Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular diagnostics in the era of COVID-19
Harindi
Jayakody
 ab,
Guy
Kiddle
b,
Semali
Perera
a,
Laurence
Tisi
*b and
Hannah S.
Leese
ab,
Guy
Kiddle
b,
Semali
Perera
a,
Laurence
Tisi
*b and
Hannah S.
Leese
 *a
*a
aMaterials for Health Lab, Department of Chemical Engineering, University of Bath, Bath, UK. E-mail: h.s.leese@bath.ac.uk
bErba Molecular, Ely, Cambridgeshire, UK. E-mail: l.tisi@erbamannheim.com
First published on 5th August 2021
Abstract
As the COVID-19 pandemic continues to escalate globally and acquires new mutations, accurate diagnostic technologies continue to play a vital role in controlling and understanding the epidemiology of this disease. A plethora of technologies have enabled the diagnosis of individuals, informed clinical management, aided population-wide screening to determine transmission rates and identified cases within the wider community and high-risk settings. This review explores the application of molecular diagnostics technologies in controlling the spread of COVID-19, and the key factors that affect the sensitivity and specificity of the tests used.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the virus responsible for causing the COVID-19 pandemic that was first identified in Wuhan, China in 2019.1 According to data published by the John Hopkins Coronavirus Research Centre, there have been over 190 million cases globally and over 4 million deaths, as of July 2021.2SARS-CoV-2 is a positive sense RNA virus made of four main structural proteins and other accessory proteins.3 The spike protein binds to the angiotensin-converting enzyme 2 (ACE2) receptor found on human host cells, enabling entry.4,5 COVID-19 most commonly spreads via direct exposure to respiratory secretions of an infected person, but it can also be spread by airborne transmission, through contact with contaminated surfaces and via the faecal-oral route.6–13 Infected individuals may present mild to severe symptoms of the infection, such as fever or chills, a persistent cough, shortness of breath, and headaches amongst others, and these are typically displayed 2–14 days following initial exposure.6,14
Testing for SARS-CoV-2 aids for a timely diagnosis and helps to control transmission within different settings, thereby preventing the re-introduction of SARS-CoV-2 into settings where the virus is under control (i.e. testing visitors travelling into a given jurisdiction).15 In addition to tests performed on individuals, environment monitoring for contamination through surface swabbing, wastewater testing, and air sampling have all proven advantageous in managing the spread of SARS-CoV-2.13–19
Sensitive molecular tests are essential to ensure that individuals infected with SARS-CoV-2 are accurately identified and quarantined quickly. Whilst molecular tests, in particular reverse transcription polymerase chain reaction (RT-PCR), are widely used due to their high sensitivity, many factors can impede the accuracy of these tests. This review explores the use of molecular tests in different contexts and discusses the technologies that have been widely deployed, exploring the key factors affecting a tests sensitivity and specificity.
2. Diagnosis
Two main types of testing are used to aid the diagnosis of COVID-19 during an active infection; molecular tests that detect the presence of the RNA genome, and antigen tests that detect the presence of viral antigens, such as the viral protein coating.20 Additionally, serological tests (antibody tests) that target the immune response of an individual post-infection, have also been deployed during this pandemic.21 As COVID-19 is still a relatively new disease, clinicians have monitored host antibody levels to determine how sustained the natural immune response is, post-infection. Such investigations have revealed that most COVID-19 patients develop antibodies against SARS-CoV-2, one to three weeks following symptom onset, with high levels of neutralising antibodies detected in severe cases.22–24 The World Health Organization (WHO) does not recommend the use of antibody tests, in isolation, for the diagnosis or clinical management of acute infection.25,26Molecular testing is conducted to identify COVID-19 positive individuals who are: symptomatic and develop mild (paucisymptomatic) to severe disease; asymptomatic (may not express symptoms); and presymptomatic (infected individuals that may develop symptoms after a positive test result). The WHO recommends a testing cascade for the clinical management of patients suspected of COVID-19 infection (Fig. 1).
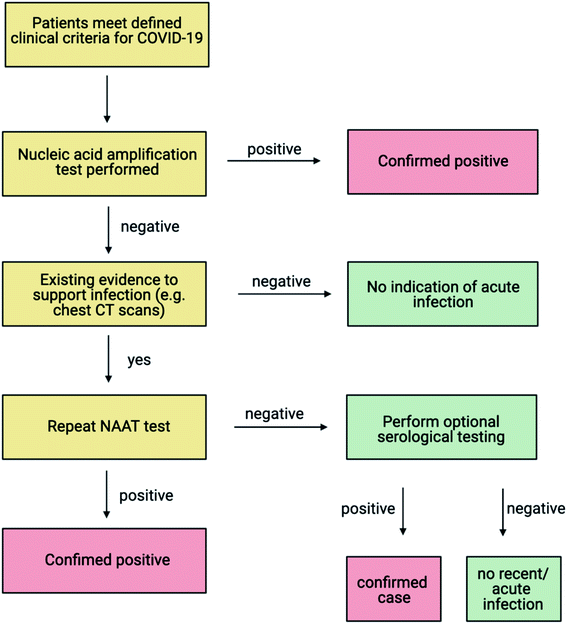 | ||
| Fig. 1 WHO testing cascade for diagnosis of COVID-19 (adapted from WHO, 2020,26 created with BioRender.com). If a patient meets the clinical criteria for COVID-19 as specified by the WHO, a NAAT (Nucleic Acid Amplification Test) test is recommended. Antigen testing can be used alongside the NAAT depending on the prevalence of disease and the sensitivity and specificity of the antigen test. Following a positive result by the NAAT, the patient is classified as a positive case. Confirmation of weak positive results either through repeat NAAT testing or in some cases, viral sequencing is recommended. Negative results do not rule out COVID-19 infection and in the event of sustained clinical suspicion due to an epidemiological link and other clinical findings such as radiological signs (chest CT scans), repeat testing is recommended. A positive result on repeat testing is identified as a confirmed case. A negative result upon retesting can be further evaluated using one serum sample collected in the acute phase and one sample collected in the convalescent phase, to explore seroconversion or a rise in antibodies.26 | ||
The management of the COVID-19 disease burden has relied upon molecular testing within specialised laboratories and the rapid transportation of these samples from collection points within communities. Nasopharyngeal (NP) swabs are by far the most routinely collected samples, although other sample types have been used. The Centers for Disease Control & Prevention (CDC) specifies the range of upper respiratory tract (URT) and lower respiratory tract (LRT) specimens that are compatible with a given commercial test kit.27 Since prolonged detection of SARS-CoV-2 RNA has been reported in faecal samples, the testing of this sample type during the later stages of this disease has been proposed by the WHO.28 Irrespective of the sample type, laden swabs are placed within viral transport medium (VTM) for storage during transportation, followed by nucleic acid extraction using commercially available kits, and then amplification and detection of RNA in the case of molecular testing.27,29 There are currently over 240 molecular amplification chemistries that have been approved for emergency use (EUA) by the U.S. Food & Drug Administration (FDA).30 Most approved assays are based on reverse transcription polymerase chain reactions (RT-PCR).
2.1 Polymerase chain reaction
All RT-PCR amplifications are initiated by a reverse transcriptase, which is an enzyme that typically converts two or more targeted regions of the SARS-CoV-2 viral RNA into complementary DNA (cDNA); this component of the reaction is performed at a constant temperature for a few minutes prior to the PCR reaction. The resultant cDNA is then exponentially amplified via a series of thermal cycles (Ct; normally 35–40), in a reaction that has the potential to double the targeted DNA load within each consecutive cycle. The increasing amount of amplified product generated during the thermal cycling is typically detected by exciting fluorophores that are coupled to target specific nucleotide sequences, which become unquenched during the amplification process.31 The quantity of initial SARS-CoV-2 RNA is inversely proportional to the Ct in which fluorescence is detected (Fig. 2). A typical RT-PCR assay will use a maximum of 40 Ct's and when fluorescence is detected above a given signal intensity, this would be regarded as a positive result.32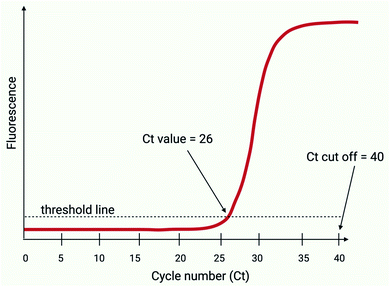 | ||
| Fig. 2 Ct thresholds (adapted from Public Health England, 2020,32 created with BioRender.com). The Ct value refers to the cycle number at which the fluorescence level exceeds a set threshold. In this case, the Ct value is 26. The Ct cut off is set at 40 Ct’s and therefore all samples yielding a Ct greater than 40 are considered as negatives. | ||
Many factors can affect the efficiency of a given RT-PCR, particularly the choice of sample and the method of extraction, as well as the timing of sample collection. Individuals experiencing prolonged viral shedding with low viral loads (Ct values greater than 40 Ct's), can be inaccurately called out as negative. Similarly, borderline qualitative positive results cannot adequately differentiate between naked, sheared or encapsulated viral RNA and these tests currently do not indicate an individual's potential to transmit the disease.33,34
RT-PCR is incredibly sensitive and enables a high throughput analysis of samples; each run can typically allow the comparison of positivity for at least 90 presumptive samples. However, this amplification is slow compared to alternative polymerase chain reactions. RT-PCR also requires experienced operators, complex automated extraction technologies and logistical pipelines. There are however variants of the PCR technology that utilise the reverse transcriptase, but do not rely on thermal cycling to amplify the cDNA but can instead be amplified at a constant temperature. These so-called ‘isothermal technologies’ all use sophisticated priming mechanisms and strand displacement polymerases and are more tolerant to sample derived inhibitors. These isothermal amplifications can normally achieve a similar sensitivity to RT-PCR with the advantage of being maintained on a simple heating block with the potential for faster turnaround times.29
2.2 Isothermal technologies
Many examples of isothermal PCR amplifications have now been developed and commercialised for SARS-CoV-2 RNA amplification: loop mediated isothermal amplification (RT-LAMP), recombinase polymerase amplification (RPA), nicking endonuclease amplification reaction (RT-NEAR), and transcription mediated amplification (RT-TMA).30,35,36 These isothermal technologies offer the advantage of shorter turnaround times compared to RT-PCR (less than 1 hour in most cases) and most can be deployed at the point-of-care (POC) and within limited resource settings.37RT-LAMP typically uses 6 primers to target 8 positions within a conserved portion of the SARS-CoV-2 genome.38 The amplification process takes place at a steady temperature and makes use of strand displacement polymerases, which are more resistant to typical PCR inhibitors and afford the invasion of the duplex DNA molecules that are primed from the cDNA.39 During the propagation of RT-LAMP, some of the primers fold over after polymerisation forming open loop structures in the amplicon that can be effortlessly re-amplified.
Several RT-LAMP based assays have been developed for the detection of SARS-CoV-2 including the rapid, affordable, easy-to-use multiplex RT-LAMP assay with a turnaround time of 1 hour among others.40–42 Despite the benefits offered, designing multiple primers for the assay is complex and therefore multiplexing the assay is difficult.29
NEAR uses a strand displacement polymerase and nicking endonucleases that exponentially amplify the SARS-CoV-2 target sequences.43 One of the main limitations of NEAR, is the formation of non-specific products, which affect the amplification efficiency and sensitivity; despite this, recent optimisations have been shown to limit the impact of nonspecific amplification.44 NEAR forms the basis of the ID NOW test, developed by Abbott Diagnostics for the detection of SARS-CoV-2 and is complete within 15 minutes.29
NEAR is similar to RPA, as this method only requires a pair of primers to amplify any given sequence, and it can be performed at lower temperatures compared to most other isothermal amplifications, with equivalent turnaround times. However, RPA suffers from unwanted priming interactions and requires multiple enzymes that can impact the specificity and complicate the manufacturing process.29 Examples of these RPA based SARS-CoV-2 assays have been published, but to date, no tests have been commercialised.45,46
TMA has been commercialised by a few companies and a few have been approved for EUA, including the Aptima SARS-CoV-2 assay developed by Hologics and Pacific Diagnostics, among others.30 The TMA based Panther Fusion assay was reported to be simpler to use and expressed higher sensitivity with 100% detection down to 5.5 × 103 copies per ml, which was an order of magnitude greater compared to RT-PCR.47
Despite the advantages that these isothermal technologies afford, RT-PCR is still more widely used for SARS-CoV-2 testing for several reasons. For instance, RT-PCR has been implemented for decades and is now considered to be the gold standard molecular amplification method. Access to RT-PCR machines is common in hospitals and centralised laboratories throughout the world, as are the consumables, support networks and infrastructure. For many of these reasons RT-PCR has dominated the testing landscape during the COVID-19 pandemic.29,48
2.3 The need for sensitive and specific diagnostic tests
Sensitive and specific molecular amplification technologies are essential and have helped to inform clinicians on the accuracy of a given respiratory diagnosis.49 Manufacturers and end-users are required to determine the analytical and clinical performance of their test with the use of a reference panel.30,50,51The limit of detection (LoD) is a measure of an RT-PCR's analytical sensitivity and is defined as the lowest concentration of the target that can be detected with a probability of detection of 95% at a stated level of confidence.52 A test with a low LoD or high analytical sensitivity will be able to detect individuals with low viral loads, thereby limiting the number of false negatives.53 Although the exact definition for a high or low viral load has not been defined for SARS-CoV-2, studies do report that the average viral load from an NP or oropharyngeal (OP) swab is approximately 105 cp per ml up to 5 days post symptom onset, with a maximum load of 7.11 × 108 cp per swab; higher viral loads have been reported for sputum samples (2.35 × 109 cp per ml).54 Another study investigating viral loads in throat and sputum samples revealed that viral loads ranged from 641 to 1011 cp per ml.55
According to the WHO and the Medicines and Healthcare products Regulatory Agency (MHRA), an analytical LoD of 1000 cp per ml for a diagnostic test for any respiratory tract specimen type is considered as an acceptable LoD, whilst an analytical LoD of less than 100 cp per ml in upper/lower respiratory tract specimens and stool samples is desirable.56,57 Another important consideration in the reporting of the LoD is that values are reported using different units such as cp per ml, genome equivalents per ml, copies/rxn.58,59 According to the target product profile for COVID-19 testing developed by the WHO, there is currently no international standard unit. Nonetheless, the use of a standard metric may enable better comparisons to be drawn between workflows.49,59 Currently, the most sensitive tests that have received FDA EUA approval include the PerkinElmer New Coronavirus Nucleic Acid Detection Kit and the Viracor SARS-CoV-2 assay, which both achieve an LoD of 180 NAAT NDU per ml (NAAT detectable units per ml).50
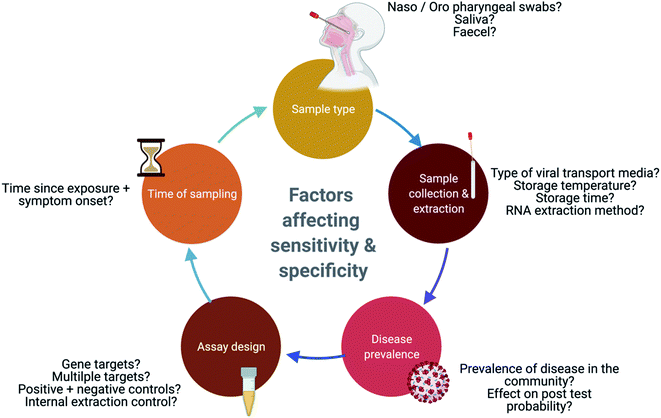
2.4 Factors affecting test sensitivity and specificity
Several factors affect the accuracy of molecular tests including the site and quality of sampling, the stage of disease, rate of viral clearance, and prevalence. In addition, design features of the molecular test are also important to consider e.g., genes targeted and the reliance on amplification from multiple targets. For these reasons, a test is never 100% accurate and the lack of a gold standard for benchmarking performance, further compounds this problem.60False negatives and false positives can be derived from the factors described in Fig. 3.61,62 False negatives can lead to infected individuals circulating within the community, unwittingly propagating the pandemic; these misleading results could arise from inadequate sample loads, sampling individuals too early or too late within the disease cycle, and/or result from the degradation of the viral genome.63 False positives, which affect a test specificity, can result from the contamination of reagents such as primers, or contamination occurring during sample collection and processing, and this in turn could lead to an overestimation of COVID-19 incidences.61,64,65 This review will now explore some of the other key factors that have been reported, which influence the sensitivity of these molecular tests.
