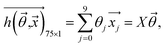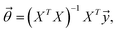Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A surface-based calibration approach to enable dynamic and accurate quantification of colorimetric assay systems†
Lin
Tong
 * and
Joshua D.
Hutcheson
* and
Joshua D.
Hutcheson

Department of Biomedical Engineering, Florida International University, 10555 W Flagler St, EC 2612, Miami, FL 33174, USA. E-mail: jhutches@fiu.edu
First published on 2nd September 2021
Abstract
Colorimetry is widely used in assay systems for its low-cost, ease-of-use, rapidity, moderate storage requirements and intuitively visible effects. However, the application is limited due to its relatively low sensitivity. Conventional colorimetric calibration methods often use a fixed incubation time that can limit the detection range, system robustness and sensitivity. In this paper, we used color saturation to measure the accumulation of product (correlation coefficient R2 = 0.9872), and we created a novel “calibration mesh” method based on an expanded sigmoid function to enhance sensitivity. The novel calibration mesh method can be adapted for a wide variety of assay systems to improve robustness and detection range, and provide a dynamic and faster output.
1. Introduction
Colorimetric analyses have been widely used in various types of assays, including tests for pregnancy,1 bacteria,2 toxicity,3 pH,4 soil,5,6 industry waste water7 and many common biomarkers in blood8 and sweat.9,10 The development of low-cost point-of-care devices, such as those that use simple paper pads,17 could help address health disparity issues related to inadequate access to care.16 Colorimetry offers a low-cost, accessible measurement technique that does not require complex imaging modalities or burdensome data storage. However, compared to fluorescence or other modern detection methods, the biggest disadvantage of colorimetry is low sensitivity and high noise. Most colorimetric data analyses are based on light absorption,11 gray-scale intensity,12 red, green, blue (RGB) color space measurements,8,9 or analyses based on the hue channel of the hue, saturation, value (HSV) color space.13 Analyses based on the saturation channel of HSV color space can outperform other channels when a colored product accumulates in an assay14 and is less dependent on ambient light conditions.15A linear or non-linear fit of measured values (e.g., intensity, absorption, impedance) to known concentrations is often used to establish a conventional calibration curve in an assay system. Readings based on this curve often require a fixed incubation time, which limits both the detection time and the detection range. Low concentration samples may require more incubation time to be distinguished from blank samples; high concentration samples may exceed the upper limit of detection of the reader after a fixed incubation time. Also, using a fixed incubation time provides only a single readout per sample. Technical replicates are often utilized to overcome single measurement inaccuracies, but this requires additional sample and reagents.
In this study, we developed a colorimetric paper pad sensing unit and analysis technique to quantify the concentration of the enzyme invertase. Our technique utilizes an innovative “calibration mesh” method that uses a temporal analysis of colorimetric saturation to replace the conventional fixed-time calibration method. To demonstrate the robustness of this method, we compared the performance of the “calibration mesh” to the conventional calibration approach.
