Open Access Article
Open Access ArticleInvestigation of the mechanical effects of targeted drugs on cancerous cells based on atomic force microscopy
Jiajing
Zhu
 a,
Yanling
Tian
a,
Yanling
Tian
 *a,
Zuobin
Wang
*a,
Zuobin
Wang
 bc,
Ying
Wang
bc,
Ying
Wang
 bc,
Wenxiao
Zhang
bc,
Wenxiao
Zhang
 bc,
Kaige
Qu
bc,
Kaige
Qu
 bc,
Zhankun
Weng
bc,
Zhankun
Weng
 bc and
Xianping
Liu
bc and
Xianping
Liu
 *a
*a
aSchool of Engineering, University of Warwick, Coventry, CV4 7AL, UK. E-mail: Y.Tian.1@warwick.ac.uk; X.Liu@warwick.ac.uk
bInternational Research Centre for Nano Handling and Manufacturing of China, Changchun University of Science and Technology, Changchun, 130022, China
cMinistry of Education Key Laboratory for Cross-Scale Micro and Nano Manufacturing, Changchun University of Science and Technology, Changchun 130022, China
First published on 28th May 2021
Abstract
Cancer is currently drawing more and more attention as the leading factor in death worldwide. However, little research has been directed towards investigating the micro/nanoscale mechanical properties of cancer cells treated by targeted drugs to evaluate the model systems of targeted drugs using atomic force microscopy (AFM) nano-indentation, especially in light of the multiple drugs targeting various cancerous cells. This paper aims to compare the mechanical effects of sorafenib tosylate and osimertinib mesylate on hepatoma carcinoma cells and lung cancerous cells using atomic force microscopy from the perspective of a model system based on nano-indentation at the micro/nanoscale, which has rarely been investigated. The Sneddon model is applied to fit the force–distance curves, and the mechanical properties, i.e., Young's moduli, can then be calculated. For the SMMC-7721 cells, osimertinib mesylate is a more effective inhibitor than sorafenib tosylate. For the A549 cells, osimertinib mesylate and sorafenib tosylate both have an obvious inhibitory effect. The experimental results may make possible contributions to the diagnosis and treatment of early-stage cancers.
1. Introduction
Cancer is currently considered to be one of the leading causes of death worldwide. Among the various cancers, lung cancer has been recognized as having the highest incidence (11.6%) and mortality (18.4%), followed by liver cancer with a 8.2% death rate.1 Therefore, the detection of cancer and identification of drugs have become promising and urgent work in a variety of disciplines, especially the biomechanics field. Atomic force microscopy (AFM) has played an important role in measurement, because it represents a milestone in nanotechnology based on its powerful visualization and multifunction operation for measuring various materials, especially biological samples, in liquids or in the atmosphere at ambient temperature with (sub-)nanometre resolution.2–8There are many antineoplastic agents and propaedeutic potential drugs for the treatment of carcinomas, including resveratrol, paclitaxel, oridonin, docetaxel, colchicine, taxol, disulfiram, Celebrex, Totamine, TPA, valproic acid, among others. Some studies have focused on highlighting the effects of a single drug on a single cell type. Iturri et al. mainly quantified the temporal cytomechanical changes in cells exposed to a 50 μM concentration of resveratrol and concluded that both the Young's modulus and adhesion force decreased at 3 h, increased at 24 h and finally decreased at 48 h.9 In addition, the anticancer activity of oridonin toward living esophageal cancer KYSE-150 cells was evaluated using AFM-based single molecule force spectroscopy (AFM-SMFS).10 Another study combined multiple techniques to evaluate the effects of a single drug on a single cell type. Docetaxel was applied to stimulate HeLa cells, and the microtubule network in the HeLa cells was found to increase due to the drug effects via atomic force microscopy (AFM), transmission electron microscopy (TEM) and chiasmal targeted blotting techniques.11 In addition, comparisons between the effects of a single drugs on various cell types have been carried out. The effect of paclitaxel on the morphology, biophysical properties and apoptosis of Ishikawa and HeLa cells was evaluated using AFM, and the results showed that the Ishikawa cells exhibited more apparent changes compared to HeLa cells after paclitaxel treatment.12 Moreover, comparisons between the effects of different drugs on a single cell type have been outlined in previous research. In one investigation, U937 cancer cells were measured using AFM before and after the treatment with colchicine and Taxol, and it was found that their Young's moduli followed the order Ecolchicine < Econtrol < Etaxol.13 Another study based on AFM also obtained the result that the Young's modulus of PC-3 cells increased after the administration of eight drugs, including disulfiram, paclitaxel, MK-2206, Celebrex, BAY, Totamine, TPA, and valproic acid.14
In general, sorafenib and osimertinib are universal drugs for anticancer therapy in actual clinical treatment. Sorafenib has been regarded as a blocker that obstructs vascular endothelial growth factor (VEGF) ligands or vascular endothelial growth factor receptor (VEGFR) signalling pathways.15,16 Osimertinib, an FDA-approved epidermal growth factor receptor (EGFR) inhibitor, has been reported to have the potential function of inhibiting the MAPK pathway.17 However, little research has been directed towards the mechanical properties of cancer cells treated by targeted drugs to evaluate the model systems of targeted drugs using AFM nano-indentation at the micro/nanoscale. It is worth noting that an increasing amount of research focuses on the effect of only one drug on one type of cancerous cells which lacks the comparison of multiple targeted drugs on various types of cancerous cells. Therefore, it is essential to investigate the mechanical effects of targeted drugs on cancerous cells using AFM. This paper aims to compare the effects of two targeted drugs on the micro/nanoscale biomechanical properties of hepatoma carcinoma cells and lung cancer cells using atomic force microscopy nano-indentation in order to provide guidance for evaluating the effects of distinct drugs on different types of cancerous cells from the perspective of the model system.