Saliva samples, which are easy to collect, enable much more simplistic self-sampling, thereby reducing the risk of transmission to HCPs; this sample type has been proposed as a more suitable alternative.68 Several studies support the use of saliva, with evidence to prove that similar sensitivities were achieved compared to NP swabs.69–71 Some reports also demonstrate that saliva was a more viable alternative compared to NP swabs for the diagnosis of severely infected patients, and for the screening of asymptomatic and presymptomatic individuals,72 whilst other reports are contradictory.68 Although the CDC recommends saliva as an acceptable sample type, the WHO does not recommend the use of saliva as the sole specimen type for diagnosis and screening of SARS-CoV-2.72
The use of faecal samples for the detection of SARS-CoV-2 has also been explored.28,54,67,73 A review by van Doorn et al.74 revealed that 43% of patients tested positive for SARS-CoV-2 via anal swabs and faecal samples, up to 70 days following symptom onset. Viable virus was found in 35% of the patients examined and positive faecal samples remained positive for up to a maximum of 33 days following negative results obtained from a respiratory tract sample.75 In line with this finding, Chinese authorities have recently adopted the use of anal swabs for mass screening of individuals as a more stringent effort to mitigate transmission.76
Given these findings, it is clear that the distribution of SARS-CoV-2 viral load varies markedly between different sample types, as well as between the collection method i.e., self-collection, clinician collection or supervised self-collection.77 For these reasons, obtaining specimens from multiple sample types may help to improve the sensitivity of testing and limit the occurrence of false negative results.67,78
The RNA extraction step is the rate-limiting step in any workflow, due to the challenges associated with liquid handling, the cost of reagents, consumables, and machines. Some examples of methods are now emerging that have the potential to allow the direct addition of crude RNA to a given amplification.91
The RNase P (RP) internal control used in the CDC SARS-CoV-2 assay amplifies both the human genomic DNA (gDNA) and reverse transcribed RNA. Therefore, in the presence of human gDNA, the RP control yields a positive signal even if the RNA extraction has failed or if the sample processing resulted in RNA degradation; in these instances, the negative SARS-CoV-2 results obtained would be interpreted as false negative results.94 The potential for these types of false negative results has prompted a redesign of the RP internal control to specifically detect human RP mRNA and not the genome.95
The inclusion of multiple target regions into an RT-PCR can help to minimise the proportion of false negatives that occur because of mutations arising in the regions of the genome that are targeted by these tests and gives information about genome integrity.96,97 For instance, the emergence of the variant (B.1.1.7; 501Y.V1), which harbours several new mutations have caused concern with regards to the sensitivity of certain diagnostic tests. In fact, the 69–70 deletion has resulted in the spike (S) gene dropout when using the TaqPath COVID-19 Combo Kit (Thermofisher). Given that the TaqPath RT-PCR test targets multiple sequences, the overall impact of the mutated spike gene is minimal.98–100 For instance, the TaqPath is currently being used by PHE to monitor the prevalent Delta variant (B.1.617.2) through detection of the S gene. Other variants of concern include the B.1.351, P.1, P.2, B.1.427, B.1.429, B.1.525, B.1.526, A.23.1 with E484K, B.1.1.7 with E484K.101 Whilst RT-PCR tests and isothermal assays tend to be less affected by the current mutations of concern, as they typically target multiple regions spanning the entire SARS-CoV-2 genome, the effect of these mutations on the rapid antigen tests may become insidious, as the virus evolves and the target antigen degenerates.102,103 The frequent occurrence of such variants also highlights the importance of sequencing technologies that rapidly identify any mutations occurring at the PCR test target region.61 Understanding the operational false positive and negative rates within centralised testing facilities is also a key determinant that affects the accurate reporting of results. In line with this, Public Health England has put forward recommendations that may help in the determination of these values.61,104
The reporting of Ct values may enable clinicians to distinguish between patients with high or low viral loads, and these results can help clinicians make patient-specific interventions, prioritise the clinical management of individuals who are likely to be more infectious, determine the need for isolation, help with contact tracing strategies to prioritise the search for contacts of cases with low Ct's and to aid epidemiologists pinpoint COVID-19 outbreaks.107
One of the main obstacles for using Ct thresholds to manage COVID-19 relates to differences in the LoD for a given workflow, as these often use different reagents and sample preparation methods (extraction and elution volumes). In addition, some assays do not report Ct values, whilst others directly report the results.32 For instance, the Cepheid Xpert Xpress SARS-CoV-2 kit and the Abbot Real Time assay have an identical workflow LoD as reported by the FDA. However, due to higher thresholds set for the Cepheid assay, several negative samples called by the Abbott assay were reported as positive with the Cepheid assay.109,110 For these reasons, some researchers recommend that Ct values are only provided to clinicians on a case-by-case basis, while others advise against the use of Ct values for patient management.108,111–114 As Ct values have also been used as a marker to determine viral loads associated with transmission, the implications of quantification will be explored below.115
All the factors highlighted above form part of the quality management system that is required when manufacturing, performing and reporting results of SARS-CoV-2 tests. Quality control of COVID-19 molecular tests is essential and must be included in every step of the diagnostic testing scheme. This includes but is not limited to the validation and verification of the molecular test using reference panels such as those provided by the FDA to ensure the test performs as intended and thereby gain regulatory approval, and the inclusion of external and internal quality controls to validate nucleic acid extraction and PCR.50 Quality control also encompasses aspects of the testing laboratory including external quality assessments to monitor the performance of the laboratory and international accreditations.116,117 The need for a standardised quality management system is clear by the efforts of the International Organisation for Standardisation (ISO) who are now drafting a quality management standard for NAATs that detect SARS-CoV-2.118
3. Viral load quantification
Whilst an understanding of viral load has proved particularly important in treating and monitoring HIV disease progression, the importance of quantifying COVID-19 has not yet been clearly defined. As discussed earlier, the presence of SARS-CoV-2 RNA in a sample does not necessarily indicate the presence of viable virus.119 Viral cultures prepared from clinical samples generate a more accurate representation of a sample's potential for viral replication; despite this, positive viral culture results do not always concur with a qualitative positive RT-PCR result.54 Due to the challenges and the obvious risks associated with routinely preparing and propagating live viral cultures, this technique is not recommended for the routine diagnosis of infectivity, and viral load determined by RT-PCR is instead regarded as the best surrogate for determining an individual's propensity to transmit the virus.26,34,120The relationship between high nasopharyngeal/saliva/plasma viral loads and disease outcomes, such as intubation, disease progression, systemic inflammation and death has been reported.121–127 Some studies demonstrate that high viral loads in nasopharyngeal swabs are not necessarily related to disease outcomes, such as hospital admission, length of oxygen support and mortality, but instead the data accumulated links mortality directly to other factors, such as an uncontrolled inflammatory response, underlying co-morbidities and a patients age.127,128 Undoubtedly, viral load quantification via RT-PCR could be a useful tool for identifying individuals with a high burden of disease, which in turn, would enable rapid triaging of patients to appropriate medical departments.
Viral load monitoring can also be used to explore the rate of secondary transmission, which may help with implementing effective control measures and inform a clinician when a patient can be released from isolation, predicting future outbreaks and determining COVID-19 incidence within communities.67,129–132
Viral load in respiratory specimens is often higher in patients with mild disease compared to patients with severe disease, whereas no such difference was reported for stool and serum samples.34 Furthermore, viral loads in respiratory samples obtained from patients with mild disease have been reported to increase in the first week after symptom onset, peaking within 2 weeks, then declining thereafter. Whereas those with severe disease had high viral loads even 3–4 weeks after symptom onset. This study also suggests that older individuals over the age of 60 have prolonged infections (viral loads of 1.40–2.19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 RNA cp per test) up to 2 weeks post symptom onset, whilst children under 10 experienced peak viral loads (2.50
log10 RNA cp per test) up to 2 weeks post symptom onset, whilst children under 10 experienced peak viral loads (2.50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 RNA cp per test) during the first 2 days following symptom onset declining rapidly thereafter.133
log10 RNA cp per test) during the first 2 days following symptom onset declining rapidly thereafter.133
Recent reports published by bodies such as the AACC (American Association for Clinical Chemistry) and PHE have outlined the challenges with using Ct values for estimating viral loads thereby clearly highlighting the need for quantitative molecular technologies for viral load monitoring, which would help to support both clinical and public health decisions (Fig. 4).113,114,129,133,134
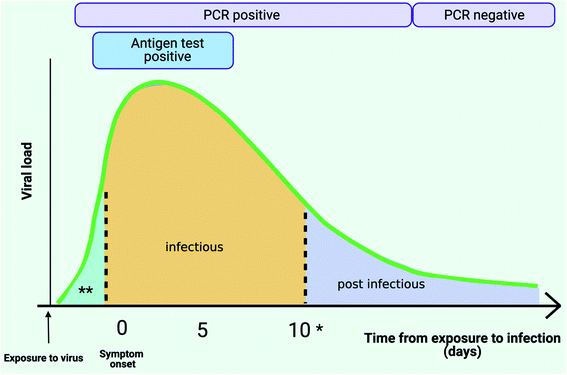 | ||
| Fig. 4 SARS-CoV-2 viral load and testing (Adapted from Cevik et al.;147 Gugleimi,166 Larremore et al.,167 created with BioRender.com). Following exposure to the virus, SARS-Cov-2 viral load in the upper respiratory tract peaks during week 1 since symptom onset. *Individuals can continue shedding infectious virus for approximately 14 days. Highly sensitive PCR tests can detect the presence of virus prior to symptom onset (** pre infectious), through the infectious phase and even post-infection. Antigen tests with their low sensitivity threshold can only detect high viral loads when individuals are likely to be most infectious. | ||
4. Mass screening
Mass screening refers to the process of testing large numbers of individuals regardless of their symptoms. The objective of this testing strategy is to identify individuals who are positive for SARS-CoV-2 and to use this information to determine the most effective intervention for managing transmission. Such testing regimes can be implemented at a household level, at an individual level (testing before international travel & returning to work), and for monitoring of disease in crowded indoor spaces such as schools and healthcare settings. Screening can also be coupled with contact tracing to inform individuals who have come into close contact with individuals who have tested positive upon screening.135The main considerations for controlling the spread of COVID-19 are to account for the possibility of asymptomatic or pre-symptomatic transmission.43,136–145 Presymptomatic transmission has been defined as transmission from a COVID-19 positive individual (index) to another before the recipient displays any symptoms for COVID-19. The potential to infect is highest when an individual is close to symptom onset (Fig. 4).54,146–150 However, the potential for asymptomatic transmission in comparison to symptomatic transmission is still unclear, some reports suggest similar transmission potentials due to similar viral loads observed between asymptomatic and symptomatic cases,148,149 whilst others suggest that the potential to transmit is low from asymptomatic individuals.151,152
Early studies investigating the transmissibility of SARS-CoV-2 among children reported that children were less susceptible to infection, and experienced milder disease compared to adults. However, more recent studies have determined that children of all ages are prone to infection, mortality, and transmission.153 Yonker et al., reported that one-third of school children displaying symptoms were positive for SARS-CoV-2.154 Although younger children are less susceptible and pose a lower risk of transmission due to lower expression of ACE2 receptors.154–157
Elements of the literature state the proportion of infections related to asymptomatic transmission can vary from 17–20%.158,159 Understanding this variation, along with reports of low rates of transmission from asymptomatic individuals, some researchers claim that mass screening for asymptomatic individuals may be a waste of resources.160 Instead, it has been suggested that more effort should be focused on rapidly identifying symptomatic individuals until further evidence is available to implicate the transmission rates from asymptomatic cases. Nonetheless, given that asymptomatic and presymptomatic individuals could equally pose a significant risk to public health, researchers have proposed the frequent use of rapid antigen tests with rapid turnaround times (15–30 min). Such rapid and accessible tests when performed frequently, do facilitate effective surveillance and tend to be more affordable and easier to use than their more sensitive molecular alternatives. Antigen tests have been recommended for use by the CDC for screening in high-risk congregate settings such as housing schemes, nursing facilities, universities, workplaces, airports among others and the FDA have currently issued EUA to 25 commercial antigen tests as of July 2021.161,162
Whilst some reports propose the adoption of antigen tests, others have expressed concern as they tend to have a reduced sensitivity compared to RT-PCR, which may result in a high rate of false negative case reporting.49,162,163 However, it is important to note that in order for mass screening to be successful, speed is as important as sensitivity and in countries with limited access to resources and testing infrastructure and those facing a strain on test supply demand, these easy-to-use, affordable rapid antigen tests have helped in controlling the spread of COVID-19 .164,165
5. Pooling
The widespread testing of individuals to contain the spread of COVID-19 puts pressure on available testing resources, and the pooling of samples from several individuals may help to relieve some of this pressure.168–171 Pooling of samples has been proposed to facilitate rapid population wide screening in regions where prevalence is low.15 Pooling refers to the practice of combining multiple patient samples and performing one laboratory test on this combined sample, thereby enabling high throughput testing, rapid turnaround times and regular testing.168,172 Pooling can be performed at different stages of the workflow i.e., during sample collection, prior to RNA extraction or by combing extracts within an RT-PCR.171,173–175 If the result of pooling is negative, then it is likely that all samples within the pool are negative. If the pool is positive, then all the samples within that pool are separated and tested individually. For these reasons, pooling is most effective in settings where the prevalence of infection is low.Important factors to consider for the pooling of samples include the limit of detection, the specificity and sensitivity of the assay, prevalence of infection, the number of samples in a pool, the efficiency of the pooling strategy, the feasibility, and adaptability to change in prevalence rates.171,176 The FDA recommends >85% positive percentage agreement between a pooling test and an individual test, in order for the pooling strategy to receive EUA and be implemented.175
Aragon-Caqueo et al.,177 propose a model to determine the optimal pool size based on the prevalence of infection and highlight that as prevalence increases, the pooling strategy loses its effectiveness. Hence, the WHO does not recommend the routine use of pooling in clinical settings or for contact tracing.26 The first EUA for a pooling test was granted by the FDA for the Quest SARS-CoV-2 RT-PCR test (Quest Diagnostics) for use of pooling up to 4 samples, in July 2020.178
