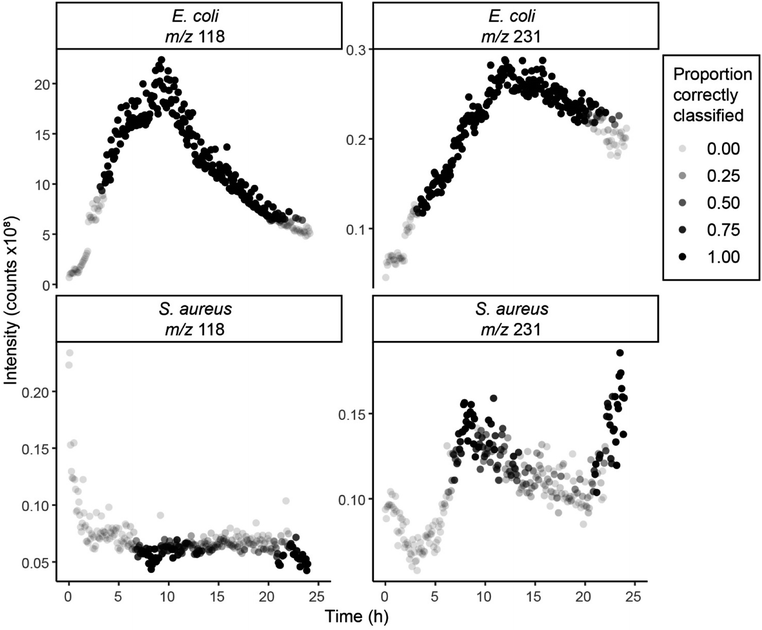
The variables included in the model were m/z 118 and m/z 231, with median signal-to-noise ratios across time points of 44.8 and 10.2, respectively, for the dataset 1 5 min samples. The coefficients for the model are in Table S1 (ESI†).The model had a mean (across times and classes) Youden's index of 0.713 for the test set and 0.897 for the blood agar set. Youden's index for each set separated by class and time is shown in ESI,† Fig. S6. A visualisation of the separation for this model is shown in Fig. 3. The model was especially accurate at identifying the different bacteria species at 24 and 48 h, with Youden's index's ≥0.947 for both test sets, only one false negative across all the sets. Most of the misclassifications were at 2–6 h or after 48 h of growth. This suggests the cause of the misclassification is the low growth of the bacteria, or the start of bacteria death. For the samples grown in blood agar, only E. coli samples analysed at 2 h were misclassified, with all samples at 6 h or after correctly identified. This indicates that this method can successfully identify bacteria after only six hours of growth, much less than the 24 hours often required for other methods.5
The model was also used to predict the class of the 24 h continuous samples. Plots of the intensity over time for m/z 118 and m/z 231 with the proportion of correctly classified resamples indicated are shown in Fig. 4. The model was able to correctly classify the 24 h continuous E. coli sample between 5–20 h, which given the differences in the methods, shows the strength of the model, and the consistency of the analysis methods. The model was less accurate at classifying the S. Aureus 24 h continuous sample; however, this is likely due to a delay in bacterial growth which seems to peak after 24 h, as shown in Fig. 2. All the misclassifications of the 24 h continuous samples were as blank agar, not the other strain of bacteria, suggesting that the model's predictive ability is very high, providing adequate bacterial growth has occurred.
As the headspace of the 24 h continuous samples was constantly being drawn into the instrument, compounds could not gather in the headspace as they would for the discrete samples, which may be why the intensities measured were lower for the continuous samples. The constant movement of air over the sample may also have had an effect on the agar integrity. Additionally, the 5 min samples were stored in an incubator when not being sampled whereas the continuous samples were in the heating block for the whole 24 hours, which may have led to differences in growth patterns. Differences between the methods should be investigated further. An adjustment of the data processing methods may be required to account for the lower intensities measured and ensure relevant changes are identified.
The data processing methods in this paper were developed specifically for the data produced by the vAPCI-MS technique. As there was no separation prior to the sample entering the mass spectrometer and the samples were expected to be biologically stable over the 5 min sampling period (any changes should be too slow to be detected over such a short period), each individual scan over the 5 min sample could be treated as a technical replicate. Fig. S1 (ESI†) shows that the measurements over the 5 min sample were not always stable, which was assumed to be due to instrument drift, transfer line contamination or other sampling/technical error. For dataset 2, the equipment was left to equilibrate for 1 minute before a reading was taken, which appears to reduce the effect of transfer line contamination. Some of the dataset 1 samples, however, have large changes in total intensity counts in the first minute before levelling off.