2. Experimental details
2.1 Chemicals
RPMI1640 medium and fetal bovine serum (FBS) were supplied by Hyclone Laboratories Inc. (Canada). Dulbecco's modified Eagle medium (DMEM), trypsin and phosphate buffered saline (PBS) were obtained from Gibco (USA). Sorafenib tosylate was purchased from Bayer Pharma (Germany) and osimertinib mesylate was obtained from Astrazeneca (Sweden).2.2 Cell culture
The hepatoma carcinoma cells (SMMC-7721 cells) and lung cancer cells (A549 cells) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The SMMC-7721 cells were cultured in RPMI 1640 medium supplemented with 10% FBS while the A549 cells were cultivated in DMEM with 10% FBS. The prepared cells were grown in an incubator (IP610, Yamato, Japan) at 37 °C with 5% CO2. Before the experiment, diluted live cells were dispersed on thin glass sheets in the culture dish with cell densities of 1.0 × 104 cells for another 24 h of cultivation after centrifuging. In order to eliminate impurities and dead cells, PBS was used to wash the experimental samples three times, and fresh medium solutions were added.2.3 Cell viability assay
The inhibition rates of the targeted drugs on cell growth were determined using the MTT assay. A microplate spectrophotometer (Epoch 2, BioTek, USA) was used to detect the apoptosis of SMMC-7721 cells and A549 cells treated with the targeted drugs at an optical density (OD) value of 492 nm. Cells in the logarithmic growth phase were seeded into 96-well plates (Kangning Co., USA) with a density of 1.0 × 104 cells with a volume of 100 μL per well and placed in an incubator for 24 h at 37 °C with 5% CO2. 100 μL aliquots of the two targeted drugs with increasing concentrations (0, 3.90625, 7.8125, 15.625, 31.25, 62.5, 125, 250, 500, 1000 μmol mL−1) were added to individual wells for another 24 h of cultivation in the incubator. Therefore, the total volume of the cell culture medium containing the targeted drugs was 200 μL in each well. After 24 h of stimulation with the targeted drugs, 20 μL of MTT (Sigma-Aldrich, America) was added to each well for another 4 h of incubation. Following the removal of the supernatant liquid, 150 μL of dimethylsulfoxide (DMSO) was added to each well, and the inhibition rates of cell proliferation were examined by MTT assay.2.4 AFM analysis
A JPK AFM system (NanoWizard®3, Germany), was employed to record the morphological and mechanical properties of the live cells. QI mode was used to measure the morphology of the cells with 256 × 256 pixels, while force spectroscopy mode was applied to obtain the force–distance curves in the nano-indentation experiments. In order to remove the influence of different culture media and serum concentrations, PBS was applied as the medium solution during the measurements using the JPK system. Before measurements, the samples were washed with PBS three times in order to eliminate dead cells and floating impurities, and then new PBS solutions were added for the following measurement. To ensure the accuracy of the experimental results, it was also essential to avoid rupture or contamination of the probe. All experiments were carried out at room temperature on a super-clean bench.A Bruker MLCT A probe with a triangular silicon nitride tip (Bruker Corporation, Santa Barbara, CA) was utilized for the AFM nano-indentation experiments. The Bruker MLCT A probe has a nominal spring constant of 0.07 N m−1 and nominal radius of 20 nm. The sharp tip is suitable for closely tracking the morphological changes of the cells. The deflection sensitivity and spring constant were optimized by pushing on the same glass base in liquid without cells for calibration of the probe using thermal tuning. The deflection sensitivity and spring constant for the MLCT A probe were calculated to be 17.82 nm V−1 and 0.072 N m−1, respectively.