The occurrence of false negatives caused by the dilution of positive samples within a pool and the impact of this on RT-PCR test sensitivity remains a concern.179,180 Lohse et al., provide evidence to support the pooling of 30 samples at a prevalence rate of 1.93%, resulting in acceptable diagnostic accuracy.179 Yelin et al., reports that a single positive sample can be detected in pools of up to 32 samples with a false negative rate of 10% and this investigation suggests including additional PCR cycles over 40 cycles to enable the detection of samples with low viral loads.181 Wacharapluesadee et al., reports that the sensitivity of the test is not affected by pooling strategies, even at a prevalence rate of 20%, with the provision that the Ct value of the individual positive sample is less than 35.182 The inclusion of more than one positive sample in a pool has also been supported in studies where the inclusion of an increasing number of positive samples reduced the Ct difference between individual and pool tests and increased the sensitivity of PCR testing.174
Barak et al., reports a highly efficient 8 and 5 sample pooling strategy, that spared 76% of RT-PCR and extraction tests upon pooling of nearly 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 samples.171 The increase in efficiency and sensitivity of their strategy has been explained by the non-random distribution of samples. Although the assumption is that samples are randomly distributed within a pool, most samples arrive in batches, with each batch containing samples from a particular university, school, community etc. As a result, samples from each batch are likely to be pooled together from geographical areas of a high or low prevalence, which is thought to improve the efficiency and sensitivity of pooling. The improvement in efficiency and sensitivity and therefore a reduction in false negatives can be explained as the inclusion of a single positive sample with a high viral load results in the detection of other low positive samples in a pool using the same number of PCR tests. The same weak positive samples would have a lower chance of being identified if they were placed on their own in a pool.171
000 samples.171 The increase in efficiency and sensitivity of their strategy has been explained by the non-random distribution of samples. Although the assumption is that samples are randomly distributed within a pool, most samples arrive in batches, with each batch containing samples from a particular university, school, community etc. As a result, samples from each batch are likely to be pooled together from geographical areas of a high or low prevalence, which is thought to improve the efficiency and sensitivity of pooling. The improvement in efficiency and sensitivity and therefore a reduction in false negatives can be explained as the inclusion of a single positive sample with a high viral load results in the detection of other low positive samples in a pool using the same number of PCR tests. The same weak positive samples would have a lower chance of being identified if they were placed on their own in a pool.171
Sensitivity is also affected by the process of sample collection and the extraction method.183 de Salazar et al., and Lone Lim et al., observe high efficiency across the different nucleic acid extraction and amplification platforms.180,184 Positive samples with low Ct values were detected in all volumes tested (25, 40, 60, 100 μl), although the detection of low viral load samples was affected at low volumes.
Other key considerations for evaluating pooling strategies include ease of use, risk of contamination due to the handling of multiple samples, and risk of sample exchange which creates the risk of false positives.168,171,184 Clear protocols that define the procedure for sample pooling and address any losses that occur through the workflow can help minimise the risk of false negatives and simplify the pooling procedure.185 Furthermore, deconvoluting large pools in the event of positive samples outweighs the time benefits offered by pooling.183 For this reason, prevalence rates must always be estimated prior to employing a pooling strategy to reap the benefits of increased testing efficiency and shorter turnaround times. Given the effect of the factors described above, all laboratories must validate their pooling strategies based on the prevalence rate and sensitivity of the test being used and could consider the use of such strategies in settings where there is a strain on testing infrastructure.186
6. Environmental contamination
The need for environmental swabbing to manage the spread of COVID-19 has been warranted upon the knowledge that SARS-CoV-2 virions remain infectious on surfaces for up to 72 hours.9,187,188 The overall objective of this type of testing is to determine the presence of SARS-CoV-2 RNA on surfaces following disinfection, whereby positive results may trigger further cleaning and negative results will confirm effective disinfection protocols.Several commercial environmental swabbing test kits are available for monitoring surface contamination in a range of different settings including, workplaces, food processing facilities, public transport hubs and vehicles, healthcare settings (dental practices and hospitals), communal areas among others.189,190
The stability of SARS-CoV-2 in aerosols and surfaces has been reported, with the longest period of viability observed on stainless steel and plastic surfaces.9 Whilst some studies have provided evidence for environmental contamination by presymptomatic, paucisymptomatic, symptomatic and asymptomatic individuals, thereby warranting proper disinfection programmes, other reports demonstrate no such environmental contamination by infected individuals.191–196
The WHO has provided guidance for surface swabbing that should sample from a surface area of no less than 25 cm3 with the use of multiple swabs.197 Parker et al., describe a protocol (using Isohelix swabs for sample collection, DNA/RNA shield preservatives, automated RNA extraction and RT-qPCR based amplification) that can be followed for environmental swabbing and report the need for a minimum of 1000 viable viral particles per 25 cm3 in order to ensure successful recovery and detection of the virus.17 This study assessed the effect of using swabs for surface sampling with different transport medium and preservatives, extraction methods, and inhibition caused by cleaning agents and components of building materials and concluded there were no significant differences in viral recovery rates between the different RNA extraction methods and debris leftover by cleaning fluids. Differences in recovery rates of viral particles were observed between different surface materials and between sample collection methods, with the highest recovery recorded when swabbing from stainless steel. SARS-CoV-2 RNA and viable virus was also detected in air samples obtained in hospitalised settings, with RNA detected at higher concentrations during the early stages of infection, thereby providing evidence for acquiring COVID-19 via inhalation.198,199 As discussed earlier, SARS-CoV-2 RNA has been detected in the faeces of infected individuals and has led to the initiation of wastewater monitoring for assessing community disease burden.200–203
Overall, these studies provide evidence for the SARS-CoV-2 to persist on surfaces, within the air and wastewater and highlight the importance of monitoring these environments to control the spread of COVID-19 and to broaden our understanding with respect to transmission pathways.
7. Commercially available molecular diagnostic tests
As of July 2021, over 250 molecular tests had received EUA by the FDA, of which over 100 were evaluated using the FDA reference panel.30,50 This section aims to compare several commercial technologies that were selected based on evidence from published literature.As described earlier, the majority of SARS-CoV-2 in vitro diagnostics (IVDs) utilise RT-PCR for the amplification of viral RNA. A performance study assessing four workflows; ePlex assay (GenMark), Simplexa (DiaSorin), Panther Fusion SARS-CoV-2 (Hologic) and a CDC assay, reported that the ePlex demonstrated a higher LoD compared to the Panther Fusion and Simplexa, which failed to detect samples with viral loads of less than 1000 cp per ml. The Simplexa workflow also offers short turnaround times (approximately 1.8 hours) compared to other workflows, such as the Allplex, Quanty (Seegene, 96 samples in 4–5 hours), and Panther Fusion (120 samples in 4 hours). For these reasons, the Panther Fusion assay, Allplex and Quanty have been proposed for use in settings that require a high throughput of samples, whilst the ePlex and Simplex assays were shown to be more suited to settings that require rapid turnaround times but have fewer sample numbers to contend with.204–206
Comparisons between the Cepheid Xpert Xpress SARS-CoV-2, the Roche Cobas® SARS-CoV-2 and the Roche LightMix assay revealed high concordance for samples with high viral loads with Ct <34, this, however, dropped to 22% for samples with Ct >34.207 For these reasons, confirmatory testing of samples that yield negatives is recommended. In contrast, other studies report excellent agreement between the Cepheid Xpert and the Roche Cobas assay and superior sensitivity for both compared to the Becton Dickinson MAX assay, the GenMark ePlex and NeuMoDx Molecular assay.208–210
Given that the SARS-CoV-2 viral load in clinical samples are reported to be greater than 105 cp per ml, most of the technologies (with the exception of the ID NOW assay) report LoD's of less than 5400 NDU per ml and therefore will be able to accurately identify positive samples for SARS-CoV-2 (Table 1).50 Whilst the technologies highlighted above offer high sensitivity, most are located in centralised laboratories, they require expensive equipment, reagents and trained staff, thereby making these tests generally inaccessible to laboratories within limited resource settings. These community-based laboratories have warranted the development of point-of-care (POC) tests that are rapid, cost-effective and robust.211–213 POC tests can improve infection control measures and enable better management of infected patients.214 POC tests are also suited to home testing, which enable GPs to decide if patients can come into clinic for consultations. This type of testing can also service nursing homes and help to determine which individuals need to isolate and engage employers to determine which employees can return to work.215
| Name | Manufacturer | Sample type(s) | LoD (NDU per ml) | Amplification | Applications | ||
|---|---|---|---|---|---|---|---|
| POC | Screening | Pooling | |||||
| a Data unavailable, HCP – healthcare provider. | |||||||
| Abbott RealTime SARS-CoV-2 assay | Abbott molecular | Nasal swabs – self-collection (SC) at healthcare location or by HCP. NP and OP swabs, and bronchoalveolar lavage (BAL) specimens collected by HCP | 2700 | RT-PCR | No | No | No |
| Accula SARS-CoV-2 test | Mesa Biotech Inc | Nasal or nasal mid-turbinate (NMT) swab samples collected by HCP/SC/clinician-supervised self-collected | Under review | RT-PCR | Yes | No | No |
| Allplex™ 2019-nCoV assay | Seegene, Inc. | NP, OP, anterior nasal swab, NMT swab, NP wash/aspirate, nasal aspirate, BAL and sputum specimens collected by HCP | —a | RT-PCR | No | No | No |
| BD SARS-CoV-2 reagents for BD MAX system | Becton, Dickinson & Company | NP, nasal, NMP, and OP swab specimens, NP wash/aspirate or nasal aspirates collected by HCP | 5400 | RT-PCR | No | No | No |
| BioFire COVID-19 test | BioFire Defense, LLC | NP swabs collected by HCP | 5400 | RT-PCR | No | No | Yes |
| BioFire respiratory panel 2.1 (RP2.1) | BioFire diagnostics, LLC | NP swabs collected by HCP | 6000 | RT, Nested multiplex PCR | Yes | No | Yes |
| Cobas SARS-CoV-2 | Roche molecular systems, Inc. | Nasal, NP and OP swab collected by HCP or clinical supervised SC | 1800 | RT-PCR | No | No | Yes |
| ePlex SARS-CoV-2 test | GenMark diagnostics, Inc. | NP swabs collected by HCP | —a | RT-PCR | No | No | No |
| ID NOW COVID-19 | Abbott diagnostics Scarborough, Inc. | Direct nasal, NP or throat swabs collected by HCP | 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
RT, isothermal amplification | Yes | No | No |
| Lucira COVID-19 all-In-one test kit | Lucira health, Inc. | nasal swab collected by SC | —a | RT-LAMP | Yes | No | No |
| NEcov19 RT-PCR assa | Nebraska Medicine clinical laboratory | NP, OP (throat), anterior nasal, NMT swabs, nasal washes, nasal aspirates and BAL specimens collected by HCP | —a | RT-PCR | No | No | No |
| NeuMoDx SARS-CoV-2 assay | NeuMoDx molecular, Inc. | Nasal, NP, OP swabs, BAL specimens collected by HCP. Saliva by clinician supervised SC | 5400 | RT-PCR | No | No | No |
| Panther Fusion SARS-CoV-2 assay | Hologic, Inc. | Upper respiratory specimens (such as nasal, mid-turbinate, nasopharyngeal, and oropharyngeal swab specimens) and sputum by HCP | 600 | RT-PCR | No | Yes | Yes |
| Simplexa COVID-19 direct assay | DiaSorin molecular LLC | NP, nasal swabs, nasal wash/aspirate, or BAL specimens collected by HCP | 6000 | RT-PCR | No | No | No |
| Xpert Xpress SARS-CoV-2 test | Cepheid | Upper respiratory specimens (i.e., NP, OP, nasal, or mid-turbinate swabs or nasal wash/aspirate) collected by HCP | 5400 | RT-PCR | Yes | No | No |
Studies evaluating the performance of the ID NOW (Abbott) have reported that the Xpert, Cobas, Simplexa, Abbott m2000 and RealTime SARS-CoV-2 assays all have better analytical and clinical performance.204 However, the ID NOW, has a rapid turnaround time (15 min) compared to the Xpert, Cobas and RealTime assays; consequently, both the ID NOW and the Xpert are often deployed at POC.204,216–221
A study assessing the performance of the BioFire Respiratory Panel 2.1-EZ (RP2.1-EZ) against the Xpert Xpress, Roche Cobas and BioFire Defense FilmArray, demonstrated a high agreement between all assays and showcased the capacity for the BioFire Respiratory Panel 2.1-EZ to detect low SARS-CoV-2 viral loads down to 103 cp per ml.222 The ID NOW and Aptima tests in contrast were not able to detect low viral load samples that yielded Ct values which were within the upper and lower 30's when tested on the Roche Cobas and NECOV-19 platform tested.223
Other POC platforms include the Accula SARS-CoV-2, which exhibit high overall agreement with a laboratory developed EUA comparator test, however failed to identify some challenging low viral load samples.224 The Lucira COVID-19 All-In-One test kit is another POC platform that has a turnaround time of 30 minutes.224,225 The COVID Nudge was rolled out rapidly in the UK and was reported to have 94% sensitivity, 100% specificity, no sample handling, rapid turnaround time (less than 90 minutes) as well as direct storage of data in the Cloud. However, the platform is limited by its low throughput, as only one sample can be processed at a time. Nonetheless, POC platforms such as the COVID Nudge have the potential to speed up decision making, enabling better clinical management of patients.226
8. Conclusion
Ascertaining the most effective strategies for testing in a global pandemic is a complicated phenomenon that requires consideration of a plethora of factors during implementation such as the effect of sample type, sample collection time, test characteristics on the sensitivity and specificity of the test. Sound test and trace strategies are also needed to ensure that individuals are tested, results are reported in a timely manner, contacts are identified quickly and placed in isolation.Testing for SARS-CoV-2 has many uses including diagnostic testing for identification and management of COVID-19 positive patients, population-wide screening and environment monitoring for controlling the spread of COVID-19, and informing public health decisions and viral load monitoring to promote the appropriate management of positive cases. The type of test used (POC, qualitative or quantitative molecular tests, antigen tests) should fit with the required need.227 For instance, rapid, affordable antigen tests despite their low sensitivity may prove to be more useful in the context of mass screening. The cost vs. benefits of strategies such as pooling and mass screening is also being debated. It is clear that there is not one strategy that is sufficient to control the spread of COVID-19. Instead, what is important is ensuring that the guidelines for each of these strategies are adapted to fit the current requirements.228,229 Furthermore, research has proved that the adoption of combinatory strategies including testing, contact tracing and isolation reduces the effective reproductive number to a greater extent than when one strategy is employed on its own.230
Hence, the molecular testing strategies described throughout this review must be implemented alongside other measures such as contact tracing, physical distancing and isolation of positive cases to limit the spread of COVID-19.