2. Experimental methods
(a) Reagents and solutions
Sucrose was purchased from VWR. Glucose oxidase (G2133-50KU, 242![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 776 U g−1), peroxidase from horseradish (P8125-25KU, 50–150 U mg−1), and invertase (I4504-250MG, ≥300 U mg−1) were purchased from Sigma. D-Trehalose, 99% anhydrous was from Acros Organics (NJ). Sodium acetate was purchased from Fisher Scientific. Phosphate buffered saline (28
776 U g−1), peroxidase from horseradish (P8125-25KU, 50–150 U mg−1), and invertase (I4504-250MG, ≥300 U mg−1) were purchased from Sigma. D-Trehalose, 99% anhydrous was from Acros Organics (NJ). Sodium acetate was purchased from Fisher Scientific. Phosphate buffered saline (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372) was purchased from Thermo Scientific. Sodium iodide (409286-1G) was purchased from Aldrich. Acetic acid was purchased from Sigma-Aldrich. Cellulose fiber sample pad strips (CFSP203000) were purchased from Millipore. Deionized water (18.0 MΩ) was used in all procedures. Details of buffer preparation are available in ESI.†
372) was purchased from Thermo Scientific. Sodium iodide (409286-1G) was purchased from Aldrich. Acetic acid was purchased from Sigma-Aldrich. Cellulose fiber sample pad strips (CFSP203000) were purchased from Millipore. Deionized water (18.0 MΩ) was used in all procedures. Details of buffer preparation are available in ESI.†
(b) Sensing paper pad procedure
Paper pad fabrication. The cellulose fiber paper was cut into round paper pads using a quarter size hole puncher (Walmart), and the edge was trimmed to remove burrs. The pad was then sealed in a plastic storage bag for future use. | (1) |
 | (2) |
 | (3) |
(c) Experiment and camera setup
The whole experiment was carried out under a stable and uniform white illumination area. All steps were as follows:Step 1: place 24 substrate-treated paper pads on a cleaned pipette tip holder as described above (8 rows and 3 columns). Fix the camera (details in ESI†) 20 cm above the paper pads.
Step 2: after focusing the image, turn off all camera automatic adjustment and balance functions, and acquire images at 5, 20, 30, 40, and 72 min.
Step 3: use an 8-channel pipette to add 100 μL invertase solution to the center of each paper pad in the first column avoiding touching or moving the paper pads.
Step 4: repeat step 3 for the second and third columns. The time interval between each addition is 20 s.
(d) Image processing
After capturing the images of paper pads, we converted the captured image to a gray scale image, an R, G, B color space image, and an H, S, V color space image (Fig. 1). | ||
| Fig. 1 Response image captured at 30 min. From left to right are raw image, gray image, red channel, green channel, blue channel, hue channel, saturation channel and value channel. | ||
To calculate the average saturation value of HSV color space for each paper pad, the pad area was selected using a circular mask, and the saturation values of regions outside of the paper pads were set to 0. The average saturation value of all pixels in a paper pad was calculated as follows:
(e) Correlation coefficients calculation
Similar to the saturation channel processing described above, we calculated both raw data and data with the background subtracted for the gray image, and all channels of R, G, B, H, and V. We then calculated the correlation coefficient between the target concentration and magnitude of each of these channels (Table 1).| R squared value | Grey | Red | Green | Blue | Hue | Saturation | Value |
|---|---|---|---|---|---|---|---|
| Average value of raw image | 0.9004 | 0.7110 | 0.9166 | 0.9588 | 0.4258 | 0.9790 | 0.7111 |
| Average value – background | 0.6414 | 0.2836 | 0.6354 | 0.9628 | 0.5953 | 0.9872 | 0.2873 |
(f) Calibration method
The data obtained from the standard concentrations of 0.1, 0.5, 1, 2.5, and 10 μg mL−1 were used as training dataset to establish conventional calibration curve or calibration mesh. The dataset obtained from the standard concentrations of 0.25 and 5 μg mL−1 were used as testing data to evaluate the performance of the algorithm and different methods. and the concentration
and the concentration  data were set as inputs. The saturation data were set as outputs
data were set as inputs. The saturation data were set as outputs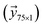 . Two different models were proposed for establishing the calibration mesh:
. Two different models were proposed for establishing the calibration mesh:
2.1. Linear regression model. In this model, we generated the prediction function as a polynomial function:
The t and c here were normalized by dividing each time and concentration by the maximum values of time and concentration, respectively.
First, we defined an input matrix:
 to
to  is a 75 × 1 vector
is a 75 × 1 vector
Then, the prediction function can be rewritten as:
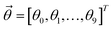 .
.