Using the JPK force spectroscopy mode, the force–distance curves were obtained by applying a stable force on the central area of cells in order to investigate the internal microstructural changes of the cells. The force–distance curves were measured using targeted drug concentrations of 0 μmol mL−1, 31.25 μmol mL−1, 62.5 μmol mL−1 and 125 μmol mL−1. Based on the AFM morphology results above, the stimulation time of the targeted drugs was finally set to 10 h for the nano-indentation test. Moreover, in order to exclude the influence of other measurement parameters, the measurement parameters of indentation force, indentation velocity, extend time, extract time and Z length were set to 1.0 nN, 2.0 μm s−1, 0.0 s, 0.0 s and 5.0 μm, respectively. The samples were cleaned with fresh PBS three times and finally, fresh PBS solutions were added at room temperature for the nano-indentation test. In order to ensure the validity and consistency of the experimental results, at least five force–depth curves were obtained for each cell and at least five cells were detected for each sample. The force–distance curves were simplified by selecting only five curves for drawing after removing the curves with random error.
2.5 Theoretical contact models
Essentially, the analysis and characterization of mechanical properties are closely related to the theoretical contact models. The fitting results of the mechanical properties may be influenced by the application of different theoretical contact models. Therefore, the selection of suitable theoretical models plays a significant role in the experimental results of the fitting. Theoretical contact models have been developed for decades since the Hertz model was proposed in 1881,18 such as the Sneddon,19 Bradley,20 JKR,21 DMT,22 M-D,23 CSLC,24–26 SLS,27–29 poroelastic30 and t-shell models.31The mechanical properties and behaviour of cells determined based on the AFM nano-indentation offer the possibility to explore the potential interior microstructure and character inside the cells. The force spectroscopy mode of the JPK instrument was applied in this experiment to characterize the mechanical properties of the cells, and their Young's moduli were calculated from the experimental force–depth curves by fitting them with the Sneddon model in case of the triangular probe. The equation of Sneddon model is expressed as:
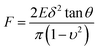 | (1) |
2.6 Data analysis
JPK SPM Data Processing software was implemented for baseline correction of the force–distance curves before the nano-indentation experiments. The morphology, height, length, width, adhesion and roughness results were obtained in QI mode (peak force tapping mode for quick imaging) during the scanning process using AFM. The Young's moduli were derived from the force–distance curves using the JPK force spectroscopy mode. The experimental results obtained from the AFM indentation were processed and extracted using JPK SPM Data Processing software. In the case of the triangular probe, the Hertz–Sneddon model was implemented for the analysis of the approach curve. Statistical analysis and graphical presentation were carried out using the software Origin Pro 9.0 and Microsoft Visio 2010. The software Matlab R2014a was used to fit the force curves obtained from the AFM nano-indentation experiments.3. Results and discussion
3.1 Cell viability
The inhibition rates of sorafenib tosylate and osimertinib mesylate on cell growth were determined by MTT assay. After cell cultivation with a density of 1.0 × 104 cells with 100 μL per well in 96 well plates for 24 h, 100 μL of sorafenib tosylate or osimertinib mesylate with increasing concentrations (0, 3.90625, 7.8125, 15.625, 31.25, 62.5, 125, 250, 500, and 1000 μmol mL−1) were added to the individual wells with for another 24 hours of cultivation in an incubator. Finally, the inhibition rates of the targeted drugs on cell growth were measured using a microplate spectrophotometer an OD value of 492 nm.The MTT experimental results (shown in Fig. 1) reflect the effects of sorafenib tosylate and osimertinib mesylate on the SMMC-7721 cells and A549 cells after stimulation for 24 h. Because the inhibition rates of the targeted drugs on cell growth were almost relatively stable above a concentration of 250 μmol mL−1, the results for concentrations of 500 μmol mL−1 and 1000 μmol mL−1 are omitted in Fig. 1. It can be observed that the survival rates of the SMMC-7721 cells and A549 cells both decrease with increasing concentrations of the targeted drugs. Thus, the inhibitory effects of sorafenib tosylate and osimertinib mesylate on the SMMC-7721 cells and A549 cells are demonstrated from the MTT assay results.
Additionally, the apparent inflection point of the decrease of the survival rate of the SMMC-7721 cells is 21.96% at 62.5 μmol mL−1 for sorafenib tosylate and 6.95% at 125 μmol mL−1 for osimertinib mesylate. For the A549 cells, the apparent inflection points of the decrease of the survival rate are 6.28% at 62.5 μmol mL−1 for sorafenib tosylate and 4.02% at 125 μmol mL−1 for osimertinib mesylate. Thus, sorafenib tosylate has a more inhibitory effect on A549 cells and a somewhat inhibitory effect on SMMC-7721 cells. Osimertinib mesylate has an obvious inhibitory effect on both SMMC-7721 cells and A549 cells. Hence, it can be concluded from the MTT assay results that osimertinib mesylate is more effective in inhibiting SMMC-7721 cells than sorafenib tosylate. However, osimertinib mesylate and sorafenib tosylate both have obvious inhibitory effects on A549 cells. The obtained knowledge will provide guidelines for specifying the effects of different drugs on different cancerous cell types in scientific research and clinical applications.