Abbreviations
| AACC | American association for clinical chemistry |
| ACE2 | Angiotensin-converting enzyme 2 |
| BAL | Bronchoalveolar lavage |
| CDC | Centers for disease control and prevention |
| cDNA | Complementary DNA |
| cp | Copies |
| CT | Computed tomography |
| Ct | Cycle threshold |
| ECDC | European center for disease control and prevention |
| EUA | Emergency use approval |
| FDA | U.S Food & Drug Administration |
| gDNA | Genomic DNA |
| HCP | Health care professional |
| ISO | International Organisation for Standardisation |
| IVD | Invitro diagnostics |
| LAMP | Loop mediated isothermal amplification |
| LoD | Limit of detection |
| LRT | Lower respiratory tract |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| MHRA | Medicines and healthcare products regulatory agency |
| MTM | Molecular transport medium |
| NAAT | Nucleic acid amplification test |
| NDU per ml | NAAT detectable units per ml |
| NEAR | Nicking endonuclease amplification reaction |
| NMT | Nasal mid-turbinate |
| NP | Nasopharyngeal |
| OP | Oropharyngeal |
| PBS | Phosphate buffered saline |
| PCR | Polymerase chain reaction |
| PHE | Public health England |
| POC | Point of care |
| PPE | Personal protective equipment |
| QCMD | Quality control molecular diagnostics |
| RNA | Ribonucleic acid |
| RP | RNase P |
| RPA | Recombinase polymerase amplification |
| RT-PCR | Reverse transcription polymerase chain reaction |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SC | Self collection |
| TMA | Transcription mediated amplification |
| URT | Upper respiratory tract |
| VTM | Viral transport medium |
| WHO | World Health Organization |
Author contributions
Conceptualisation: HJ and HSL; writing – original draft HJ, GK and HSL, writing – review & editing, all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the reviewers for their suggestions on the manuscript.References
- Novel Coronavirus – China, https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/, accessed 11 Feb, 2021 Search PubMed.
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at, Johns Hopkins University (JHU), https://coronavirus.jhu.edu/map.html, accessed 20 Jul, 2021 Search PubMed.
- R. A. Khailany, M. Safdar and M. Ozaslan, Gen. Rep., 2020, 19, 100682 Search PubMed.
- I. Astuti and Y. Ysrafil, Diabetes, Metab. Syndr. Obes.: Targets Ther., 2020, 14, 407–412 CrossRef PubMed.
- J. Shang, Y. Wan, C. Luo, G. Ye, Q. Geng, A. Auerbach and F. Li, Proc. Natl. Acad. Sci., 2020, 117, 11727–11734 CrossRef CAS PubMed.
- How COVID-19 spreads, https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html, accessed 11 Feb, 2021 Search PubMed.
- P. Y. Chia, K. K. Coleman, Y. K. Tan, S. W. X. Ong, M. Gum, S. K. Lau, X. F. Lim, A. S. Lim, S. Sutjipto, P. H. Lee, T. T. Son, B. E. Young, D. K. Milton, G. C. Gray, S. Schuster, T. Barkham, P. P. De, S. Vasoo, M. Chan, B. S. P. Ang, B. H. Tan, Y.-S. Leo, O.-T. Ng, M. S. Y. Wong, K. Marimuthu, D. C. Lye, P. L. Lim, C. C. Lee, L. M. Ling, L. Lee, T. H. Lee, C. S. Wong, S. Sadarangani, R. J. Lin, D. H. L. Ng, M. Sadasiv, T. W. Yeo, C. Y. Choy, G. S. E. Tan, F. Dimatatac, I. F. Santos, C. J. Go, Y. K. Chan, J. Y. Tay, J. Y.-L. Tan, N. Pandit, B. C. H. Ho, S. Mendis, Y. Y. C. Chen, M. Y. Abdad and D. Moses, Singapore Novel Coronavirus Outbreak Research, Nat. Commun., 2020, 11, 2800 CrossRef CAS PubMed.
- J. F.-W. Chan, S. Yuan, K.-H. Kok, K. K.-W. To, H. Chu, J. Yang, F. Xing, J. Liu, C. C.-Y. Yip, R. W.-S. Poon, H.-W. Tsoi, S. K.-F. Lo, K.-H. Chan, V. K.-M. Poon, W.-M. Chan, J. D. Ip, J.-P. Cai, V. C.-C. Cheng, H. Chen, C. K.-M. Hui and K.-Y. Yuen, Lancet, 2020, 395, 514–523 CrossRef CAS.
- N. van Doremalen, T. Bushmaker, D. H. Morris, M. G. Holbrook, A. Gamble, B. N. Williamson, A. Tamin, J. L. Harcourt, N. J. Thornburg, S. I. Gerber, J. O. Lloyd-Smith, E. de Wit and V. J. Munster, N. Engl. J. Med., 2020, 382, 1564–1567 CrossRef.
- I. Ghinai, T. D. McPherson, J. C. Hunter, H. L. Kirking, D. Christiansen, K. Joshi, R. Rubin, S. Morales-Estrada, S. R. Black, M. Pacilli, M. J. Fricchione, R. K. Chugh, K. A. Walblay, N. S. Ahmed, W. C. Stoecker, N. F. Hasan, D. P. Burdsall, H. E. Reese, M. Wallace, C. Wang, D. Moeller, J. Korpics, S. A. Novosad, I. Benowitz, M. W. Jacobs, V. S. Dasari, M. T. Patel, J. Kauerauf, E. M. Charles, N. O. Ezike, V. Chu, C. M. Midgley, M. A. Rolfes, S. I. Gerber, X. Lu, S. Lindstrom, J. R. Verani, J. E. Layden, S. Brister, K. Goldesberry, S. Hoferka, D. Jovanov, D. Nims, L. Saathoff-Huber, C. Hoskin Snelling, H. Adil, R. Ali, E. Andreychak, K. Bemis, M. Frias, P. Quartey-Kumapley, K. Baskerville, E. Murphy, E. Murskyj, Z. Noffsinger, J. Vercillo, A. Elliott, U. S. Onwuta, D. Burck, G. Abedi, R. M. Burke, R. Fagan, J. Farrar, A. M. Fry, A. J. Hall, A. Haynes, C. Hoff, S. Kamili, M. E. Killerby, L. Kim, S. A. Kujawski, D. T. Kuhar, B. Lynch, L. Malapati, M. Marlow, J. R. Murray, B. Rha, S. K. K. Sakthivel, S. E. Smith-Jeffcoat, E. Soda, L. Wang, B. L. Whitaker and T. M. Uyeki, Lancet, 2020, 395, 1137–1144 CrossRef CAS.
- Z.-D. Guo, Z.-Y. Wang, S.-F. Zhang, X. Li, L. Li, C. Li, Y. Cui, R.-B. Fu, Y.-Z. Dong, X.-Y. Chi, M.-Y. Zhang, K. Liu, C. Cao, B. Liu, K. Zhang, Y.-W. Gao, B. Lu and W. Chen, Emerging Infect. Dis., 2020, 26, 1583–1591 CrossRef PubMed.
- L. Hamner, P. Dubbel, I. Capron, A. Ross, A. Jordan, J. Lee, J. Lynn, A. Ball, S. Narwal, S. Russell, D. Patrick and H. Leibrand, Morb. Mortal. Wkly. Rep., 2020, 69, 606–610 CrossRef CAS PubMed.
- Y. Liu, Z. Ning, Y. Chen, M. Guo, Y. Liu, N. K. Gali, L. Sun, Y. Duan, J. Cai, D. Westerdahl, X. Liu, K. Xu, K.-f. Ho, H. Kan, Q. Fu and K. Lan, Nature, 2020, 582, 557–560 CrossRef CAS.
- Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions, https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions, accessed 1 Mar, 2021 Search PubMed.
- COVID-19 Testing Strategies and Objectives, https://www.ecdc.europa.eu/sites/default/files/documents/TestingStrategy_Objective-Sept-2020.pdf, accessed 11 Feb, 2021 Search PubMed.
- B. Pastorino, F. Touret, M. Gilles, X. de Lamballerie and R. N. Charrel, Emerging Infect. Dis., 2020, 26, 2256–2257 CrossRef CAS.
- C. W. Parker, N. Singh, S. Tighe, A. Blachowicz, J. M. Wood, A. Seuylemezian, P. Vaishampayan, C. Urbaniak, R. Hendrickson, P. Laaguiby, K. Clark, B. G. Clement, N. B. O'Hara, M. Couto-Rodriguez, D. Bezdan, C. E. Mason and K. Venkateswaran, mSystems, 2020, 5, e00771-00720 CrossRef.
- Monitoring the Presence and Infection Risk of SARS-CoV-2 in the Environment: Approaches, Limitations and Interpretation Transmission of Covid-19 in the Wider Environment Group, gov.uk, 2020 Search PubMed.
- Science Brief, SARS-CoV-2 and Potential Airborne Transmission, https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-sars-cov-2.html, accessed 11 Feb, 2021 Search PubMed.
- M. Touma, J. Mol. Med., 2020, 98, 947–954 CrossRef CAS PubMed.
- A. La Marca, M. Capuzzo, T. Paglia, L. Roli, T. Trenti and S. M. Nelson, Reprod. BioMed. Online, 2020, 41, 483–499 CrossRef CAS.
- What We Know about the COVID-19 Immune Response, World Health Organization, who.int, 2020 Search PubMed.
- J. Seow, C. Graham, B. Merrick, S. Acors, S. Pickering, K. J. A. Steel, O. Hemmings, A. O'Byrne, N. Kouphou, R. P. Galao, G. Betancor, H. D. Wilson, A. W. Signell, H. Winstone, C. Kerridge, I. Huettner, J. M. Jimenez-Guardeño, M. J. Lista, N. Temperton, L. B. Snell, K. Bisnauthsing, A. Moore, A. Green, L. Martinez, B. Stokes, J. Honey, A. Izquierdo-Barras, G. Arbane, A. Patel, M. K. I. Tan, L. O'Connell, G. O'Hara, E. MacMahon, S. Douthwaite, G. Nebbia, R. Batra, R. Martinez-Nunez, M. Shankar-Hari, J. D. Edgeworth, S. J. D. Neil, M. H. Malim and K. J. Doores, Nat. Microbiol., 2020, 5, 1598–1607 CrossRef CAS.
- F. J. Ibarrondo, J. A. Fulcher, D. Goodman-Meza, J. Elliott, C. Hofmann, M. A. Hausner, K. G. Ferbas, N. H. Tobin, G. M. Aldrovandi and O. O. Yang, N. Engl. J. Med., 2020, 383, 1085–1087 CrossRef.
- COVID-19 Testing Overview, accessed 11 Feb, 2021 Search PubMed.
- Diagnostic testing for SARS-CoV-2, https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2, accessed 11 Feb, 2021 Search PubMed.
- Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing, https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html, accessed 11 Feb, 2021 Search PubMed.
- S. C. Ng, F. K. L. Chan and P. K. S. Chan, Lancet Gastroenterol Hepatol, 2020, 5, 642–643 CrossRef PubMed.
- T. Kilic, R. Weissleder and H. Lee, iScience, 2020, 23, 101406 CrossRef CAS.
- In Vitro Diagnostics EUAs – Molecular Diagnostic Tests for SARS-CoV-2, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2#individual-molecular, accessed 20 July, 2021 Search PubMed.
- K. B. Mullis, Sci. Am., 1990, 262, 56–65 CrossRef CAS PubMed.
- Understanding cycle threshold (Ct) in SARS-CoV-2 RT-PCR, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/926410/Understanding_Cycle_Threshold__Ct__in_SARS-CoV-2_RT-PCR_.pdf Search PubMed.
- N. Sethuraman, S. S. Jeremiah and A. Ryo, JAMA, 2020, 323, 2249–2251 CrossRef CAS PubMed.
- S. Zheng, J. Fan, F. Yu, B. Feng, B. Lou, Q. Zou, G. Xie, S. Lin, R. Wang, X. Yang, W. Chen, Q. Wang, D. Zhang, Y. Liu, R. Gong, Z. Ma, S. Lu, Y. Xiao, Y. Gu, J. Zhang, H. Yao, K. Xu, X. Lu, G. Wei, J. Zhou, Q. Fang, H. Cai, Y. Qiu, J. Sheng, Y. Chen and T. Liang, BMJ, 2020, 369, m1443 CrossRef.
- Hologic SARS-CoV-2 Assays, https://www.hologic.com/hologic-products/diagnostic-solutions/hologic-sars-cov-2-assays, accessed 28 April, 2021 Search PubMed.
- Lucira COVID-19 All-In-One Test Kit, fda.gov, 2020 Search PubMed.
- Antigen and Molecular Tests for COVID-19, https://www.centerforhealthsecurity.org/covid-19TestingToolkit/molecular-based-tests/current-molecular-and-antigen-tests.html, accessed 11 Feb, 2021 Search PubMed.
- Y. Mori, K. Nagamine, N. Tomita and T. Notomi, Biochem. Biophys. Res. Commun., 2001, 289, 150–154 CrossRef CAS PubMed.
- D. Thompson and Y. Lei, Sensors and Actuators Reports, 2020, 2, 100017 CrossRef.
- G.-S. Park, K. Ku, S.-H. Baek, S.-J. Kim, S. I. Kim, B.-T. Kim and J.-S. Maeng, J. Mol. Diagn., 2020, 22, 729–735 CrossRef CAS.
- L. Yu, S. Wu, X. Hao, X. Dong, L. Mao, V. Pelechano, W.-H. Chen and X. Yin, Clin. Chem., 2020, 66, 975–977 CrossRef.
- X. Zhu, X. Wang, L. Han, T. Chen, L. Wang, H. Li, S. Li, L. He, X. Fu, S. Chen, M. Xing, H. Chen and Y. Wang, Biosens. Bioelectron., 2020, 166, 112437 CrossRef CAS PubMed.
- C. Qian, R. Wang, H. Wu, F. Ji and J. Wu, Anal. Chim. Acta, 2019, 1050, 1–15 CrossRef CAS PubMed.
- L. Wang, C. Qian, H. Wu, W. Qian, R. Wang and J. Wu, Analyst, 2018, 143, 1444–1453 RSC.
- S. Xia and X. Chen, Cell Discovery, 2020, 6, 37 CrossRef CAS PubMed.
- E.-T. Mohamed, B. Haim and S. Jinzhao, A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry, 2020 Search PubMed.
- A. J. Gorzalski, H. Tian, C. Laverdure, S. Morzunov, S. C. Verma, S. VanHooser and M. W. Pandori, J. Clin. Virol., 2020, 129, 104501 CrossRef CAS.
- R. Howard, The Future of Molecular Diagnostics: Will Isothermal Amplification Techniques Steal PCR's Crown in the Close-To-Patient Era?, https://www.ttp.com/blog/the-future-of-molecular-diagnostics-will-isothermal-amplification-techniques-steal-pcrs-crown-in-the-close-to-patient-era, accessed 11 Feb, 2021 Search PubMed.
- R. Arnaout, R. A. Lee, G. R. Lee, C. Callahan, C. F. Yen, K. P. Smith, R. Arora and J. E. Kirby, bioRxiv, 2020 DOI:10.1101/2020.06.02.131144.
- SARS-CoV-2 Reference Panel Comparative Data, https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data, accessed 20 July, 2021 Search PubMed.
- V. Matheeussen, V. M. Corman, O. Donoso Mantke, E. McCulloch, C. Lammens, H. Goossens, D. Niemeyer, P. S. Wallace, P. Klapper, H. G. Niesters, C. Drosten, M. Ieven, R. project and n. collaborating, Eurosurveillance, 2020, 25, 2001223 CrossRef CAS PubMed.
- Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures, https://clsi.org/standards/products/method-evaluation/documents/ep17/, 2nd edn Search PubMed.
- Importance of COVID-19 test limit of detection, https://www.zymoresearch.com/blogs/blog/importance-of-covid-19-test-limit-of-detection, accessed 11 Feb, 2021 Search PubMed.
- R. Wölfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Müller, D. Niemeyer, T. C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brünink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten and C. Wendtner, Nature, 2020, 581, 465–469 CrossRef PubMed.
- Y. Pan, D. Zhang, P. Yang, L. L. M. Poon and Q. Wang, Lancet Infect. Dis., 2020, 20, 411–412 CrossRef CAS.
- Target Product Profile: Laboratory-Based SARS-CoV-2 Viral Detection Tests, https://www.gov.uk/government/publications/how-tests-and-testing-kits-for-coronavirus-covid-19-work/target-product-profile-laboratory-based-sars-cov-2-viral-detection-tests, accessed 11 Feb, 2021 Search PubMed.
- COVID-19 Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.1.0, https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1, accessed 11 Feb, 2021 Search PubMed.
- Find Evaluation Update: Sars-Cov-2 Molecular Diagnostics, https://www.finddx.org/covid-19-old/sarscov2-eval-molecular/, accessed 15 Mar, 2021 Search PubMed.
- Y. Sohni, Lab. Med., 2021, 52(2), 107–115 CrossRef.
- J. Watson, P. F. Whiting and J. E. Brush, BMJ, 2020, 369, m1808 CrossRef.
- Impact of False-Positives and False-Negatives in the UK's COVID-19 RT-PCR Testing Programme, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/895843/S0519_Impact_of_false_positives_and_negatives.pdf, accessed 3 May, 2021 Search PubMed.
- B. Healy, A. Khan, H. Metezai, I. Blyth and H. Asad, Clin. Med., 2021, 21, e54–e56 CrossRef.
- S. Woloshin, N. Patel and A. S. Kesselheim, N. Engl. J. Med., 2020, 383, e38 CrossRef CAS.
- J. F. Huggett, V. Benes, S. A. Bustin, J. A. Garson, K. Harris, M. Kammel, M. Kubista, T. D. McHugh, J. Moran-Gilad, T. Nolan, M. W. Pfaffl, M. Salit, G. Shipley, P. M. Vallone, J. Vandesompele, C. Wittwer and H. Zeichhardt, Clin. Chem., 2020, 66, 1369–1372 CrossRef.
- E. Surkova, V. Nikolayevskyy and F. Drobniewski, Lancet Respir. Med., 2020, 8, 1167–1168 CrossRef CAS.
- Diagnostic testing and screening for SARS-CoV-2, https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing, accessed 12 Feb 2021 Search PubMed.
- W. Wang, Y. Xu, R. Gao, R. Lu, K. Han, G. Wu and W. Tan, JAMA, 2020, 323, 1843–1844 CAS.
- M. C. Lisa Caulley, L. Eapen, J. Whelan, J. B. Angel, K. Antonation, N. Bastien, G. Poliquin and S. Johnson-Obaseki, Ann. Intern. Med., 2021, 174, 131–133 CrossRef.
- L. M. Czumbel, S. Kiss, N. Farkas, I. Mandel, A. Hegyi, Á. Nagy, Z. Lohinai, Z. Szakács, P. Hegyi, M. C. Steward and G. Varga, Frontiers in Medicine, 2020, 7, 465 CrossRef.
- E. Williams, K. Bond, B. Zhang, M. Putland and D. A. Williamson, J. Clin. Microbiol., 2020, 58, e00776-00720 Search PubMed.
- E. Pasomsub, S. P. Watcharananan, K. Boonyawat, P. Janchompoo, G. Wongtabtim, W. Suksuwan, S. Sungkanuparph and A. Phuphuakrat, Clin. Microbiol. Infect., 2021, 27, 285.e281–285.e284 Search PubMed.
- A. L. Wyllie, J. Fournier, A. Casanovas-Massana, M. Campbell, M. Tokuyama, P. Vijayakumar, J. L. Warren, B. Geng, M. C. Muenker, A. J. Moore, C. B. F. Vogels, M. E. Petrone, I. M. Ott, P. Lu, A. Venkataraman, A. Lu-Culligan, J. Klein, R. Earnest, M. Simonov, R. Datta, R. Handoko, N. Naushad, L. R. Sewanan, J. Valdez, E. B. White, S. Lapidus, C. C. Kalinich, X. Jiang, D. J. Kim, E. Kudo, M. Linehan, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takahashi, M. Taura, O.-E. Weizman, P. Wong, Y. Yang, S. Bermejo, C. D. Odio, S. B. Omer, C. S. Dela Cruz, S. Farhadian, R. A. Martinello, A. Iwasaki, N. D. Grubaugh and A. I. Ko, N. Engl. J. Med., 2020, 383, 1283–1286 CrossRef PubMed.
- Y. Chen, L. Chen, Q. Deng, G. Zhang, K. Wu, L. Ni, Y. Yang, B. Liu, W. Wang, C. Wei, J. Yang, G. Ye and Z. Cheng, J. Med. Virol., 2020, 92, 833–840 CrossRef CAS PubMed.
- A. S. van Doorn, B. Meijer, C. M. A. Frampton, M. L. Barclay and N. K. H. de Boer, Aliment. Pharmacol. Ther., 2020, 52, 1276–1288 CAS.
- V. Kipkorir, I. Cheruiyot, B. Ngure, M. Misiani and J. Munguti, J. Med. Virol., 2020, 92, 2328–2331 CrossRef CAS PubMed.
- Chinese Cities Using Anal Swabs to Screen COVID-19 Infections, https://www.reuters.com/article/us-health-coronavirus-china-testing-idUSKBN29W1RN, accessed 11 Feb, 2021 Search PubMed.
- N. Kojima, F. Turner, V. Slepnev, A. Bacelar, L. Deming, S. Kodeboyina and J. D. Klausner, Clin. Infect. Dis., 2020 DOI:10.1093/cid/ciaa1589.
- J. A. Lieberman, G. Pepper, S. N. Naccache, M.-L. Huang, K. R. Jerome and A. L. Greninger, J. Clin. Microbiol., 2020, 58, e00821-00820 CrossRef.
- B. Ford, F. Lam, J. Wilson and M. Moubarak, Role of Viral Transport Media in Sustaining COVID-19 Testing, https://www.medlabmag.com/article/1700 Search PubMed.
- L. Garnett, A. Bello, K. N. Tran, J. Audet, A. Leung, Z. Schiffman, B. D. Griffin, N. Tailor, D. Kobasa and J. E. Strong, J. Virol. Methods, 2020, 285, 113947 CrossRef CAS.
- K. G. Rodino, M. J. Espy, S. P. Buckwalter, R. C. Walchak, J. J. Germer, E. Fernholz, A. Boerger, A. N. Schuetz, J. D. Yao and M. J. Binnicker, J. Clin. Microbiol., 2020, 58, e00590-00520 Search PubMed.
- A. A. Rogers, R. E. Baumann, G. A. Borillo, R. M. Kagan, H. J. Batterman, M. M. Galdzicka and E. M. Marlowe, J. Clin. Microbiol., 2020, 58, e00708–00720 CAS.
- What is the Difference between Standard Viral and Molecular Transport Media?, https://www.ekfdiagnostics.com/Molecular-Transport-Media-VS-Viral-Transport-Media.html, accessed 11 Feb, 2021 Search PubMed.
- S. R. Welch, K. A. Davies, H. Buczkowski, N. Hettiarachchi, N. Green, U. Arnold, M. Jones, M. J. Hannah, R. Evans, C. Burton, J. E. Burton, M. Guiver, P. A. Cane, N. Woodford, C. B. Bruce, A. D. G. Roberts and M. J. Killip, J. Clin. Microbiol., 2020, 58, e01713-01720 CrossRef PubMed.
- P. D. Kirkland and M. J. Frost, Pathology, 2020, 52, 811–814 CrossRef CAS PubMed.
- L. M. Kucirka, S. A. Lauer, O. Laeyendecker, D. Boon and J. Lessler, Ann. Intern. Med., 2020, 173, 262–267 CrossRef PubMed.
- X. He, E. H. Y. Lau, P. Wu, X. Deng, J. Wang, X. Hao, Y. C. Lau, J. Y. Wong, Y. Guan, X. Tan, X. Mo, Y. Chen, B. Liao, W. Chen, F. Hu, Q. Zhang, M. Zhong, Y. Wu, L. Zhao, F. Zhang, B. J. Cowling, F. Li and G. M. Leung, Nat. Med., 2020, 26, 672–675 CrossRef CAS PubMed.
- C. Ambrosi, C. Prezioso, P. Checconi, D. Scribano, M. Sarshar, M. Capannari, C. Tomino, M. Fini, E. Garaci, A. T. Palamara, G. De Chiara and D. Limongi, J. Virol. Methods, 2021, 287, 114008 CrossRef CAS.
- E. M. Ransom, R. F. Potter, M. A. Wallace, K. F. Mitchell, M. L. Yarbrough, C.-A. D. Burnham, N. W. Anderson and B. A. Parikh, J. Clin. Microbiol., 2020, 58, e01622-01620 Search PubMed.