Repeated random resampling of the ‘technical replicates’ allows for the effects of instrument drift and sampling error to be minimised and artificially increases the sample size, reducing the chance of overfitting a model. This makes significance values irrelevant, as they can be made smaller simply by increasing the number of resamples. However, ensuring that the test sets are completely independent to the data the model is trained on, and judging the performance of the model based on its ability to correctly classify the test sets, ensures that the model is not overfitting and can be applied to other samples. This has been shown to work previously on breath samples analysed using vAPCI-MS.22 In this study, the model performed well across all the test sets, even on data collected by different researchers and/or using different protocols.
Conclusions
Analysing bacterial culture headspace with a vAPCI-MS can be used to detect markers of bacteria and differentiate between different sample types. This system can provide rapid results, is non-destructive so can be used to analyse changes over time, and is smaller than many traditional bench-top mass spectrometers.These methods have potential clinical applications, to identify bacterial infections within a relevant time frame; however, substantially more research is required and, if using the traditional culturing methods, this is unlikely to replace existing diagnostic tools such as MALDI-TOF-MS. Further research will examine whether bacterial infection can be identified and monitored by sampling the headspace of a wound directly, or a breath sample in the case of lung infections, which this system in particular is ideal for, as it can analyse directly in real-time. Additionally, these methods will be used to study antimicrobial resistance, by detecting differences between strains or monitoring changes in response to an antibiotic. This is a relatively new area for MS research, with few studies performed to differentiate resistant and non-resistant strains by headspace VOC profiles.36 Nevertheless, differences have been detected,37 demonstrating the potential, with standardised and robust methods, to develop a clinical diagnostic method for antimicrobial resistance or further the understanding of antimicrobial resistant bacteria.
Author contributions
KR: data curation, formal analysis, investigation, methodology, software, visualization, writing – original draft, review, and editing. EH: conceptualization, formal analysis, methodology, supervision, writing – review and editing. ER: methodology, supervision, writing – review and editing. MRL: supervision, writing – review and editing. JL: investigation. JRH: investigation. MAT: conceptualization, funding acquisition, methodology, resources, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dr Preeti Holland for her help on early experiments, and Dr Rowena Jenkins and Professor Rose Cooper for providing insightful early discussions. The authors would like to thank and acknowledge CAMS-UK for the funding and support to this project.References
- K. L. Tang, N. P. Caffrey, D. B. Nóbrega, S. C. Cork, P. E. Ronksley, H. W. Barkema, A. J. Polachek, H. Ganshorn, N. Sharma, J. D. Kellner and W. A. Ghali, Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis, Lancet Planet. Heal., 2017, 1, e316–e327 CrossRef.
- M. Riera, E. Moreno-Ruiz, S. Goyard, C. d'Enfert and G. Janbon, in Host-Fungus Interactions: Methods and Protocols, ed. A. C. Brand and D. M. MacCallum, Humana Press, Totowa, NJ, 2012, pp. 369–377 Search PubMed.
- A. M. Caliendo, D. N. Gilbert, C. C. Ginocchio, K. E. Hanson, L. May, T. C. Quinn, F. C. Tenover, D. Alland, A. J. Blaschke, R. A. Bonomo, K. C. Carroll, M. J. Ferraro, L. R. Hirschhorn, W. P. Joseph, T. Karchmer, A. T. MacIntyre, L. B. Reller, A. F. Jackson and Infectious Diseases Society of America (IDSA), Better tests, better care: improved diagnostics for infectious diseases, Clin. Infect. Dis., 2013, 57(suppl 3), 139–170 CrossRef PubMed.
- P. E. Fournier, M. Drancourt, P. Colson, J. M. Rolain, B. La Scola and D. Raoult, Modern clinical microbiology: new challenges and solutions, Nat. Rev. Microbiol., 2013, 11, 574–585 CrossRef CAS PubMed.
- Y. Hadano, M. Isoda, K. Ishibashi and T. Kakuma, Validation of blood culture gram staining for the detection of Staphylococcus aureus by the ‘oozing sign’ surrounding clustered gram-positive cocci: a prospective observational study, BMC Infect. Dis., 2018, 18, 4–6 CrossRef PubMed.
- J. D. Bard and F. Lee, Why Can't We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing, Clin. Microbiol. Newsl., 2018, 40, 87–95 CrossRef PubMed.
- L. Simon, E. Ughetto, A. Gaudart, N. Degand, R. Lotte and R. Ruimy, Direct identification of 80 percent of bacteria from blood culture bottles by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a 10-minute extraction protocol, J. Clin. Microbiol., 2019, 57, 1–9 CrossRef PubMed.
- A. Küntzel, P. Oertel, S. Fischer, A. Bergmann, P. Trefz, J. Schubert, W. Miekisch, P. Reinhold and H. Köhler, Comparative analysis of volatile organic compounds for the classification and identification of mycobacterial species, PLoS One, 2018, 13, 1–18 CrossRef PubMed.
- W. L. Wood, D. J. Higbee, M. Gooldy, S. Glogowski, R. Fitzpatrick, R. J. Karalus, T. D. Wood and D. J. Mangino, Analysis of volatile bacterial metabolites by gas chromatography-mass spectrometry, Spectroscopy, 2006, 21, 20–28 CAS.
- W. Filipiak, A. Sponring, M. M. Baur, C. Ager, A. Filipiak, H. Wiesenhofer, M. Nagl, J. Troppmair and A. Amann, Characterization of volatile metabolites taken up by or released from Streptococcus pneumoniae and Haemophilus influenzae by using GC-MS, Microbiology, 2012, 158, 3044–3053 CrossRef CAS PubMed.
- P. Trefz, H. Koehler, K. Klepik, P. Moebius, P. Reinhold, J. K. Schubert and W. Miekisch, Volatile Emissions from Mycobacterium avium subsp. paratuberculosis Mirror Bacterial Growth and Enable Distinction of Different Strains, PLoS One, 2013, 8, 1–10 CrossRef PubMed.
- E. Tait, J. D. Perry, S. P. Stanforth and J. R. Dean, Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS, J. Chromatogr. Sci., 2014, 52, 363–373 CAS.
- F. A. Franchina, G. Purcaro, A. Burklund, M. Beccaria and J. E. Hill, Evaluation of different adsorbent materials for the untargeted and targeted bacterial VOC analysis using GC×GC-MS, Anal. Chim. Acta, 2019, 1066, 146–153 CrossRef CAS PubMed.
- Y. Jin, D. Li, M. Ai, Q. Tang, J. Huang, X. Ding, C. Wu and R. Zhou, Correlation between volatile profiles and microbial communities: a metabonomic approach to study Jiang-flavor liquor Daqu, Food Res. Int., 2019, 121, 422–432 CrossRef CAS PubMed.
- H. Antolak, H. Jeleń, A. Otlewska and D. Kręgiel, Volatile compounds associated with growth of Asaia bogorensis and Asaia lannensis-unusual spoilage bacteria of functional beverages, Food Res. Int., 2019, 121, 379–386 CrossRef CAS PubMed.
- S. Ramírez-Guízar, H. Sykes, J. D. Perry, E. C. Schwalbe, S. P. Stanforth, M. C. I. Perez-Perez and J. R. Dean, A chromatographic approach to distinguish Gram-positive from Gram-negative bacteria using exogenous volatile organic compound metabolites, J. Chromatogr. A, 2017, 1501, 79–88 CrossRef PubMed.
- P. Perez-Hurtado, E. Palmer, T. Owen, C. Aldcroft, M. H. Allen, J. Jones, C. S. Creaser, M. R. Lindley, M. A. Turner and J. C. Reynolds, Direct analysis of volatile organic compounds in foods by headspace extraction atmospheric pressure chemical ionisation mass spectrometry, Rapid Commun. Mass Spectrom., 2017, 31, 1947–1956 CrossRef CAS PubMed.
- V. Shestivska, A. Nemec, P. Dřevínek, K. Sovová, K. Dryahina and P. Spaněl, Quantification of methyl thiocyanate in the headspace of Pseudomonas aeruginosa cultures and in the breath of cystic fibrosis patients by selected ion flow tube mass spectrometry, Rapid Commun. Mass Spectrom., 2011, 25, 2459–2467 CrossRef CAS PubMed.
- M. Bunge, N. Araghipour, T. Mikoviny, J. Dunkl, R. Schnitzhofer, A. Hansel, F. Schinner, A. Wisthaler, R. Margesin and T. D. Märk, On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry, Appl. Environ. Microbiol., 2008, 74, 2179–2186 CrossRef CAS PubMed.
- M. O'Hara and C. A. Mayhew, A preliminary comparison of volatile organic compounds in the headspace of cultures of Staphylococcus aureus grown in nutrient, dextrose and brain heart bovine broths measured using a proton transfer reaction mass spectrometer, J. Breath Res., 2009, 3, 027001 CrossRef PubMed.
- L. M. Heaney, D. M. Ruszkiewicz, K. L. Arthur, A. Hadjithekli, C. Aldcroft, M. R. Lindley, C. P. Thomas, M. A. Turner and J. C. Reynolds, Real-time monitoring of exhaled volatiles using atmospheric pressure chemical ionization on a compact mass spectrometer, Bioanalysis, 2016, 8, 1325–1336 CrossRef CAS PubMed.
- K. Rosenthal, D. M. Ruszkiewicz, H. Allen, M. R. Lindley, M. A. Turner and E. Hunsicker, Breath selection methods for compact mass spectrometry breath analysis, J. Breath Res., 2019, 13, 046013 CrossRef CAS PubMed.
- S. Rankin-Turner, M. A. Turner, P. F. Kelly, R. S. P. King and J. C. Reynolds, Transforming presumptive forensic testing: in situ identification and age estimation of human bodily fluids, Chem. Sci., 2019, 10, 1064–1069 RSC.
- J. Chong, O. Soufan, C. Li, I. Caraus, S. Li, G. Bourque, D. S. Wishart and J. Xia, MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis, Nucleic Acids Res., 2018, 46, W486–W494 CrossRef CAS PubMed.
- J. P. C. Pereira, P. J. T. Verheijen and A. J. J. Straathof, Growth inhibition of S. cerevisiae, B. subtilis, and E. coli by lignocellulosic and fermentation products, Appl. Microbiol. Biotechnol., 2016, 100, 9069–9080 CrossRef CAS PubMed.
- J. M. Scotter, R. A. Allardyce, V. S. Langford, A. Hill and D. R. Murdoch, The rapid evaluation of bacterial growth in blood cultures by selected ion flow tube-mass spectrometry (SIFT-MS) and comparison with the BacT/ALERT automated blood culture system, J. Microbiol. Methods, 2006, 65, 628–631 CrossRef CAS PubMed.
- R. A. Allardyce, V. S. Langford, A. L. Hill and D. R. Murdoch, Detection of volatile metabolites produced by bacterial growth in blood culture media by selected ion flow tube mass spectrometry (SIFT-MS), J. Microbiol. Methods, 2006, 65, 361–365 CrossRef CAS PubMed.
- J. Zhu, H. D. Bean, Y. M. Kuo and J. E. Hill, Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry, J. Clin. Microbiol., 2010, 48, 4426–4431 CrossRef CAS PubMed.
- R Core Team, and R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2019, https://www.R-project.org/ Search PubMed.
- J. A. Kirwan, R. J. M. Weber, D. I. Broadhurst and M. R. Viant, Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control, Sci. Data, 2014, 1, 1–13 Search PubMed.
- E. Zelena, W. B. Dunn, D. Broadhurst, S. Francis-McIntyre, K. M. Carroll, P. Begley, S. O'Hagan, J. D. Knowles, A. Halsall, I. D. Wilson and D. B. Kell, Development of a robust and repeatable UPLC – MS method for the long-term metabolomic study of human serum, Anal. Chem., 2009, 81, 1357–1364 CrossRef CAS PubMed.
- W. B. Dunn, D. Broadhurst, P. Begley, E. Zelena, S. Francis-Mcintyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall, J. N. Haselden, A. W. Nicholls, I. D. Wilson, D. B. Kell and R. Goodacre, Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry, Nat. Protoc., 2011, 6, 1060–1083 CrossRef CAS PubMed.
- B. D. Ripley and W. N. Venables, Package ‘ nnet ’, https://cran.r-project.org/web/packages/nnet/nnet.pdf, accessed 12 October 2021 Search PubMed.
- L. D. J. Bos, P. J. Sterk and M. J. Schultz, Volatile Metabolites of Pathogens: a Systematic Review, PLoS Pathog., 2013, 9, 1–8 Search PubMed.
- Q. Zhong, F. Cheng, J. Liang, X. Wang, Y. Chen, X. Fang, L. Hu and Y. Hang, Profiles of volatile indole emitted by Escherichia coli based on CDI-MS, Sci. Rep., 2019, 9, 1–6 Search PubMed.
- N. Drabińska, B. de Lacy Costello, K. Hewett, A. Smart and N. Ratcliffe, From fast identification to resistance testing: volatile compound profiling as a novel diagnostic tool for detection of antibiotic susceptibility, TrAC, Trends Anal. Chem., 2019, 115, 1–12 CrossRef.
- H. Li and J. Zhu, Differentiating Antibiotic-Resistant Staphylococcus aureus Using Secondary Electrospray Ionization Tandem Mass Spectrometry, Anal. Chem., 2018, 90, 12108–12115 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ay01555a |
| This journal is © The Royal Society of Chemistry 2021 |