Furthermore, concentrations of 0 μmol mL−1, 31.25 μmol mL−1, 62.5 μmol mL−1 and 125 μmol mL−1 were selected for the following AFM measurements in order to analyse the morphological and mechanical changes before and after the irritation of SMMC-7721 cells and A549 cells with sorafenib tosylate and osimertinib mesylate.
3.2 Morphological and mechanical properties of cells
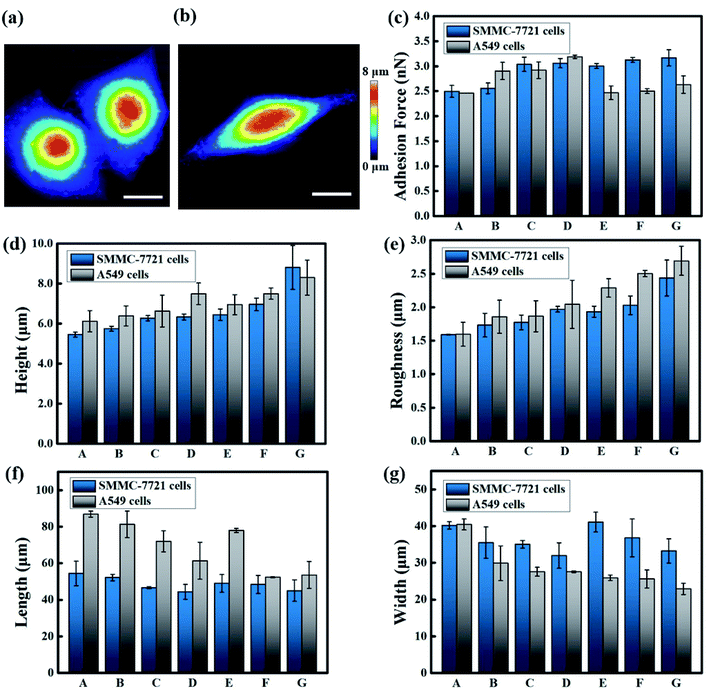
A JPK system (NanoWizard®3, Germany) was also utilized to characterize the effects of sorafenib tosylate and osimertinib mesylate on the morphological and mechanical properties of the cells. Based on the MTT assay results, four concentrations, namely, 0 μmol mL−1, 31.25 μmol mL−1, 62.5 μmol mL−1 and 125 μmol mL−1 were selected to analyse the morphological and adhesion changes before and after the irritation of the SMMC-7721 and A549 cells with sorafenib tosylate and osimertinib mesylate using the QI mode of AFM with 256 × 256 pixel imaging. In order to maintain consistency with the MTT assay results, the same concentrations of the targeted drugs were selected. Preliminary experiments with stimulation times of 10 h, 12 h, 24 h and 48 h were carried out in the early stage. However, the cells were almost in a state of suspension and became unsuitable for measurement at times longer than 10 hours according to the prior experimental results. For these reasons, the stimulation time was finally set to 10 h for the AFM test.Fig. 2–4 demonstrate the effects of the different concentrations of sorafenib tosylate and osimertinib mesylate on the morphology and adhesion characterizations based on the AFM images for the SMMC-7721 cells and A549 cells. The height map images, adhesion map images and cross-section images for the stimulation of the SMMC-7721 cells and A549 cells with sorafenib tosylate and osimertinib mesylate (with concentrations of 0 μmol mL−1, 31.25 μmol mL−1, 62.5 μmol mL−1 and 125 μmol mL−1) are presented in Fig. 2 and 3. The morphology and mechanical characterization results of over five samples are summarized in Fig. 4 based on the AFM images acquired by scanning in QI mode, which include the adhesion force, height, roughness, length and width changes.
As shown in Fig. 2, 3 and 4a, the SMMC-7721 cells exhibit an elliptical shape, whereas the A549 cells are spindle shaped in appearance. After stimulation with sorafenib tosylate and osimertinib mesylate, the lamellipodium on the edges of the cells were destroyed and gradually shrivelled and disappeared. As the concentrations of the drugs were increased, the filamentous and disordered ridge-like microstructure also became blurred and almost invisible. The shapes of the SMMC-7721 cells and A549 cells were more round in the drug groups with drug stimulation. Particularly for the SMMC-7721 cells, the morphology of the cells became oval under stimulation by sorafenib tosylate, while the morphology of cells became more circular with increased concentration of osimertinib mesylate. It can be speculated that osimertinib mesylate is more effective in inhibiting SMMC-7721 cells than sorafenib tosylate.