- A. Petrich, J. Mahony, S. Chong, G. Broukhanski, F. Gharabaghi, G. Johnson, L. Louie, K. Luinstra, B. Willey, P. Akhaven, L. Chui, F. Jamieson, M. Louie, T. Mazzulli, R. Tellier, M. Smieja, W. Cai, M. Chernesky and S. E. Richardson, J. Clin. Microbiol., 2006, 44, 2681 CrossRef CAS PubMed.
- E. A. Bruce, M.-L. Huang, G. A. Perchetti, S. Tighe, P. Laaguiby, J. J. Hoffman, D. L. Gerrard, A. K. Nalla, Y. Wei, A. L. Greninger, S. A. Diehl, D. J. Shirley, D. G. B. Leonard, C. D. Huston, B. D. Kirkpatrick, J. A. Dragon, J. W. Crothers, K. R. Jerome and J. W. Botten, PLoS Biol., 2020, 18, e3000896 CrossRef CAS.
- SARS-COV-2 RT-PCR controls, https://perkinelmer-appliedgenomics.com/2020/05/11/sars-cov-2-rt-pcr-controls/, accessed 11 Feb, 2021 Search PubMed.
- A. Tahamtan and A. Ardebili, Expert Rev. Mol. Diagn., 2020, 20, 453–454 CrossRef CAS.
- A. P. Rosebrock, Clin. Chem., 2020, 67, 435–437 CrossRef.
- R. J. Dekker, W. A. Ensink, S. van Leeuwen, H. Rauwerda and T. M. Breit, bioRxiv, 2020 DOI:10.1101/2020.06.12.147819.
- M. C. Chang, J. Hur and D. Park, Am. J. Phys. Med., 2020, 99, 583–585 CrossRef.
- FDA Issues Alert Regarding SARS-CoV-2 Viral Mutation to Health Care Providers and Clinical Laboratory Staff, https://www.fda.gov/news-events/press-announcements/fda-issues-alert-regarding-sars-cov-2-viral-mutation-health-care-providers-and-clinical-laboratory, accessed 28 Mar 2021 Search PubMed.
- Solutions for Surveillance of the S Gene Mutation in the B.1.1.7 (501Y.V1) SARS-CoV-2 Strain Lineage, https://www.thermofisher.com/blog/behindthebench/solutions-for-surveillance-of-the-s-gene-mutation-in-the-b117-501yv1-sars-cov-2-strain-lineage/, accessed 13 May 2021 Search PubMed.
- Genetic Variants of SARS-CoV-2 May Lead to False Negative Results with Molecular Tests for Detection of SARS-CoV-2 - Letter to Clinical Laboratory Staff and Health Care Providers, https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2, accessed 11 Feb, 2021 Search PubMed.
- S. E. Galloway, P. Paul, D. R. MacCannell, M. A. Johansson, J. T. Brooks, A. MacNeil, R. B. Slayton, S. Tong, B. J. Silk, G. L. Armstrong, M. Biggerstaff and V. G. Dugan, Morb. Mortal. Wkly. Rep., 2021, 70, 95–99 CrossRef CAS.
- SARS-CoV-2 Variant Classifications and Definitions, https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html, accessed 26 July, 2021 Search PubMed.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci., 2020, 117, 24450–24458 CrossRef CAS.
- E. Mahase, BMJ, 2020, 371, m4469 CrossRef.
- J. N. Kanji, N. Zelyas, C. MacDonald, K. Pabbaraju, M. N. Khan, A. Prasad, J. Hu, M. Diggle, B. M. Berenger and G. Tipples, Virol. J., 2021, 18, 13 CrossRef CAS.
- T. Nolan, J. Huggett and E. Sanchez, Good practice guide for the application of quantitative PCR (qPCR), LGC, 2013 Search PubMed.
- M. W. Pfaffl, in A-Z of Quantitative PCR, International University Line (IUL), La Jolla, CA, USA, 2004, ch. 3, pp. 87–112 Search PubMed.
- M. R. Tom and M. J. Mina, Clin. Infect. Dis., 2020, 71, 2252–2254 CrossRef CAS PubMed.
- M. L. Landry, Discussion from a Hospital Laboratory Perspective, https://medicine.yale.edu/labmed/sections/virology/COVID-19%20Ct%20values_YNHH%20Aug.%202020%20_395430_36854_v1.pdf, accessed 11 Feb, 2021 Search PubMed.
- A. Basu, T. Zinger, K. Inglima, K.-M. Woo, O. Atie, L. Yurasits, B. See and M. E. Aguero-Rosenfeld, J. Clin. Microbiol., 2020, 58, e01136-01120 CrossRef PubMed.
- W. Wodarg and M. Yeadon, European Medicines Agency Petition Pfizer Trial, 2020 Search PubMed.
- D. Rhoads, D. R. Peaper, R. C. She, F. S. Nolte, C. M. Wojewoda, N. W. Anderson and B. S. Pritt, Clin. Infect. Dis., 2021, 72(10), e685–e686 CrossRef CAS PubMed.
- M. J. Binnicker, J. Clin. Microbiol., 2020, 58, e01695-01620 CrossRef PubMed.
- SARS-CoV-2 Cycle Threshold: A Metric That Matters (or Not), https://www.aacc.org/cln/cln-stat/2020/december/3/sars-cov-2-cycle-threshold-a-metric-that-matters-or-not, accessed 12 Feb, 2021 Search PubMed.
- AACC Recommendation for Reporting SARS-CoV-2 Cycle Threshold (CT) Values, https://www.aacc.org/science-and-research/covid-19-resources/statements-on-covid-19-testing/aacc-recommendation-for-reporting-sars-cov-2-cycle-threshold-ct-values, accessed 21 July, 2021 Search PubMed.
- B. La Scola, M. Le Bideau, J. Andreani, V. T. Hoang, C. Grimaldier, P. Colson, P. Gautret and D. Raoult, Eur. J. Clin. Microbiol. Infect. Dis., 2020, 39, 1059–1061 CrossRef CAS PubMed.
- Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases,%20https://www.who.int/publications/i/item/10665-331501 Search PubMed.
- Recommendations for National SARS-CoV-2 Testing Strategies and Diagnostic Capacities, https://www.who.int/publications/i/item/WHO-2019-nCoV-lab-testing-2021.1-eng, accessed 19 July, 2021 Search PubMed.
- ISO/WD TS 5798- Quality Practice for detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by nucleic acid amplification methods, https://www.iso.org/standard/81712.html, accessed 21 July, 2021 Search PubMed.
- T. Jefferson, E. A. Spencer, J. Brassey and C. Heneghan, Clin. Infect. Dis., 2020, ciaa1764 CrossRef.
- G. M. Joynt and W. K. Wu, Lancet Infect. Dis., 2020, 20, 635–636 CrossRef CAS.
- R. Magleby, L. F. Westblade, A. Trzebucki, M. S. Simon, M. Rajan, J. Park, P. Goyal, M. M. Safford and M. J. Satlin, Clin. Infect. Dis., 2020, ciaa851 CrossRef.
- E. Pujadas, F. Chaudhry, R. McBride, F. Richter, S. Zhao, A. Wajnberg, G. Nadkarni, B. S. Glicksberg, J. Houldsworth and C. Cordon-Cardo, Lancet Respir. Med., 2020, 8, e70 CrossRef CAS.
- A. Shlomai, H. Ben-Zvi, A. Glusman Bendersky, N. Shafran, E. Goldberg and E. H. Sklan, Critical Care, 2020, 24, 539 CrossRef PubMed.
- L. F. Westblade, G. Brar, L. C. Pinheiro, D. Paidoussis, M. Rajan, P. Martin, P. Goyal, J. L. Sepulveda, L. Zhang, G. George, D. Liu, S. Whittier, M. Plate, C. B. Small, J. H. Rand, M. M. Cushing, T. J. Walsh, J. Cooke, M. M. Safford, M. Loda and M. J. Satlin, Cancer Cell, 2020, 38, 661–671 CrossRef CAS.
- J. Silva, C. Lucas, M. Sundaram, B. Israelow, P. Wong, J. Klein, M. Tokuyama, P. Lu, A. Venkataraman, F. Liu, T. Mao, J. E. Oh, A. Park, A. Casanovas-Massana, C. B. F. Vogels, M. C. Muenker, J. Zell, J. B. Fournier, M. Campbell, M. Chiorazzi, E. R. Fuentes, M. E. Petrone, C. C. Kalinich, I. M. Ott, A. Watkins, A. J. Moore, M. Nakahata, N. D. Grubaugh, S. Farhadian, C. D. Cruz, A. I. Ko, W. L. Schulz, A. Ring, S. Ma, S. Omer, A. L. Wyllie and A. Iwasaki, medRxiv, 2021 DOI:10.1101/2021.01.04.21249236.
- J. Fajnzylber, J. Regan, K. Coxen, H. Corry, C. Wong, A. Rosenthal, D. Worrall, F. Giguel, A. Piechocka-Trocha, C. Atyeo, S. Fischinger, A. Chan, K. T. Flaherty, K. Hall, M. Dougan, E. T. Ryan, E. Gillespie, R. Chishti, Y. Li, N. Jilg, D. Hanidziar, R. M. Baron, L. Baden, A. M. Tsibris, K. A. Armstrong, D. R. Kuritzkes, G. Alter, B. D. Walker, X. Yu, J. Z. Li, B. A. Abayneh, P. Allen, D. Antille, A. Balazs, J. Bals, M. Barbash, Y. Bartsch, J. Boucau, S. Boyce, J. Braley, K. Branch, K. Broderick, J. Carney, J. Chevalier, M. C. Choudhary, N. Chowdhury, T. Cordwell, G. Daley, S. Davidson, M. Desjardins, L. Donahue, D. Drew, K. Einkauf, S. Elizabeth, A. Elliman, B. Etemad, J. Fallon, L. Fedirko, K. Finn, J. Flannery, P. Forde, P. Garcia-Broncano, E. Gettings, D. Golan, K. Goodman, A. Griffin, S. Grimmel, K. Grinke, C. A. Hartana, M. Healy, H. Heller, D. Henault, G. Holland, C. Jiang, H. Jordan, P. Kaplonek, E. W. Karlson, M. Karpell, C. Kayitesi, E. C. Lam, V. LaValle, K. Lefteri, X. Lian, M. Lichterfeld, D. Lingwood, H. Liu, J. Liu, K. Lopez, Y. Lu, S. Luthern, N. L. Ly, M. MacGowan, K. Magispoc, J. Marchewka, B. Martino, R. McNamara, A. Michell, I. Millstrom, N. Miranda, C. Nambu, S. Nelson, M. Noone, L. Novack, C. O'Callaghan, C. Ommerborn, M. Osborn, L. C. Pacheco, N. Phan, S. Pillai, F. A. Porto, Y. Rassadkina, A. Reissis, F. Ruzicka, K. Seiger, K. Selleck, L. Sessa, A. Sharpe, C. Sharr, S. Shin, N. Singh, S. Slaughenhaupt, K. S. Sheppard, W. Sun, X. Sun, E. Suschana, O. Talabi, H. Ticheli, S. T. Weiss, V. Wilson and A. Zhu, The Massachusetts Consortium for Pathogen, Nat. Commun., 2020, 11, 5493 CrossRef CAS.
- C. Prebensen, P. L. Myhre, C. Jonassen, A. Rangberg, A. Blomfeldt, M. Svensson, T. Omland and J.-E. Berdal, Clin. Infect. Dis., 2020 DOI:10.1093/cid/ciaa1338.
- K. V. Argyropoulos, A. Serrano, J. Hu, M. Black, X. Feng, G. Shen, M. Call, M. J. Kim, A. Lytle, B. Belovarac, T. Vougiouklakis, L. H. Lin, U. Moran, A. Heguy, A. Troxel, M. Snuderl, I. Osman, P. Cotzia and G. Jour, Am. J. Pathol., 2020, 190, 1881–1887 CrossRef CAS PubMed.
- P. Foladori, F. Cutrupi, N. Segata, S. Manara, F. Pinto, F. Malpei, L. Bruni and G. La Rosa, Sci. Total Environ., 2020, 743, 140444 CrossRef CAS.
- W. Randazzo, P. Truchado, E. Cuevas-Ferrando, P. Simón, A. Allende and G. Sánchez, Water Res., 2020, 181, 115942 CrossRef CAS PubMed.
- F. Xiao, M. Tang, X. Zheng, Y. Liu, X. Li and H. Shan, Gastroenterology, 2020, 158, 1831–1833 CrossRef CAS.
- H. Kawasuji, Y. Takegoshi, M. Kaneda, A. Ueno, Y. Miyajima, K. Kawago, Y. Fukui, Y. Yoshida, M. Kimura, H. Yamada, I. Sakamaki, H. Tani, Y. Morinaga and Y. Yamamoto, PLoS One, 2020, 15, e0243597 CrossRef CAS.
- P. Bustos, C. Tambley, A. Acevedo, W. Andrade, G. Leal, D. Vidal, F. Roldán, R. Fasce and E. Ramírez, J. Med. Virol., 2021, 93, 2439–2445 CrossRef CAS.
- S. Rao, D. Manissero, V. Steele and J. Pareja, J. Infect. Dis. Ther., 2020, 9, 573–586 CrossRef.
- Population-wide Testing of SARS-CoV-2: Country Experiences and Potential Approaches in the EU/EEA and the United Kingdom, https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-population-wide-testing-country-experiences.pdf, accessed 12 Feb, 2021 Search PubMed.
- Z.-D. Tong, A. Tang, K.-F. Li, P. Li, H.-L. Wang, J.-P. Yi, Y.-L. Zhang and J.-B. Yan, Emerging Infect. Dis., 2020, 26, 1052–1054 CrossRef CAS.
- W. E. Wei, Z. Li, C. J. Chiew, S. E. Yong, M. P. Toh and V. J. Lee, Morb. Mortal. Wkly. Rep., 2020, 69, 411–415 CrossRef CAS.