Next, we defined the cost function as:
The goal was to solve the coefficient  to make the predicted output saturation value
to make the predicted output saturation value 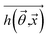 as close as possible to the training saturation values
as close as possible to the training saturation values  by finding the minimum of
by finding the minimum of  Since the dataset was not large, we used a normal equation approach to solve for theta:
Since the dataset was not large, we used a normal equation approach to solve for theta:
A regularization was applied to prevent over fitting. Then,
D is a 10 by 10 matrix, which is similar to the identity matrix except that the first element is 0.
2.2. Sigmoid model. Plotting the saturation value vs. concentration and time (Fig. S4, S5† and 2) revealed a sigmoidal curve. Therefore, we expanded the basic sigmoid model
 as follows:
as follows:where
 is a 7 × 1 vector of [k(1), k(2), …, k(7)]
is a 7 × 1 vector of [k(1), k(2), …, k(7)]
The  function retains a sigmoid shape but is shifted and stretched by the coefficients
function retains a sigmoid shape but is shifted and stretched by the coefficients . To extend the sigmoidal function (s) to a surface, we define z(t,c) to capture the relationship between time and concentration.
. To extend the sigmoidal function (s) to a surface, we define z(t,c) to capture the relationship between time and concentration.
Next, we defined the cost function:
The training goal was to find an optimal solution of  to minimize the cost of
to minimize the cost of  . Since the training dataset was discrete, we used the fminsearch function within Matlab.
. Since the training dataset was discrete, we used the fminsearch function within Matlab.
For both the linear regression model and the sigmoid model, once the optimal solution for  or
or  was obtained, we could build the calibration mesh within the given range of time (0–74 min) and concentration (0–10 μg mL−1).
was obtained, we could build the calibration mesh within the given range of time (0–74 min) and concentration (0–10 μg mL−1).
(g) Performance evaluation
 within the respective cost function, we compared the total errors or difference between the predicted values and the output values (Table 2).
within the respective cost function, we compared the total errors or difference between the predicted values and the output values (Table 2).
 | ||
| Fig. 3 Calibration mesh with average of training data set. Linear regression model (A) vs. sigmoid model (B). The yellow dots were the average training data set. | ||
| Calibration Mesh Model | Linear Regression | Sigmoid |
|---|---|---|
| Error or cost | 1.7067 × 10−4 | 4.7337 × 10−5 |
For the calibration mesh, we used the saturation data obtained from 0.25 and 5 μg mL−1 concentrations measured at 20 min, 30 min 40 min and 72 min in the calibration mesh model and used our algorithm presented in Fig. 4 for mapping the predicted concentration.
Absolute relative difference (ARD) for each of these methods was then calculated as:
3. Results and discussion
(a) Saturation channel performance
Rather than working in the orthogonal RGB space, which depends on linear combinations of red, green, and blue values to describe a color, saturation data obtained in the HSV space does not depend on the color produced in a reaction (Fig. S7†). The accumulation of any color product yields a saturation value that can be directly related to concentration.By observing the histogram and normal distribution of pixel saturation (Fig. S1 to S3†) and the comparison between the raw image and the data channels shown in Fig. 1, we found that the average saturation value of all pixels from a paper pad represents the color change of the paper pad well.
The correlation coefficient values in Table 1 show that the saturation channel had the best correlation with concentration among all channels. Also, the saturation data with background subtracted gave a higher correlation coefficient than the raw data. Therefore, we selected the average saturation value of the raw image minus background as our training or testing saturation data in this study.
(b) Calibration curve and calibration mesh development
A conventional calibration curve (Fig. 5) was established using a linear fit based on the 30 minutes training dataset. These data yielded the following fit:| y = 0.0159x + 0.0075 |
Next, we developed the first calibration mesh model using a linear regression technique. After training, the optimal solution of  was [−0.0017, –0.0986, −0.0442, 0.3161, 0.8425, 0.0059, −0.1326, −0.1963, −0.3172, 0.0067]. After plotting the mesh (Fig. 3A), we observed that the average training dataset (“yellow dots”) did not fit the calibration mesh well.
was [−0.0017, –0.0986, −0.0442, 0.3161, 0.8425, 0.0059, −0.1326, −0.1963, −0.3172, 0.0067]. After plotting the mesh (Fig. 3A), we observed that the average training dataset (“yellow dots”) did not fit the calibration mesh well.
From the contour plot of average saturation raw data vs. time and concentration (Fig. 2), we observed that when both time and concentration are small (left bottom corner) or large (top right corner), the distance between contour lines are relatively large, while the density of contour lines in the middle is higher. This indicates that the slope in the middle is higher than the lower left or the upper right regions of the plot. Based on the slopes observed in the contour plot data, we hypothesized that a sigmoidal function may provide a more appropriate fit for our calibration mesh.
After training the sigmoid model, the optimal solution of  was [0.5294 1.4278 1.2644, 5.0479, −0.6219, 0.7222, −0.0043]. The sigmoid model calibration mesh fit the average training saturation data well (Fig. 3B, “yellow dots”). As shown in Table 2, the error of the sigmoid model (4.7337 × 10−5) was smaller than the error obtained using the linear regression model (1.7067 × 10−4). Therefore, we concluded that the sigmoid model outperformed the linear regression model in this study.
was [0.5294 1.4278 1.2644, 5.0479, −0.6219, 0.7222, −0.0043]. The sigmoid model calibration mesh fit the average training saturation data well (Fig. 3B, “yellow dots”). As shown in Table 2, the error of the sigmoid model (4.7337 × 10−5) was smaller than the error obtained using the linear regression model (1.7067 × 10−4). Therefore, we concluded that the sigmoid model outperformed the linear regression model in this study.
The linear regression model, however, has some advantages that should be considered. First, as a common method, the linear regression model could be used for general situations, especially when the detection range of interest demonstrates a linear relationship between saturation and both concentration and time. Second, the linear regression model does not require optimization to account for variations in mesh shape. Third, training by solving the normal equation is more cost-effective than “fminsearch”. In this study, the training time (averaged over 10 runs) for the linear regression model was 0.0002 s, while the average training time of the sigmoid model was 0.0323 s. Therefore, the linear regression model may be more appropriate for platforms in which computational effort is a major concern.
(c) Calibration mesh utilization
For the conventional calibration method, we can map a saturation value at a fixed time (e.g., 30 min) on the calibration curve to determine the associated concentration. Similarly, for the “calibration mesh”, we can map a saturation value (s) at a time (t) on the “calibration mesh” and find the associated concentration (c). The added benefit of the calibration mesh method is the ability to capture images at multiple time points and increase the accuracy of the concentration measurement (Fig. 4).(d) Performance of the conventional calibration curve vs. calibration mesh models
As shown in Table 3, although the conventional calibration curve method did well in identifying concentration for 5 μg mL−1 (ARD = 0.48%), it was unable to accurately identify the 0.25 μg mL−1 condition (ARD = 410.22%). The calibration mesh using the sigmoid model accurately calculated both concentrations (ARDs were 0.67% and 0%, respectively).| Model | Conventional calibration method | Calibration mesh via linear regression model | Calibration mesh via sigmoid model | |
|---|---|---|---|---|
| For concentration | Prediction | 0.0490 μg mL−1 | 0.2167 μg mL−1 | 0.25 μg mL−1 |
| 0.25 μg mL−1 testing | ARD | 410.22% | 13.32% | 0% |
| For concentration | Prediction | 4.9761 μg mL−1 | 5.6667 μg mL−1 | 4.9667 μg mL−1 |
| 5 μg mL−1 testing | ARD | 0.48% | 11.77% | 0.67% |
(e) Merits of calibration mesh
In this study, the conventional calibration curve was constructed based on a fixed time (30 min). However, from the calibration mesh (Fig. 3), we can see that a higher concentration sample has a faster chemical reaction and a rapid increase in saturation value, which requires a shorter time to be distinguished from the blank. On the other hand, as shown in the results presented above, the conventional calibration curve may be inaccurate for low concentration prediction. The calibration mesh can overcome this limitation by extending the reaction time so that the lower concentration samples would eventually exhibit and increase in saturation to improve the signal-to-noise ratio. Using this method, an investigator does not need to determine the incubation time a priori. Further, if multiple samples with large concentration differences are being assayed simultaneously (or perhaps several biomarkers with drastically different concentrations in a multiplex assay), the calibration mesh technique can provide reliable quantification without the need to perform separate incubation times for each sample or biomarker. The designed algorithm for utilizing the calibration mesh can give a robust and accurate output by setting conversion criteria for the predicted concentration over time. The calibration mesh can expand the detection range and provide an earlier output.4. Conclusions
We have demonstrated that color saturation has a higher correlation to accumulated product in a paper pad assay system. We have also shown that the calibration mesh method has an improved dynamic range and accuracy compared to a conventional calibration curve. The utilize of calibration mesh is simple and can be adapted for a wide variety of assay systems.There are several things we can do to improve the performance of the calibration mesh method in future iterations. First, increasing the standard concentrations and automating image capture with higher frequency could provide better model inputs and achieve higher resolution calibration mesh in both concentration and temporal axis. Second, instead of using saturation only, we could explore using a combination of gray, R, G, B, hue, saturation, and value data to obtain a combined output, which may further enhance the correlation between the imaged data and the concentration. Third, since temperature changes can affect chemical reactions, we can also integrate temperature as a variable in the training model. Fourth, for the calibration mesh utilization algorithm, we can also introduce a factor that changes the weight of a single measurement based on the ARD at particular time points in the assay. Fifth, we can utilize the slope of calibration mesh to eliminate potential background error. Utilization and iterative improvements of the calibration mesh technique could yield a robust algorithm that will enhance the accuracy and sensitivity of colorimetric assays.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the NSF Engineering Research Center, Precise Advanced Technologies and Health Systems for Underserved Populations (PATHS-UP) – award no. 1648451. The authors also acknowledge helpful discussions with Daniel Chaparro and other members of the Cardiovascular Matrix Remodeling Laboratory.References
- S. L. Cohen, A Method for the Rapid Colorimetric Assay of Total Estrins in Pregnancy Urine1, J. Clin. Endocrinol. Metab., 1966, 26(9), 994–1004, DOI:10.1210/jcem-26-9-994.
- K. Methee, X. Zhu and E. Almeide, et al., Paper based point-of-care testing disc for multiplex whole cell bacteria analysis, Biosens. Bioelectron., 2011, 26(11), 4342–4348, DOI:10.1016/j.bios.2011.04.035.
- X. Zhu, E. Hondroulis, W. Liu and C. Li, Biosensing Approaches for Rapid Genotoxicity and Cytotoxicity Assays upon Nanomaterial Exposure, Small, 2013, 9(9–10), 1821–1830, DOI:10.1002/smll.201201593.
- A. R. C. Haas, Colorimetric Determination of the Hydrogen Ion Concentration in Small Quantities of Solution, J. Biol. Chem., 1919, 38(1), 49–55, DOI:10.1016/s0021-9258(18)87373-2.
- S. R. Dickman and R. H. Bray, Colorimetric Determination of Phosphate, Ind. Eng. Chem., Anal. Ed., 1940, 12(11), 665–668, DOI:10.1021/ac50151a013.
- W. E. Baethgen and M. M. Alley, A manual colorimetric procedure for measuring ammonium nitrogen in soil and plant Kjeldahl digests, Commun. Soil Sci. Plant Anal., 1989, 20(9–10), 961–969, DOI:10.1080/00103628909368129.
- K. Shrivas, S. Sahu, G. K. Patra, N. K. Jaiswal and R. Shankar, Localized surface plasmon resonance of silver nanoparticles for sensitive colorimetric detection of chromium in surface water, industrial waste water and vegetable samples, Anal. Methods, 2016, 8(9), 2088–2096, 10.1039/c5ay03120f.
- C. M. McGeough and S. O'Driscoll, Camera Phone-Based Quantitative Analysis of C-Reactive Protein ELISA, IEEE Trans. Biomed. Circuits Syst., 2013, 7(5), 655–659, DOI:10.1109/TBCAS.2012.2234122.
- A. Koh, D. Kang and Y. Xue, et al., A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat, Sci. Transl. Med., 2016, 8(366), 366ra165, DOI:10.1126/scitranslmed.aaf2593.
- R. Ghaffari, J. A. Rogers and T. R. Ray, Recent progress, challenges, and opportunities for wearable biochemical sensors for sweat analysis, Sens. Actuators, B, 2021, 332(October 2020), 129447, DOI:10.1016/j.snb.2021.129447.
- J. Riemer, H. H. Hoepken, H. Czerwinska, S. R. Robinson and R. Dringen, Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells, Anal. Biochem., 2004, 331(2), 370–375, DOI:10.1016/j.ab.2004.03.049.
- J. C. Jokerst, J. A. Adkins, B. Bisha, M. M. Mentele, L. D. Goodridge and C. S. Henry, Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens, Anal. Chem., 2012, 84(6), 2900–2907, DOI:10.1021/ac203466y.
- S. T. Krauss, A. Q. Nauman, G. T. Garner and J. P. Landers, Color manipulation through microchip tinting for colorimetric detection using hue image analysis, Lab Chip, 2017, 17(23), 4089–4096, 10.1039/C7LC00796E.
- K. Su, Q. Zou and J. Zhou, et al., High-sensitive and high-efficient biochemical analysis method using a bionic electronic eye in combination with a smartphone-based colorimetric reader system, Sens. Actuators, B, 2015, 216, 134–140, DOI:10.1016/j.snb.2015.04.052.
- B. Coleman, C. Coarsey and W. Asghar, Cell phone based colorimetric analysis for point-of-care settings, Analyst, 2019, 144(6), 1935–1947, 10.1039/c8an02521e.
- K. R. King, L. P. Grazette and D. N. Paltoo, et al., Point-of-Care Technologies for Precision Cardiovascular Care and Clinical Research: National Heart, Lung, and Blood Institute Working Group, J. Am. Coll. Cardiol. Basic Trans. Science, 2016, 1(1–2), 73–86, DOI:10.1016/j.jacbts.2016.01.008.
- Z. S. Ballard, H. A. Joung and A. Goncharov, et al., Deep learning-enabled point-of-care sensing using multiplexed paper-based sensors, NPJ Digit. Med., 2020, 3(1), 1–8, DOI:10.1038/s41746-020-0274-y.
- AAT Bioquest I. Acetate Buffer (pH 3.6 to 5.6) Preparation. https://www.aatbio.com/resources/buffer-preparations-and-recipes/acetate-buffer-ph-3-6-to-5-6.
- E. Roemer, M. K. Schorp, J. J. Piadé, J. I. Seeman, D. E. Leyden and H. J. Haussmann, Scientific assessment of the use of sugars as cigarette tobacco ingredients: A review of published and other publicly available studies, Crit. Rev. Toxicol., 2012, 42(3), 244–278, DOI:10.3109/10408444.2011.650789.
- E. P. Golikova, N. V. Lakina, O. V. Grebennikova, V. G. Matveeva and E. M. Sulman, A study of biocatalysts based on glucose oxidase, Faraday Discuss., 2017, 202, 303–314, 10.1039/C7FD00042A.
- E. L. Thomas and T. M. Aune, Cofactor Role of Iodide in Peroxidase Antimicrobial Action Against Escherichia coli, Antimicrob. Agents Chemother., 1978, 13(6), 1000–1005, DOI:10.1128/AAC.13.6.1000.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ay01130h |
| This journal is © The Royal Society of Chemistry 2021 |