As the concentrations of sorafenib tosylate and osimertinib mesylate is increased, the length and width of the SMMC-7721 cells and A549 cells decrease. The highest length values are exhibited in the control groups, with values of 54.473 ± 6.667 μm for the SMMC-7721 cells and 86.880 ± 1.649 μm for the A549 cells, respectively. As the concentration of sorafenib tosylate is increased, the length values change to 52.162 ± 1.816 μm, 46.635 ± 0.625 μm, and 44.383 ± 4.023 μm for SMMC-7721 cells and 81.277 ± 7.255 μm, 71.990 ± 5.781 μm, and 61.387 ± 10.118 μm for the A549 cells. As the concentrations of osimertinib mesylate are increased, the length values change to 49.005 ± 4.859 μm, 48.405 ± 4.943 μm, and 45.012 ± 5.886 μm for SMMC-7721 cells and 77.925 ± 1.131 μm, 52.340 ± 0.269 μm, and 53.580 ± 7.389 μm for A549 cells, respectively. Likewise, an increasing trend also emerges for the width values with increasing concentrations of sorafenib tosylate and osimertinib mesylate. As the concentrations of sorafenib tosylate are increased, the width values change from 40.218 ± 1.004 μm for 0 μmol mL−1 to 35.520 ± 4.295 μm for 31.25 μmol mL−1, 35.067 ± 1.031 μm for 62.5 μmol mL−1, and 31.998 ± 3.481 μm for 125 μmol mL−1 for the SMMC-7721 cells while the width values vary from 40.445 ± 1.482 μm for 0 μmol mL−1 to 29.905 ± 4.732 μm for 31.25 μmol mL−1, 27.563 ± 1.216 μm for 62.5 μmol mL−1, and 27.560 ± 0.204 μm for 125 μmol mL−1 for the A549 cells, respectively. Moreover, the width values change to 41.105 ± 2.701 μm, 36.812 ± 5.163 μm, and 33.246 ± 3.310 μm for the SMMC-7721 cells and 25.930 ± 0.743 μm, 25.590 ± 2.458 μm, and 22.907 ± 1.523 μm for the A549 cells as the concentration of osimertinib mesylate is increased from 31.25 μmol mL−1 to 125 μmol mL−1.
Additionally, increasing concentrations of the two drugs cause the height of the nucleus area to increase gradually. The lowest height values are exhibited in the control group, with values of 5.450 ± 0.122 μm for the SMMC-7721 cells and 6.125 ± 0.531 μm for the A549 cells, respectively. As the concentrations of sorafenib tosylate are increased, the height values change to 5.750 ± 0.122 μm, 6.275 ± 0.132 μm, and 6.333 ± 0.147 μm for the SMMC-7721 cells and 6.387 ± 0.500 μm, 6.632 ± 0.794 μm, and 7.490 ± 0.544 μm for the A549 cells. As the concentrations of osimertinib mesylate are increased, the height values change to 6.447 ± 0.288 μm, 6.960 ± 0.317 μm, and 8.810 ± 1.094 μm for the SMMC-7721 cells and 6.950 ± 0.500 μm, 7.500 ± 0.283 μm, and 8.300 ± 0.874 μm for the A549 cells.
Additionally, it can be observed that the roughness and adhesion force of the SMMC-7721 cells and A549 cells both show increasing trends with increasing concentrations of sorafenib tosylate and osimertinib mesylate. The lowest roughness values are exhibited in the control groups for both the SMMC-7721 cells and A549 cells. As the concentration of sorafenib tosylate is increased, the roughness values change from 1.592 ± 0.005 μm for 0 μmol mL−1 to 1.732 ± 0.177 μm for 31.25 μmol mL−1, 1.773 ± 0.111 μm for 62.5 μmol mL−1, and 1.970 ± 0.042 μm for 125 μmol mL−1 for the SMMC-7721 cells while the values change from 1.596 ± 0.179 μm for 0 μmol mL−1 to 1.858 ± 0.246 μm for 31.25 μmol mL−1, 1.866 ± 0.233 μm for 62.5 μmol mL−1, and 2.044 ± 0.359 μm for 125 μmol mL−1 for the A549 cells. As the concentration of osimertinib mesylate is increased, the roughness values change to 1.935 ± 0.081 μm, 2.028 ± 0.140 μm, and 2.436 ± 0.269 μm for the SMMC-7721 cells and 2.288 ± 0.137 μm, 2.503 ± 0.047 μm, and 2.693 ± 0.218 μm for the A549 cells at concentrations of 31.25 μmol mL−1, 62.5 μmol mL−1, and 125 μmol mL−1. Moreover, the lowest adhesion force values are exhibited in the control group, with values of 2.497 ± 0.119 nN for the SMMC-7721 cells and 2.462 ± 0.001 nN for the A549 cells, respectively. As the concentration of sorafenib tosylate is increased, the adhesion force values change to 2.557 ± 0.107 nN, 3.038 ± 0.142 nN, and 3.062 ± 0.093 nN for the SMMC-7721 cells. For the A549 cells, the values change to 2.905 ± 0.171 nN, 2.92 ± 0.166 nN, and 3.190 ± 0.033 nN with increasing concentration of sorafenib tosylate. As the concentration of osimertinib mesylate is increased, the adhesion force values change to 3.004 ± 0.049 nN, 3.127 ± 0.049 nN, and 3.168 ± 0.162 nN for the SMMC-7721 cells. For the A549 cells, the values change to 2.466 ± 0.135 nN, 2.502 ± 0.048 nN, and 2.631 ± 0.174 nN with increasing concentration of osimertinib mesylate.
Based on previous research, the shrivelled morphology, membrane roughness and adhesion force are considered to be signals of cellular apoptosis.34–36 It is assumed that the inhibition effects of sorafenib tosylate and osimertinib mesylate induce the acceleration of the cell apoptosis process.
3.3 Fitting results
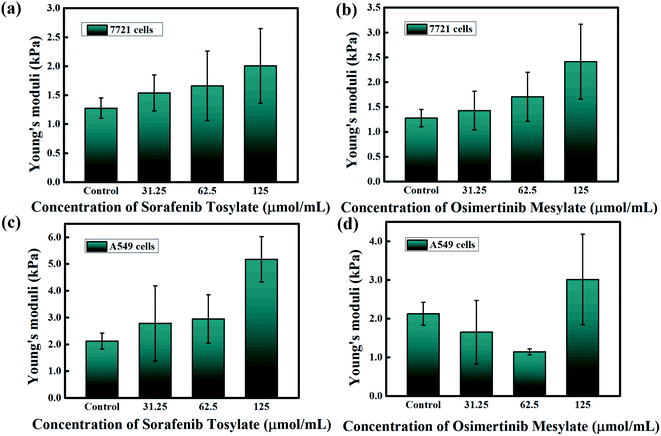
Nano-indentation tests are generally considered to be a popular tool to reveal the effects of drugs on the biophysical properties of cancerous cells. A probe with a triangular silicon nitride tip (MLCT-A, with a nominal spring constant of 0.07 N m−1) was utilized for the AFM nano-indentation experiments. Sinusoidal and the triangular waveforms are the most common waveforms used to modulate the tip-sample displacement, which might involve pre-loading steps and dwell times.38 Sinusoidal waveforms were applied for the force–distance curve measurements with a modulation frequency of 50 Hz using JPK force spectroscopy mode.The force–distance curves before and after the stimulation of the SMMC-7721 cells and A549 cells with sorafenib tosylate and osimertinib mesylate are presented in Fig. 5 and 6, respectively. In the case of the tip sharpness of triangular probes, the Sneddon model is applied to fit the approach force–distance curves and employed for the calculation of the Young's moduli. The Young's moduli were evaluated from the approach curves (shown as blue lines) in Fig. 5 and 6. As the concentrations of sorafenib tosylate and osimertinib mesylate were increased, the slopes of the force curves changed simultaneously. This phenomenon can be explained by the external stimulation with targeted drugs, which has been suggested to influence the cell stiffness with potential distribution variation of the actin filaments and cytoskeleton. For instance, chemotherapy exposure has been revealed to be a factor that increases leukemia cell stiffness.37 Although the force–distance graphs are intuitive, it is still difficult to compare the large number of force–distance curves. Thus, the final calculation results were extracted and are summarized in Fig. 7.
The Young's moduli derived from the force–distance curves based on the JPK force spectroscopy mode for the SMMC-7721 cells and A549 cells are shown in Fig. 7. The figure illustrates that the average Young's modulus values for the control groups of the SMMC-7721 cells and A549 cells are 1.276 ± 0.174 kPa and 2.122 ± 0.298 kPa, respectively. It can be observed that the Young's modulus of the SMMC-7721 cells increases with increasing concentrations of sorafenib tosylate and osimertinib mesylate. After the stimulation of the SMMC-7721 cells with sorafenib tosylate, their average Young's modulus varies from 1.276 ± 0.174 kPa for 0 μmol mL−1 to 1.53942 ± 0.3089 kPa for 31.25 μmol mL−1, 1.66058 ± 0.60221 kPa for 62.5 μmol mL−1, and 2.00379 ± 0.64333 kPa for 125 μmol mL−1, respectively. The effects of osimertinib mesylate on the SMMC-7721 cells result in a change in the average Young's modulus from 1.276 ± 0.174 kPa for 0 μmol mL−1 to 1.42821 ± 0.38798 kPa for 31.25 μmol mL−1, 1.7063 ± 0.49526 kPa for 62.5 μmol mL−1, and 2.41239 ± 0.75401 kPa for 125 μmol mL−1, respectively. The results indicate that the Young's modulus of the SMMC-7721 cells changed more significantly under stimulation with osimertinib mesylate. Thus, it can be speculated that the osimertinib mesylate is more effective in inhibiting SMMC-7721 cells than sorafenib tosylate, which is consistent with the MTT assay results.
For the A549 cells, the Young's modulus increases with increasing concentration of sorafenib tosylate, while the Young's modulus first decreases and then increases with increasing concentration of osimertinib mesylate. After stimulation of the A549 cells with sorafenib tosylate, the average Young's modulus varies from 2.122 ± 0.298 kPa for 0 μmol mL−1 to 2.78293 ± 1.40221 kPa for 31.25 μmol mL−1, 2.94556 ± 0.90315 kPa for 62.5 μmol mL−1, and 5.17427 ± 0.85226 kPa for 125 μmol mL−1, respectively. As the concentration of osimertinib mesylate is increased, the average Young's modulus of the A549 cells changes from 2.122 ± 0.298 kPa for 0 μmol mL−1 to 1.64879 ± 0.82043 kPa for 31.25 μmol mL−1, 1.1395 ± 0.0786 kPa for 62.5 μmol mL−1, and 3.01264 ± 1.16897 kPa for 125 μmol mL−1, respectively. The results demonstrate the influence of osimertinib mesylate and sorafenib tosylate on the Young's moduli of the A549 cells. Thus, osimertinib mesylate and sorafenib tosylate both have an obvious inhibitory effect on A549 cells, which is consistent with the MTT assay results. The Young's moduli of different cells under stimulation with different targeted drugs stimulation have different trends, which may because sorafenib has been regarded as a VEGF/VEGFR inhibitor while osimertinib is generally considered to be an EGFR inhibitor.15–17
4. Conclusions
In this paper, we focused mainly on comparing the mechanical effects of sorafenib tosylate and osimertinib mesylate on hepatoma carcinoma cells and lung cancer cells using atomic force microscopy in order to evaluate the model system of the targeted drugs based on nano-indentation experiments, which has rarely been discussed to date. The Sneddon model was applied to fit the force–distance curves, and then the mechanical properties, i.e., Young's moduli, could be calculated. The inhibitory effects of sorafenib tosylate and osimertinib mesylate on the SMMC-7721 cells and A549 cells are consistent with the MTT assay and AFM measurements. With increasing concentrations of the targeted drugs, the survival rates of both types of cancerous cells decrease. For the SMMC-7721 cells, osimertinib mesylate is more effective at inhibition than sorafenib tosylate. The Young's moduli of the SMMC-7721 cells increased with increasing concentrations of sorafenib tosylate and osimertinib mesylate. For the A549 cells, osimertinib mesylate and sorafenib tosylate both have an obvious inhibitory effect. The Young's moduli increase with increasing concentration of sorafenib tosylate, while the Young's moduli first decreases and then increases with increasing concentration of osimertinib mesylate. These results may make contributions to the early diagnosis and treatment of cancer.Abbreviations
| AFM | Atomic force microscopy |
| VEGF | Vascular endothelial growth factor |
| VEGFR | Vascular endothelial growth factor receptor |
| EGFR | Epidermal growth factor receptor |
| SMMC-7721 | Hepatoma carcinoma cells |
| A549 | Lung cancer cells |
| PBS | Phosphate buffered saline |
| FBS | Fetal bovine serum |
| DMSO | Dimethylsulfoxide |
| JKR model | Johnson–Kendall–Roberts model |
| DMT model | Derjaguin–Müller–Toporov model |
| M–D model | Maugis–Dugdale model |
| CSLC model | Cortical shell liquid core model |
| SLS model | Standard linear solid model |
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was supported by National Key R&D Program of China (No. 2017YFE0112100 and No. 2017YFB1104700), EU H2020 Program (MNR4SCell No. 734174), Program of International S&T Cooperation (No. 2016YFE0112100), Jilin Provincial Science and Technology Program (Nos. 20180414002GH, 20180414081GH, 20180520203JH, 20190702002GH and 20200901011SF) and “111 Project” of China (D17017).References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed
.
- G. Binnig, C. F. Quate and C. Gerber, Phys. Rev. Lett., 1986, 56, 930–934 CrossRef PubMed
.
- Y. F. Dufrêne, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin and A. Engel, Nat. Nanotechnol., 2017, 12, 295–307 CrossRef PubMed
.
- A. Alessandrini and P. Facci, Meas. Sci. Technol., 2005, 16, 65–92 CrossRef
.
- T. G. Kuznetsova, M. N. Starodubtseva, N. I. Yegorenkov, S. A. Chizhik and R. I. Zhdanov, Micron, 2007, 38, 824–833 CrossRef CAS PubMed
.
-
S. Moreno-Flores and J. L. Toca-Herrera, Hybridizing Surface Probe Microscopies Toward a Full Description of the Meso- and Nanoworlds, CRC Press, Boca Raton, 2012 Search PubMed
.
- G. Massimiliano, G. L. Tang, S. B. Chandra, J. L. Zhao, S. J. Chen and J. S. Florian, Nat. Commun., 2018, 9, 3584 CrossRef PubMed
.
- R. Garcia, Chem. Soc. Rev., 2020, 49, 5850–5884 RSC
.
- J. Iturri, A. Weber, A. Moreno-Cencerrado, M. D. Vivanco, B. Rafael and S. Leporatti, Int. J. Mol. Sci., 2019, 20, 13 Search PubMed
.
- J. Pi, H. Jin, J. Jiang, F. Yang, H. Cai and P. Yang, Pharmacol. Res., 2017, 119, 479–489 CrossRef CAS
.
- X. Yun, M. Tang, Z. Yang, J. J. Wilksch, P. Xiu and H. Gao, RSC Adv., 2017, 7, 43764–43771 RSC
.
- K. K. Sook, C. C. Hoon, P. E. Kuk, J. Min-Hyung, Y. Kyung-Sik and P. Hun-Kuk, PLoS One, 2012, 7, e30066 CrossRef
.
- M. S. Hung and M. F. Tsai, BioNanoScience, 2015, 5, 156–161 CrossRef
.
- J. Ren, H. Huang, Y. Liu, X. Zheng and Q. Zou, PLoS One, 2015, 10, 1–14 Search PubMed
.
- S. F. Razavi, F. F. Bamoharram, T. Hashemi, K. Shahrokhabadi and A. Davoodnia, Toxicol. In Vitro, 2020, 68, 104917 CrossRef CAS PubMed
.
- T. Kamba and D. M. McDonald, Br. J. Cancer, 2007, 96, 1788–1795 CrossRef CAS PubMed
.
- C. Chen, C. D. Cheng and H. Wu,
et al.
, Acta Pharmacol. Sin., 2021, 42, 108–114 CrossRef CAS PubMed
.
- H. Hertz, Journal für die reine und angewandte Mathematik, 1881, 92, 156–171 Search PubMed
.
- I. N. Sneddon, Int. J. Eng. Sci., 1865, 3(1), 47–57 CrossRef
.
- R. S. Bradley, The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science, 1932, 7, 853–862 CrossRef
.
- K. L. Johnson, K. Kendall and A. D. Roberts, Proc. R. Soc. London, Ser. A, 1971, 324, 301–313 CAS
.
- B. V. Derjaguin, V. M. Muller and Y. P. Toporov, J Colloid Interf Sci, 1975, 53, 314–326 CrossRef CAS
.
- D. Maugis, J Colloid Interf Sci, 1992, 150, 243–269 CrossRef CAS
.
- M. Krieg, Y. Arboleda-Estudillo, P. H. Puech, J. Kafer, F. Graner, D. J. Muller and C. P. Heisenberg, Nat. Cell Biol., 2008, 10, 429–436 CrossRef CAS PubMed
.
- E. B. Lomakina, C. M. Spillmann, M. R. King and R. E. Waugh, Biophys. J., 2004, 87, 4246–4258 CrossRef CAS PubMed
.
- M. J. Rosenbluth, W. A. Lam and D. A. Fletcher, Biophys. J., 2006, 90, 2994–3003 CrossRef CAS PubMed
.
- Y. M. Efremov, W. H. Wang, S. D. Hardy, R. L. Geahlen and A. Raman, Sci. Rep., 2017, 7, 1541 CrossRef PubMed
.
- J. S. De Sousa, J. A. C. Santos, E. B. Barros, L. M. R. Alencar, W. T. Cruz, M. V. Ramos and J. M. Filho, J. Appl. Phys., 2017, 121, 034901 CrossRef
.
- E. M. Darling, S. Zauscher and F. Guilak, Osteoarthritis and Cartilage, 2006, 14, 571–579 CrossRef CAS PubMed
.
- E. Moeendarbary, L. Valon, M. Fritzsche, A. R. Harris, D. A. Moulding, A. J. Thrasher, E. Stride, L. Mahadevan and G. T. Charras, Nat. Mater., 2013, 12, 253–261 CrossRef CAS PubMed
.
- I. L. Ivanovska, P. J. De-Pablo, B. Ibarra, G. Sgalari, F. C. MacKintosh, J. L. Carrascosa, C. F. Schmidt and G. J. L. Wuite, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7600–7605 CrossRef CAS PubMed
.
- C. Rianna and M. Radmacher, AIP Conf. Proc., 2016, 1760, 020057 CrossRef
.
- D. Kirmizis and S. Logothetidis, Int. J. Nanomed., 2010, 5, 137–145 CrossRef PubMed
.
- D. C. Wang, K. Y. Chen, C. H. Tsai, G. Y. Chen and C. H. Chen, J. Biomech., 2011, 44(16), 2790–2794 CrossRef PubMed
.
- P. Lazar, S. Zhang, S. Klára, Q. Li, J. P. Froning, J. Granatier, P. Hobza, R. Zboril, F. Besenbacher, M. D. Dong and M. Otyepka, ACS Nano, 2013, 7(2), 1646–1651 CrossRef CAS PubMed
.
- S. Xu, M. Dong, X. Liu, K. A. Howard, J. Kjems and F. Besenbacher, Biophys. J., 2007, 93(3), 952–959 CrossRef CAS PubMed
.
- W. A. Lam, M. J. Rosenbluth and D. A. Fletcher, Blood, 2007, 109(8), 3505–3508 CrossRef CAS PubMed
.
- R. Garcia, Chem. Soc. Rev., 2020, 49, 5850–5884 RSC
.
| This journal is © The Royal Society of Chemistry 2021 |