- G. Qian, N. Yang, A. H. Y. Ma, L. Wang, G. Li, X. Chen and X. Chen, Clin. Infect. Dis., 2020, 71, 861–862 CrossRef CAS PubMed.
- S. H. Bae, H. Shin, H.-Y. Koo, S. W. Lee, J. M. Yang and D. K. Yon, Emerging Infect. Dis., 2020, 26, 2705 CrossRef CAS.
- M. M. Arons, K. M. Hatfield, S. C. Reddy, A. Kimball, A. James, J. R. Jacobs, J. Taylor, K. Spicer, A. C. Bardossy, L. P. Oakley, S. Tanwar, J. W. Dyal, J. Harney, Z. Chisty, J. M. Bell, M. Methner, P. Paul, C. M. Carlson, H. P. McLaughlin, N. Thornburg, S. Tong, A. Tamin, Y. Tao, A. Uehara, J. Harcourt, S. Clark, C. Brostrom-Smith, L. C. Page, M. Kay, J. Lewis, P. Montgomery, N. D. Stone, T. A. Clark, M. A. Honein, J. S. Duchin, J. A. Jernigan, H. S. Public, C. King and C. C.-I. Team, N. Engl. J. Med., 2020, 382, 2081–2090 CrossRef CAS PubMed.
- M. Gandhi, D. S. Yokoe and D. V. Havlir, N. Engl. J. Med., 2020, 382, 2158–2160 CrossRef CAS PubMed.
- T. Q. M. Le, T. Takemura, M. L. Moi, T. Nabeshima, L. K. H. Nguyen, V. M. P. Hoang, T. H. T. Ung, T. T. Le, V. S. Nguyen, H. Q. A. Pham, T. N. Duong, H. T. Nguyen, D. N. Ngu, C. K. Nguyen, K. Morita, F. Hasebe and D. A. Dang, Emerging Infect. Dis., 2020, 26, 1624–1626 CrossRef.
- D. W. Al-Sadeq and G. K. Nasrallah, Int. J. Infect. Dis., 2020, 98, 372–380 CrossRef CAS.
- Y. Bai, L. Yao, T. Wei, F. Tian, D.-Y. Jin, L. Chen and M. Wang, JAMA, 2020, 323, 1406–1407 CrossRef CAS.
- C. Rothe, M. Schunk, P. Sothmann, G. Bretzel, G. Froeschl, C. Wallrauch, T. Zimmer, V. Thiel, C. Janke, W. Guggemos, M. Seilmaier, C. Drosten, P. Vollmar, K. Zwirglmaier, S. Zange, R. Wölfel and M. Hoelscher, N. Engl. J. Med., 2020, 382, 970–971 CrossRef PubMed.
- M. Cevik, M. Tate, O. Lloyd, A. E. Maraolo, J. Schafers and A. Ho, Lancet Microbe, 2021, 2, e13–e22 CrossRef CAS PubMed.
- M. Cevik, K. Kuppalli, J. Kindrachuk and M. Peiris, BMJ, 2020, 371, m3862 CrossRef.
- L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, M. Kang, Y. Song, J. Xia, Q. Guo, T. Song, J. He, H.-L. Yen, M. Peiris and J. Wu, N. Engl. J. Med., 2020, 382, 1177–1179 CrossRef PubMed.
- D. He, S. Zhao, Q. Lin, Z. Zhuang, P. Cao, M. H. Wang and L. Yang, Int. J. Infect. Dis., 2020, 94, 145–147 CrossRef CAS.
- K. K.-W. To, O. T.-Y. Tsang, W.-S. Leung, A. R. Tam, T.-C. Wu, D. C. Lung, C. C.-Y. Yip, J.-P. Cai, J. M.-C. Chan, T. S.-H. Chik, D. P.-L. Lau, C. Y.-C. Choi, L.-L. Chen, W.-M. Chan, K.-H. Chan, J. D. Ip, A. C.-K. Ng, R. W.-S. Poon, C.-T. Luo, V. C.-C. Cheng, J. F.-W. Chan, I. F.-N. Hung, Z. Chen, H. Chen and K.-Y. Yuen, Lancet Infect. Dis., 2020, 20, 565–574 CrossRef CAS.
- Y. Chen, A. H. Wang, B. Yi, K. Q. Ding, H. B. Wang, J. M. Wang, H. B. Shi, S. J. Wang and G. Z. Xu, Chinese journal of Epidemiology, 2020, 41, 667–671 CAS.
- X. Qiu, A. I. Nergiz, A. E. Maraolo, I. I. Bogoch, N. Low and M. Cevik, medRxiv, 2021, 27(4), 511–519 CAS.
- A. de Siqueira Alves Lopes, S. C. Fontes Vieira, R. Lima Santos Porto, V. Santana Santos, D. C. Fontes Leite, L. Eduardo Cuevas and R. Queiroz Gurgel, Trop. Med. Int. Health, 2021, 26, 115–119 CrossRef CAS PubMed.
- L. M. Yonker, A. M. Neilan, Y. Bartsch, A. B. Patel, J. Regan, P. Arya, E. Gootkind, G. Park, M. Hardcastle, A. St. John, L. Appleman, M. L. Chiu, A. Fialkowski, D. De la Flor, R. Lima, E. A. Bordt, L. J. Yockey, P. D'Avino, S. Fischinger, J. E. Shui, P. H. Lerou, J. V. Bonventre, X. G. Yu, E. T. Ryan, I. V. Bassett, D. Irimia, A. G. Edlow, G. Alter, J. Z. Li and A. Fasano, J. Pediatr., 2020, 227, 45–52 CrossRef CAS PubMed.
- K. Danis, O. Epaulard, T. Bénet, A. Gaymard, S. Campoy, E. Botelho-Nevers, M. Bouscambert-Duchamp, G. Spaccaferri, F. Ader, A. Mailles, Z. Boudalaa, V. Tolsma, J. Berra, S. Vaux, E. Forestier, C. Landelle, E. Fougere, A. Thabuis, P. Berthelot, R. Veil, D. Levy-Bruhl, C. Chidiac, B. Lina, B. Coignard, C. Saura and I. Team, Clin. Infect. Dis., 2020, 71, 825–832 CrossRef CAS.
- K. Shen, Y. Yang, T. Wang, D. Zhao, Y. Jiang, R. Jin, Y. Zheng, B. Xu, Z. Xie, L. Lin, Y. Shang, X. Lu, S. Shu, Y. Bai, J. Deng, M. Lu, L. Ye, X. Wang, Y. Wang and L. Gao, D. China National Clinical Research Center for Respiratory, B. C. National Center for Children’s Health, C. P. S. C. M. A. Group of Respirology, P. Chinese Medical Doctor Association Committee on Respirology, P. China Medicine Education Association Committee on, P. Chinese Research Hospital Association Committee on, P. Chinese Non-government Medical Institutions Association Committee on, C. o. C. s. H. China Association of Traditional Chinese Medicine, R. Medicine, C. o. C. s. S. M. China News of Drug Information Association and A. Global Pediatric Pulmonology, World J. Pediatr., 2020, 16, 223–231 CrossRef CAS PubMed.
- COVID-19 in Children and the Role of School Settings in Transmission – First Update, European Centre for Disease Prevention and Control, www.ecdc.europa.eu, 2020 Search PubMed.
- D. Buitrago-Garcia, D. Egli-Gany, M. J. Counotte, S. Hossmann, H. Imeri, A. M. Ipekci, G. Salanti and N. Low, PLoS Med., 2020, 17, e1003346 CrossRef CAS PubMed.
- O. Byambasuren, M. Cardona, K. Bell, J. Clark, M.-L. McLaws and P. Glasziou, Official Journal of the Association of Medical Microbiology and Infectious Disease Canada, 2020, 5, 223–234 CrossRef.
- A. M. Pollock and J. Lancaster, BMJ, 2020, 371, m4851 CrossRef.
- In Vitro Diagnostics EUAs – Antigen Diagnostic Tests for SARS-CoV-2, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-antigen-diagnostic-tests-sars-cov-2, accessed 20 July, 2021 Search PubMed.
- A. Prinzi, Screening versus Diagnostic Tests for COVID-19, What's the Difference?, https://asm.org/Articles/2020/December/Screening-Versus-Diagnostic-Tests-for-COVID-19,-Wh Search PubMed.
- COVID-19 Test Uses: FAQs on Testing for SARS-CoV-2, https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/covid-19-test-uses-faqs-testing-sars-cov-2, accessed 11 Feb, 2021 Search PubMed.
- J. Frnda and M. Durica, Infect Dis Rep, 2021, 13, 45–57 CrossRef.
- G. Guglielmi, Rapid Coronavirus Tests: a Guide for the Perplexed, https://www.nature.com/articles/d41586-021-00332-4 Search PubMed.
- G. Guglielmi, Fast Coronavirus Tests: what They Can and Can't Do, https://www.nature.com/articles/d41586-020-02661-2?utm_content=141196099%26utm_medium=social%26utm_source=facebook%26hss_channel=fbp-157821580915772, accessed 3 May 2021 Search PubMed.
- D. B. Larremore, B. Wilder, E. Lester, S. Shehata, J. M. Burke, J. A. Hay, M. Tambe, M. J. Mina and R. Parker, Sci. Adv., 2021, 7, eabd5393 CrossRef CAS PubMed.
- A. Fogarty, A. Joseph and D. Shaw, Lancet Respir. Med., 2020, 8, 1078–1080 CrossRef CAS PubMed.
- C. A. Hogan, M. K. Sahoo and B. A. Pinsky, JAMA, 2020, 323, 1967–1969 CrossRef CAS PubMed.
- G. A. Perchetti, K.-W. Sullivan, G. Pepper, M.-L. Huang, N. Breit, P. Mathias, K. R. Jerome and A. L. Greninger, J. Clin. Virol., 2020, 131, 104570 CrossRef CAS PubMed.
- N. Barak, R. Ben-Ami, T. Sido, A. Perri, A. Shtoyer, M. Rivkin, T. Licht, A. Peretz, J. Magenheim, I. Fogel, A. Livneh, Y. Daitch, E. Oiknine-Djian, G. Benedek, Y. Dor, D. G. Wolf and M. Yassour, medRxiv, 2020 DOI:10.1101/2020.10.16.20213405.
- Interim Guidance for Use of Pooling Procedures in SARS-CoV-2 Diagnostic, Screening, and Surveillance Testing, https://www.cdc.gov/coronavirus/2019-ncov/lab/pooling-procedures.html, accessed 11 Feb, 2021 Search PubMed.
- S. Deka and D. Kalita, J. Lab. Physicians, 2020, 12, 212–218 CrossRef CAS PubMed.
- E. Gupta, A. Padhi, A. Khodare, R. Agarwal, K. Ramachandran, V. Mehta, M. Kilikdar, S. Dubey, G. Kumar and S. K. Sarin, PLoS One, 2020, 15, e0236859 CrossRef CAS PubMed.
- Pooled Sample Testing and Screening Testing for COVID-19, https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/pooled-sample-testing-and-screening-testing-covid-19, accessed 12 Feb, 2021 Search PubMed.
- J. Garg, V. Singh, P. Pandey, A. Verma, M. Sen, A. Das and J. Agarwal, J. Med. Virol., 2021, 93, 1526–1531 CrossRef CAS PubMed.
- D. Aragón-Caqueo, J. Fernández-Salinas and D. Laroze, J. Med. Virol., 2020, 92, 1988–1994 CrossRef.
- Coronavirus (COVID-19) Update: FDA Issues First Emergency Authorization for Sample Pooling in Diagnostic Testing, https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-authorization-sample-pooling-diagnostic, accessed 11 Feb, 2021 Search PubMed.
- S. Lohse, T. Pfuhl, B. Berkó-Göttel, J. Rissland, T. Geißler, B. Gärtner, S. L. Becker, S. Schneitler and S. Smola, Lancet Infect. Dis., 2020, 20, 1231–1232 CrossRef CAS.
- A. de Salazar, A. Aguilera, R. Trastoy, A. Fuentes, J. C. Alados, M. Causse, J. C. Galán, A. Moreno, M. Trigo, M. Pérez-Ruiz, C. Roldán, M. J. Pena, S. Bernal, E. Serrano-Conde, G. Barbeito, E. Torres, C. Riazzo, J. L. Cortes-Cuevas, N. Chueca, A. Coira, J. M. Sanchez-Calvo, E. Marfil, F. Becerra, M. J. Gude, Á. Pallarés, M. L. Pérez Del Molino and F. García, Clin. Microbiol. Infect., 2020, 26, 1687.e1681–1687.e1685 Search PubMed.
- I. Yelin, N. Aharony, E. S. Tamar, A. Argoetti, E. Messer, D. Berenbaum, E. Shafran, A. Kuzli, N. Gandali, O. Shkedi, T. Hashimshony, Y. Mandel-Gutfreund, M. Halberthal, Y. Geffen, M. Szwarcwort-Cohen and R. Kishony, Clin. Infect. Dis., 2020, 71, 2073–2078 CrossRef CAS PubMed.
- S. Wacharapluesadee, T. Kaewpom, W. Ampoot, S. Ghai, W. Khamhang, K. Worachotsueptrakun, P. Wanthong, C. Nopvichai, T. Supharatpariyakorn, O. Putcharoen, L. Paitoonpong, G. Suwanpimolkul, W. Jantarabenjakul, P. Hemachudha, A. Krichphiphat, R. Buathong, T. Plipat and T. Hemachudha, J. Med. Virol., 2020, 92, 2193–2199 CrossRef CAS PubMed.
- B. Mishra, B. Behera, M. Mohanty, A. Ravindra and J. Ranjan, Lancet Infect. Dis., 2020, 20, 1233 CrossRef CAS.
- K. L. Lim, N. A. Johari, S. T. Wong, L. T. Khaw, B. K. Tan, K. K. Chan, S. F. Wong, W. L. E. Chan, N. H. Ramzi, P. K. C. Lim, S. L. Hakim and K. Voon, PLoS One, 2020, 15, e0238417 CrossRef CAS PubMed.
- R. Ben-Ami, A. Klochendler, M. Seidel, T. Sido, O. Gurel-Gurevich, M. Yassour, E. Meshorer, G. Benedek, I. Fogel, E. Oiknine-Djian, A. Gertler, Z. Rotstein, B. Lavi, Y. Dor, D. G. Wolf, M. Salton, Y. Drier, A. Klochendler, A. Eden, A. Klar, A. Geldman, A. Arbel, A. Peretz, B. Shalom, B. L. Ochana, D. Avrahami-Tzfati, D. Neiman, D. Steinberg, D. Ben Zvi, E. Shpigel, G. Atlan, H. Klein, H. Chekroun, H. Shani, I. Hazan, I. Ansari, I. Magenheim, J. Moss, J. Magenheim, L. Peretz, L. Feigin, M. Saraby, M. Sherman, M. Bentata, M. Avital, M. Kott, M. Peyser, M. Weitz, M. Shacham, M. Grunewald, N. Sasson, N. Wallis, N. Azazmeh, N. Tzarum, O. Fridlich, R. Sher, R. Condiotti, R. Refaeli, R. Ben Ami, R. Zaken-Gallili, R. Helman, S. Ofek, S. Tzaban, S. Piyanzin, S. Anzi, S. Dagan, S. Lilenthal, T. Sido, T. Licht, T. Friehmann, Y. Kaufman, A. Pery, A. Saada, A. Dekel, A. Yeffet, A. Shaag, A. Michael-Gayego, E. Shay, E. Arbib, H. Onallah, K. Ben-Meir, L. Levinzon, L. Cohen-Daniel, L. Natan, M. Hamdan, M. Rivkin, M. Shwieki, O. Vorontsov, R. Barsuk, R. Abramovitch, R. Gutorov, S. Sirhan, S. Abdeen, Y. Yachnin and Y. Daitch, Clin. Microbiol. Infect., 2020, 26, 1248–1253 CrossRef CAS PubMed.
- B. Abdalhamid, C. R. Bilder, E. L. McCutchen, S. H. Hinrichs, S. A. Koepsell and P. C. Iwen, Am. J. Clin. Pathol., 2020, 153, 715–718 CrossRef CAS PubMed.
- A. W. H. Chin, J. T. S. Chu, M. R. A. Perera, K. P. Y. Hui, H.-L. Yen, M. C. W. Chan, M. Peiris and L. L. M. Poon, Lancet Microbe, 2020, 1, e10 CrossRef CAS.
- Y. Liu, T. Li, Y. Deng, S. Liu, D. Zhang, H. Li, X. Wang, L. Jia, J. Han, Z. Bei, L. Li and J. Li, J Hosp Infect, 2021, 107, 105–107 CrossRef CAS PubMed.
- COVID-19 Surface Swabbing and Detection, https://www.als-testing.co.uk/services/new-covid-19-surface-swabbing-and-detection, accessed 11 Feb, 2021 Search PubMed.
- COVID-19 on Environmental Surfaces (Swab), https://www.eurofins.sg/our-services/covid-19-testing/covid-19-environmental-swab/, accessed 11 Feb, 2021 Search PubMed.
- F.-C. Jiang, X.-L. Jiang, Z.-G. Wang, Z.-H. Meng, S.-F. Shao, B. D. Anderson and M.-J. Ma, Emerging Infect. Dis., 2020, 26, 2162–2164 CrossRef CAS PubMed.
- S. W. X. Ong, Y. K. Tan, P. Y. Chia, T. H. Lee, O. T. Ng, M. S. Y. Wong and K. Marimuthu, JAMA, 2020, 323, 1610–1612 CrossRef CAS PubMed.
- L. Luo, D. Liu, H. Zhang, Z. Li, R. Zhen, X. Zhang, H. Xie, W. Song, J. Liu, Q. Huang, J. Liu, X. Yang, Z. Chen and C. Mao, PLoS Neglected Trop. Dis., 2020, 14, e0008570 CrossRef CAS PubMed.
- C. F. Yung, K.-Q. Kam, M. S. Y. Wong, M. Maiwald, Y. K. Tan, B. H. Tan and K. C. Thoon, Ann. Intern. Med., 2020, 173, 240–242 CrossRef PubMed.
- M. Suzuki, H. Kamiya, K. Okamoto, T. Yamagishi, K. Kakimoto, M. Takeda, S. Matsuyama, K. Shirato, N. Nao, H. Hasegawa, T. Kageyama, I. Takayama, S. Saito, T. Wakita, M. Ohnishi, K. Wada, R. Fujita, Y. Gu, N. Matsunaga, M. Sakaguchi, R. N. Taichi Tajima, N. Ohmagari, S. Takaya, H. Saito, K. Okinaka, M. Griffith, A. E. Parry, M. M. Plucinski, B. Barnetson and J. Leonard, medRxiv, 2020 DOI:10.1101/2020.05.02.20088567.
- J. Cai, W. Sun, J. Huang, M. Gamber, J. Wu and G. He, Emerging Infect. Dis., 2020, 26, 1343 CrossRef CAS.
- Surface Sampling of Coronavirus Disease (COVID-19): A Practical “How to” Protocol for Health Care and Public Health Professionals https://apps.who.int/iris/bitstream/handle/10665/331058/WHO-2019-nCoV-Environment_protocol-2020.1-eng.pdf, accessed 11 Feb, 2021 Search PubMed.
- G. Birgand, N. Peiffer-Smadja, S. Fournier, S. Kerneis, F.-X. Lescure and J.-C. Lucet, JAMA Network Open, 2020, 3, e2033232 CrossRef PubMed.
- J. A. Lednicky, M. Lauzardo, Z. H. Fan, A. Jutla, T. B. Tilly, M. Gangwar, M. Usmani, S. N. Shankar, K. Mohamed, A. Eiguren-Fernandez, C. J. Stephenson, M. M. Alam, M. A. Elbadry, J. C. Loeb, K. Subramaniam, T. B. Waltzek, K. Cherabuddi, J. G. Morris Jr and C.-Y. Wu, Int. J. Infect. Dis., 2020, 100, 476–482 CrossRef CAS PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins, J. W. O'Brien, P. M. Choi, M. Kitajima, S. L. Simpson, J. Li, B. Tscharke, R. Verhagen, W. J. M. Smith, J. Zaugg, L. Dierens, P. Hugenholtz, K. V. Thomas and J. F. Mueller, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Environ. Sci. Technol. Lett., 2020, 7, 511–516 CrossRef CAS.
- Y. Xu, X. Li, B. Zhu, H. Liang, C. Fang, Y. Gong, Q. Guo, X. Sun, D. Zhao, J. Shen, H. Zhang, H. Liu, H. Xia, J. Tang, K. Zhang and S. Gong, Nat. Med., 2020, 26, 502–505 CrossRef CAS PubMed.
- J. Zhang, S. Wang and Y. Xue, J. Med. Virol., 2020, 92, 680–682 CrossRef CAS PubMed.
- W. Zhen, E. Smith, R. Manji, D. Schron and G. J. Berry, J. Clin. Microbiol., 2020, 58, e00783-00720 Search PubMed.
- W. Zhen, R. Manji, E. Smith and G. J. Berry, J. Clin. Microbiol., 2020, 58, e00743-00720 Search PubMed.
- F. M. Liotti, G. Menchinelli, S. Marchetti, G. A. Morandotti, M. Sanguinetti, B. Posteraro and P. Cattani, Eur. J. Clin. Microbiol. Infect. Dis., 2020, 40, 269–277 CrossRef PubMed.
- C. F. Lowe, N. Matic, G. Ritchie, T. Lawson, A. Stefanovic, S. Champagne, V. Leung and M. G. Romney, J. Clin. Virol., 2020, 128, 104387 CrossRef CAS PubMed.
- K. Dust, A. Hedley, K. Nichol, D. Stein, H. Adam, J. A. Karlowsky, J. Bullard, P. Van Caeseele and D. C. Alexander, J. Virol. Methods, 2020, 285, 113970 CrossRef CAS PubMed.
- A. Moran, K. G. Beavis, S. M. Matushek, C. Ciaglia, N. Francois, V. Tesic and N. Love, J. Clin. Microbiol., 2020, 58, e00772-00720 CrossRef PubMed.
- H. H. Mostafa, J. Hardick, E. Morehead, J.-A. Miller, C. A. Gaydos and Y. C. Manabe, J. Clin. Virol., 2020, 130, 104578 CrossRef CAS PubMed.
- N. J. Beeching, T. E. Fletcher and M. B. J. Beadsworth, BMJ, 2020, 369, m1403 CrossRef PubMed.
- T. Nguyen, D. Duong Bang and A. Wolff, Micromachines, 2020, 11, 306 CrossRef.
- C. Sheridan, Nat. Biotechnol., 2020, 38, 515–518 CrossRef CAS.
- N. J. Brendish, S. Poole, V. V. Naidu, C. T. Mansbridge, N. J. Norton, H. Wheeler, L. Presland, S. Kidd, N. J. Cortes, F. Borca, H. Phan, G. Babbage, B. Visseaux, S. Ewings and T. W. Clark, Lancet Respir. Med., 2020, 8, 1192–1200 CrossRef CAS PubMed.
- C. P. Price and A. St John, BMJ Innov., 2021, 7, 3–5 CrossRef.
- N. M. Moore, H. Li, D. Schejbal, J. Lindsley and M. K. Hayden, J. Clin. Microbiol., 2020, 58, e00938-00920 CrossRef PubMed.
- A. Harrington, B. Cox, J. Snowdon, J. Bakst, E. Ley, P. Grajales, J. Maggiore and S. Kahn, J. Clin. Microbiol., 2020, 58, e00798-00720 CrossRef PubMed.
- V. D. Serei, R. Cristelli, K. Joho, G. Salaru, T. Kirn, M. O. Carayannopoulous and P. Uprety, Diagn. Microbiol. Infect. Dis., 2021, 99, 115208 CrossRef CAS PubMed.
- Customer Update on Our ID NOW COVID-19 Test, https://www.abbott.com/corpnewsroom/diagnostics-testing/customer-update-on-our-idnow-covid-19-test.html, accessed 11 Feb, 2021 Search PubMed.
- P. R. Lephart, M. A. Bachman, W. LeBar, S. McClellan, K. Barron, L. Schroeder and D. W. Newton, Diagn. Microbiol. Infect. Dis., 2021, 99, 115200 CrossRef CAS PubMed.
- M. C. Smithgall, I. Scherberkova, S. Whittier and D. A. Green, J. Clin. Virol., 2020, 128, 104428 CrossRef CAS.
- H. M. Creager, B. Cabrera, A. Schnaubelt, J. L. Cox, A. M. Cushman-Vokoun, S. M. Shakir, K. D. Tardif, M.-L. Huang, K. R. Jerome, A. L. Greninger, D. Drobysheva, U. Spaulding, M. Rogatcheva, K. M. Bourzac, S. H. Hinrichs, M. J. Broadhurst and P. D. Fey, J. Clin. Virol., 2020, 129, 104538 CrossRef CAS PubMed.
- E. J. Eckbo, K. Locher, M. Caza, L. Li, V. Lavergne and M. Charles, Diagn. Microbiol. Infect. Dis., 2021, 99, 115260 CrossRef CAS PubMed.
- C. A. Hogan, N. Garamani, A. S. Lee, J. K. Tung, M. K. Sahoo, C. Huang, B. Stevens, J. Zehnder and B. A. Pinsky, J. Clin. Microbiol., 2020, 58, e01072-01020 Search PubMed.
- A Study to Evaluate the Performance of the Lucira Health All-In-One COVID-19 Test Kit vs. Hologic Panther Fusion, https://clinicaltrials.gov/ct2/show/NCT04720794, accessed 11 Feb, 2021 Search PubMed.
- M. M. Gibani, C. Toumazou, M. Sohbati, R. Sahoo, M. Karvela, T.-K. Hon, S. De Mateo, A. Burdett, K. Y. F. Leung, J. Barnett, A. Orbeladze, S. Luan, S. Pournias, J. Sun, B. Flower, J. Bedzo-Nutakor, M. Amran, R. Quinlan, K. Skolimowska, C. Herrera, A. Rowan, A. Badhan, R. Klaber, G. Davies, D. Muir, P. Randell, D. Crook, G. P. Taylor, W. Barclay, N. Mughal, L. S. P. Moore, K. Jeffery and G. S. Cooke, Lancet Microbe, 2020, 1, e300–e307 CrossRef CAS.
- W. Feng, A. M. Newbigging, C. Le, B. Pang, H. Peng, Y. Cao, J. Wu, G. Abbas, J. Song, D.-B. Wang, M. Cui, J. Tao, D. L. Tyrrell, X.-E. Zhang, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 10196–10209 CrossRef CAS PubMed.
- E. Songok, Lancet Microbe, 2020, 1, e197 CrossRef CAS.
- J. P. Skittrall, M. D. Fortune, H. Jalal, H. Zhang, D. A. Enoch, N. M. Brown and A. Swift, Lancet Reg Health Eur, 2021, 1, 100002 CrossRef PubMed.
- A. J. Kucharski, P. Klepac, A. J. K. Conlan, S. M. Kissler, M. L. Tang, H. Fry, J. R. Gog, W. J. Edmunds, J. C. Emery, G. Medley, J. D. Munday, T. W. Russell, Q. J. Leclerc, C. Diamond, S. R. Procter, A. Gimma, F. Y. Sun, H. P. Gibbs, A. Rosello, K. van Zandvoort, S. Hué, S. R. Meakin, A. K. Deol, G. Knight, T. Jombart, A. M. Foss, N. I. Bosse, K. E. Atkins, B. J. Quilty, R. Lowe, K. Prem, S. Flasche, C. A. B. Pearson, R. M. G. J. Houben, E. S. Nightingale, A. Endo, D. C. Tully, Y. Liu, J. Villabona-Arenas, K. O'Reilly, S. Funk, R. M. Eggo, M. Jit, E. M. Rees, J. Hellewell, S. Clifford, C. I. Jarvis, S. Abbott, M. Auzenbergs, N. G. Davies and D. Simons, Lancet Infect. Dis., 2020, 20, 1151–1160 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |