Open Access Article
Open Access ArticleNonspecific nuclear uptake of anti-MUC1 aptamers by dead cells: the role of cell viability monitoring in aptamer targeting of membrane-bound protein cancer biomarkers†
Shane Patrick
Flanagan
 a,
Ronen
Fogel
a,
Ronen
Fogel
 a,
Adrienne Lesley
Edkins
a,
Adrienne Lesley
Edkins
 b,
Lance St. John
Ho
b,
Lance St. John
Ho
 a and
Janice
Limson
a and
Janice
Limson
 *a
*a
aBiotechnology Innovation Centre, Rhodes University, Makhanda, South Africa. E-mail: j.limson@ru.ac.za
bBiomedical Biotechnology Research Unit (BioBRU), Department of Biochemistry and Microbiology, Rhodes University, Makhanda, South Africa
First published on 19th February 2021
Abstract
Most aptamers targeting cell-expressed antigens are intended for in vivo application, however, these sequences are commonly generated in vitro against synthetic oligopeptide epitopes or recombinant proteins. As these in vitro analogues frequently do not mimic the in vivo target within an endogenous environment, the evolved aptamers are often prone to nonspecific binding. The presence of dead cells and cellular debris further complicate aptamer targeting, due to their high nonspecific affinities to single-stranded DNA. Despite these known limitations, assessment of cell viability and/or the removal of dead cells is rarely applied as part of the methodology during in vivo testing of aptamer binding. Furthermore, the extent and route(s) by which dead cells uptake existing aptamers remains to be determined in the literature. For this purpose, the previously reported aptamer sequences 5TR1, 5TR4, 5TRG2 and S22 – enriched against the MUC1 tumour marker of the mucin glycoprotein family – were used as model sequences to evaluate the influence of cell viability and the presence of nontarget cell-expressed protein on aptamer binding to the MUC1 expressing human cancer cell lines MCF-7, Hs578T, SW480, and SW620. From fluorescence microscopy analysis, all tested aptamers demonstrated extensive nonspecific uptake within the nuclei of dead cells with compromised membrane integrities. Using fluorescent-activated cell sorting (FACS), the inclusion of excess double-stranded DNA as a blocking agent showed no effect on nonspecific aptamer uptake by dead cells. Further nonspecific binding to cell-membrane bound and intracellular protein was evident for each aptamer sequence, as assessed by southwestern blotting and FACS. These factors likely contributed to the ∼120-fold greater binding response of the 5TR1 aptamer to dead MCF-7 cells over equivalent live cell populations. The identification of dead cells and cellular debris using viability stains and the subsequent exclusion of these cells from FACS analysis was identified as an essential requirement for the evaluation of aptamer binding specificity to live cell populations of the cancer cell lines MCF-7, Hs578T and SW480. The research findings stress the importance of dead cell uptake and more comprehensive cell viability screening to validate novel aptamer sequences for diagnostic and therapeutic application.
Introduction
The mucin glycoprotein MUC1 is of clinical interest as both a diagnostic indicator and a therapeutic target in carcinoma and haematological malignancies.1 MUC1 expression is restricted to the apical surface of normal epithelia, and is present as a heterodimer of N- and C-terminal subunits.2,3 Following expression, the MUC1 heterodimer is repeatedly internalised and successively modified by the sequential glycosyltransferase addition of N- and O-linked glycans to its Variable Number of Tandem Repeat (VNTR) domain.4,5 After MUC1's maturation to a fully glycosylated state, re-internalisation continues until the MUC1 heterodimer is either degraded intracellularly, or the MUC1 N-terminal subunit is shed into the extracellular matrix.6,7Compared to normally-expressed MUC1, cancer-associated isoforms are characterised by their overexpression in malignant tissues, polarized distribution to basolateral cell membranes, and truncations in O-glycosylation during post-translational modification.8,9 These aberrant features of cancer-associated MUC1 are implicated in the survival, progression, and metastasis of tumours.8,10–13 Epitope exposure due to the altered O-glycosylation patterns in the extracellular VNTR domain of cancer-associated MUC1 has allowed for the generation of several antibodies specific to this isoform.14–21 Antibody-based detection of the MUC1 N-terminus shed into circulation is of prognostic value, and provides an indication of cancer occurrence, progression, and response to therapy.22–26
Similar to antibodies, aptamers represent a promising class of targeting vectors. Aptamers consist of synthetic DNA or RNA oligonucleotides able to fold into unique three-dimensional conformations exhibiting high affinity and specificity to a target ligand.27 Given the clinical utility of MUC1 as a tumour marker, several aptamers specific to the extracellular MUC1 VNTR have been previously reported.28–31 As with antibody generation against MUC1, the selection of MUC1-targeting aptamers via Systematic Evolution of Ligands by EXponential enrichment (SELEX) has relied on structural features of the VNTR which differentiate normal and cancer-associated MUC1 glycoforms.28–31
Ferreira and colleagues first described the isolation and selection of single-stranded DNA (ssDNA) aptamer sequences 5TR1, 5TR4, and S22 which bound to nonglycosylated oligopeptides containing the immunodominant proline–aspartate–threonine–arginine–proline (PDTRP) epitope of the MUC1 VNTR.28,29 Subsequently, ssDNA aptamers including 5TRG2 were reported to bind a VNTR peptide enzymatically O-glycosylated with N-acetylgalactosamine: this target was designed to generate aptamers specific to glycopeptide antigens exclusive to cancer-associated MUC1.30 The ability of the former aptamers to bind in vitro MUC1 analogues (nonglycosylated peptides of the MUC1 VNTR) with high affinity and specificity was demonstrated in several studies.32–35
However, unlike the uniform nature of in vitro targets, aptamer binding to cancer-associated MUC1 in vivo presents additional challenges. In particular, the complex O-glycosylation profile of the MUC1 VNTR results in a wide variety of glycoforms, many of which may prevent aptamer binding due to steric or conformational constraints.14,15,17,19–21,36–41 Additionally, other nontarget considerations may prevent aptamer binding e.g. nonspecific adherence to free genomic DNA or cell-membrane components of MUC1-expressing cells were also identified in in vivo studies.39,42 Despite these findings, the capability of the aptamers S22, 5TR1 and 5TRG2 to bind endogenous MUC1 have been proposed by aptamer internalisation within cancer cell lines expressing MUC1, and by their in vivo localisation to associated cancer cell line xenografts.30,43–53 While these studies support the specific recognition of cancer-associated MUC1, often by comparing aptamer binding to control cell lines lacking MUC1 expression,30,39,54,55 few reports specifically examine nonspecific aptamer uptake or nonspecific adherence to MUC1 expressing cells.
The disconnect between the condition of a target used in SELEX-based aptamer generation and the physiological state of the in vivo target is not unique to aptamers enriched against MUC1. While developments in cell-56 and crossover-SELEX57 produce aptamers with improved target binding specificity, overcoming the complexity of cell expressed targets remains a challenge. The vast majority of aptamers continue to be produced from more conventional peptide- or recombinant protein-based SELEX.58–60 These target analogues may differ substantially from their physiological counterparts in their folded conformations, post-translational modifications, and extra- or intracellular environments.61,62 Moreover, few studies42,61,63–66 determine aptamer binding affinity and specificity to the physiological target following SELEX. Under these circumstances, aptamers generated from synthetic oligopeptides or recombinant protein analogues may not recognise the intended target in vivo or may non-specifically adhere to surrounding cell and extracellular components.67
The heterogeneous nature of cell cultures may further complicate in vivo studies of aptamer binding to cell-expressed targets. Due to their high nonspecific affinity for ssDNA, dead cells may sequester large amounts of aptamers.68–71 While not well-understood, the mechanism of nonspecific uptake of aptamers by dead cells may relate to the compromised cell membrane integrity, which allows diffusing aptamers access to intracellular DNA-binding structures and molecules.71–73 Preparative removal of dead cells before aptamer analysis has been proposed; methods include cell sorting by flow cytometry,69 microbead removal of dead cells,70 and the addition of blocking agents to saturate nonspecific DNA-binding sites.67 While these methods are utilised during cell-SELEX, nonspecific aptamer adherence to dead cells is rarely acknowledged or accounted for in studies that examine aptamer affinity after SELEX enrichment.
The failure to evaluate the physiological relevance of aptamers can lead to inaccurate diagnoses and ineffective therapeutic targeting, further delaying their commercialisation and clinical use. Moreover, the use of heterogeneous cell samples, specifically those including even minor proportions of dead cells during binding assays, is a contributing factor for inaccurate estimates of binding affinity during aptamer-target screening. To address the above, this report details a case study examining the extent, route and cause of nonspecific uptake of aptamers by dead cells. Methods reported to minimise the influence of dead cell subpopulations in targeted cell samples were evaluated. Flow cytometry-based assays, those common in cell–aptamer studies, were utilised to determine the binding capabilities of aptamers to cell–membrane targets once dead cells are considered. To achieve these outcomes, this study evaluates previously characterised MUC1-targeting aptamers applied to MUC1-expressing human cancer cell lines.
Materials and methods
Preparation of aptamer and control oligonucleotides
All ssDNA aptamer sequences, including the randomized RND1 control, were obtained from Integrated DNA Technologies (USA) and sourced with HPLC purification. Oligonucleotides were resuspended to 100 μM in TE (10 mM Tris, 0.1 mM EDTA) buffer at pH 8.0, as per manufacturer instructions. All sequences were 68 nucleotides in length: each was comprised of identical primer binding sites denoted as P1: 5′-GAGACAAGAATAAACGCTCAA-3′ and P2: 5′-TTCGACAGGAGGCTCACAACAG-3′. The binding regions of individual sequences are presented in Table 1.28–30 Aptamer-conjugated 5′ reporter agents include biotin, which was visualised using streptavidin-conjugated horseradish peroxidase (streptavidin-HRP, N100, Thermo-Fisher Scientific, USA), and the fluorophores fluorescein isothiocyanate (FITC) and cyanine-5 (Cy5).| Aptamer | Binding region sequence (5′ → 3′) | Total GC (%) | ΔG° (kcal mol−1) |
|---|---|---|---|
| 5TR1 | P1-GAAGTGAAAATGACAGAACACAACA-P2 | 36 | −2.95 |
| 5TR4 | P1-TACTGCATGCACACCACTTCAACTA-P2 | 44 | −1.07 |
| 5TRG2 | P1-GGCTATAGCACATGGGTAAAACGAC-P2 | 46 | −4.37 |
| S22 | P1-GCAGTTGATCCTTTGGATACCCTGG-P2 | 52 | −7.31 |
| RND1 control | P1-CAACATATCATCACACAGTATAACA-P2 | 32 | −0.30 |
Before binding assays, all oligonucleotide sequences were prepared for binding as previously reported: dissolved in a 100 mM NaCl and 5 mM MgCl2 salt solution initially utilised for aptamer SELEX enrichment, heated to 95 °C for 5 minutes and thereafter gradually cooled to 25 °C.28–30 After heat denaturation and refolding, aptamer binding was evaluated at 25 °C in a 100 mM NaCl and 5 mM MgCl2 supplemented 10 mM sodium phosphate buffer pH 7.4 (denoted as SPB buffer) to maintain the MUC1 binding conformation of each aptamer.
Antibodies, proteins and peptides
Three primary antibodies were used in this study: SM3, an anti-human MUC1 N-terminal monoclonal primary antibody raised in mouse (Ab22711); MH-1, an Armenian hamster anti-human MUC1 C-terminal monoclonal primary antibody (Ab80952) both sourced from Abcam (USA); and the control M5284, a mouse IgG1 isotype control monoclonal antibody obtained from MilliporeSigma (USA). Where indicated, the primary antibodies were detected using species-specific fluorophore-labelled secondary antibodies: Dylight650-labelled goat anti-mouse IgG H&L (Ab96882), Dylight488-labelled goat anti-mouse IgG H&L (Ab96871) or FITC-labelled goat anti-Armenian hamster IgG H&L (Ab5739), from Abcam (USA).A synthetic oligopeptide with a sequence similar to the VNTR regions of the MUC1 glycoprotein (hereafter referred to as the MUC1 peptide) was sourced as a filter-sterilized 0.05 mg ml−1 stock in 1× PBS pH 7.4 (137 mM NaCl, 1 mM KH2PO4, 8 mM Na2HPO4, and 3 mM KCl) and 0.1 mM EDTA solution (Vault BioIndustries, ZA). This unglycosylated MUC1 peptide comprised three 20-amino acid repeat sequences of the MUC1 N-terminal VNTR, coupled to a hexahistidine tag at the N-terminus:
NH2-MGHHHHHHPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSAPDTRPAPG-COOH.
Cell lines and tissue culture
Adherent human breast carcinoma cell lines: MCF-7 (ATCC: HTB-22) and Hs578T (ATCC: HTB-126), and the human lung carcinoma cell line A549 (ATCC: CCL-185) were sourced from American Type Culture Collection (ATCC, USA). The human colon carcinoma cell lines: SW480 (ECACC: 87092801) and SW620 (ECACC: 87051203) were obtained from the European Collection of Authenticated Cell Cultures (ECACC, UK). MCF-7 and Hs578T cells were cultured in DMEM, supplemented with 5% v/v fetal calf serum (FCS) and 1% w/v penicillin–streptomycin (P/S) within vented culture flasks at 37 °C in 10% CO2. The SW480 and SW620 cells were cultured in RPMI growth media, supplemented with 10% v/v FCS and 1% w/v P/S in sealed culture flask at 37 °C, in normal air without additional CO2. Adherent cells were maintained by regular replacement of supplemented growth media until the cultures reached 80% confluency, at which point, the cells were subcultured to T75 tissue culture flasks (75 cm2, Corning, USA). All cell culture was carried out using aseptic technique in a class II safety cabinet and routinely monitored by PCR for the absence of mycoplasma infection.75Chemical reagents
Buffer salts were obtained from MilliporeSigma (USA) and Merck (DE), with solvent reagents from Merck (DE). Unless specified, all other reagent grade chemicals were obtained from MilliporeSigma (USA) and consumables from Eppendorf (DE).Enzyme-linked oligonucleotide assays (ELONA)
ELONAs were conducted similar to previous studies.76 Briefly, the surfaces of 96-well microtiter plate wells (Greiner BioOne, USA) were coated by applying 500 ng of the MUC1 peptide dissolved in 200 μl 100 mM NaHCO3 pH 8.5 to each well and incubating the plates at 4 °C overnight. Subsequently, plates were washed three times with Tris-Buffered Saline buffer (TBS, 25 mM Tris–HCl pH 7.4, 144 mM NaCl and 0.05% v/v Nonidet P-40, NP-40) and blocked with 5% w/v skim milk powder in TBS for 1 hour at 4 °C. After rinsing three times with TBS, plate wells were incubated with biotin-labelled aptamers for 2 hours at 25 °C in 100 μl of 100 mM NaCl and 5 mM MgCl2 solution. Aptamer additions of 1.5 pmol, 5 pmol, 25 pmol, 50 pmol, 100 pmol, and 150 pmol per well were tested. Wells were washed three times with TBS and incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of 1 mg ml−1 streptavidin-HRP (ThermoFisher, USA) in TBS for 2 hours at 25 °C. After a further three washes with TBS, aptamer binding was detected by monitoring the 450 nm absorbance of 3,3′,5,5′-tetramethylbenzidine (TMB, ThermoFisher, USA) oxidation, terminating the reaction after 15 minutes with 1 M H2SO4.
1000 dilution of 1 mg ml−1 streptavidin-HRP (ThermoFisher, USA) in TBS for 2 hours at 25 °C. After a further three washes with TBS, aptamer binding was detected by monitoring the 450 nm absorbance of 3,3′,5,5′-tetramethylbenzidine (TMB, ThermoFisher, USA) oxidation, terminating the reaction after 15 minutes with 1 M H2SO4.
Fluorescence microscopy
Fluorescent micrographs were acquired using a Zeiss Axiovert A1F1 LED epifluorescent microscope (Carl Zeiss Microscopy, DE), under 400× magnification. Sample fluorescence was visualised at emission/excitation wavelengths of 358 nm/461 nm (samples stained with 4,6-diamidino-2-phenylindole, DAPI) and 494 nm/518 nm (samples stained with Dylight 488- or FITC-conjugated reporters), respectively. For the quantification of fluorescent response, images were processed using ZEN 2 (Zen 2 version 2.0.0.0, Carl Zeiss Microscopy, DE) and ImageJ-Fiji software.77Each cell line was cultured to ∼30% confluency on the surfaces of sterile 18 mm glass coverslips in 12-well microplates. Prior to staining, the live cell samples adhered to the coverslip surface were gently washed twice with SPB. For the preparation of dead cell samples, cultured cells were fixed to coverslips by ice-cold methanol and air-dried.78,79 The fixed cells were washed twice with 10 mM phosphate buffer pH 7.4, supplemented with 100 mM NaCl, 5 mM MgCl2 and 0.05% w/v NP-40, denoted as SPB-N.
FITC-labelled aptamer localisation in live and dead MUC1-expressing cells
Both live and dead cell samples were incubated with 100 pmol FITC-labelled aptamers, dissolved in 1 ml SPB, for 2 hours at 25 °C. After three washes in SPB, live and dead cells were counterstained with 1 μg ml−1 DAPI in SPB, rinsed with SPB, and mounted to microscopy slides containing 1 μl Fluoroshield reagent (MilliporeSigma, USA).SM3 antibody localisation in dead MUC1-expressing cells
For antibody staining, fixed cells were first blocked with SPB–N containing 5% w/v BSA for 1 hour at 25 °C, washed twice with SPB–N and incubated with 1 μg (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution) of either the SM3 or isotype control antibody for 1 hour at 25 °C in SPB–N buffer supplemented with 0.1% w/v BSA. Cells were subsequently washed with 0.1% w/v BSA in SPB–N buffer before staining with Dylight488-labelled secondary antibody for 1 hour at 25 °C (1
500 dilution) of either the SM3 or isotype control antibody for 1 hour at 25 °C in SPB–N buffer supplemented with 0.1% w/v BSA. Cells were subsequently washed with 0.1% w/v BSA in SPB–N buffer before staining with Dylight488-labelled secondary antibody for 1 hour at 25 °C (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution). The antibody-stained cells were washed twice with SPB–N containing 0.1% w/v BSA before counterstaining with 1 μg ml−1 DAPI and mounted on microscopy slides containing Fluoroshield as above.
500 dilution). The antibody-stained cells were washed twice with SPB–N containing 0.1% w/v BSA before counterstaining with 1 μg ml−1 DAPI and mounted on microscopy slides containing Fluoroshield as above.
Southwestern blotting
Confluent cell samples were washed three times with 1× PBS pH 7.4 and detached from a T-75 culture flask by mechanical cell scraping. These were resuspended at 4 °C in 1 ml of RIPA buffer (50 mM Tris–HCl buffer, pH 7.6, containing 150 mM NaCl, 1% w/v NP-40 and 2% w/v SDS) and lysed by sonication at 4 °C for five cycles of a 30 second 60 W ultrasonication and 30 second pause. The lysed samples were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 × g for 15 minutes, with the entire clarified supernatant retained as lysate stored at −80 °C until use. Samples of 1 μl Pierce™ Prestained Protein Molecular weight marker (26612, Thermo-Fisher Scientific, USA) and 5 μl cell line lysates (1 mg ml−1) were heated to 95 °C for 10 minutes in Laemmli buffer, electrophoresed by SDS-PAGE on 15% w/v Tris-glycine pH 8.8 polyacrylamide gels, and transferred onto nitrocellulose at a constant current of 67 mA cooled to 4 °C for 2 hours.
500 × g for 15 minutes, with the entire clarified supernatant retained as lysate stored at −80 °C until use. Samples of 1 μl Pierce™ Prestained Protein Molecular weight marker (26612, Thermo-Fisher Scientific, USA) and 5 μl cell line lysates (1 mg ml−1) were heated to 95 °C for 10 minutes in Laemmli buffer, electrophoresed by SDS-PAGE on 15% w/v Tris-glycine pH 8.8 polyacrylamide gels, and transferred onto nitrocellulose at a constant current of 67 mA cooled to 4 °C for 2 hours.
Each membrane was incubated with a single biotin-labelled aptamer, using 10 ml of 0.1 μM solutions of the aptamer in SPB. Exposure to the aptamer solutions proceeded for 2 hours at 25 °C and rinsed three times with SPB. A membrane containing the electro-transferred cell lysates, but untreated with an aptamer, served as a negative control. Following exposure to the biotinylated aptamers, membranes were subsequently blocked with 5% w/v milk powder in SPB for 1 hour at 25 °C, washed three times with SPB–N, incubated with streptavidin-HRP (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 in SPB–N) for 1 hour at 25 °C, and washed three times with SPB–N. Aptamer staining with streptavidin-HRP was visualized by chemiluminescence using luminol (Western Blotting kit, Roche Applied Science, Switzerland).
5000 in SPB–N) for 1 hour at 25 °C, and washed three times with SPB–N. Aptamer staining with streptavidin-HRP was visualized by chemiluminescence using luminol (Western Blotting kit, Roche Applied Science, Switzerland).
Flow cytometry
Before flow cytometry assays, culture media was removed and the adherent cells washed with 1× PBS pH 7.4. Adherent cells were lifted using 10 mM EDTA in 1× PBS for 10 minutes at 37 °C, centrifuged at 400 × g for 2 minutes, and the supernatant discarded. Detachment by EDTA over conventional enzymatic treatment was used to preserve cell surface markers.80 The pelleted cells were resuspended to 106 cells per 200 μl sample in SPB. Where necessary, dead cell samples were prepared by fixing 106 cells in 1× PBS containing 0.01% v/v formaldehyde on ice for 10 minutes. The fixed cells were collected by centrifugation at 1800 × g and incubated in SPB containing 0.1% w/v Triton™-X100 at 4 °C for 15 minutes. After centrifugation at 1800 × g, permeabilised cells were resuspended in 200 μl SPB.After aptamer or antibody staining, 200 μl of the prepared cell suspensions in SPB were filtered through a cell strainer (35 μm pore, BD Biosciences, USA), retaining unicellular filtrates in BD Falcon flow cytometry tubes (BD Biosciences, USA). Flow cytometry assays were performed on a BD FACSAria II flow cytometer using FACSDiva acquisition software (BD Biosciences, USA). Cytometric data was analysed using Flowing software 2 (version 2.5.1, Turku Centre for Biotechnology University of Turku, Finland) and FlowJo software (version 10.0.7r2, Treestar Inc., USA). All cell suspensions were kept in the dark until their analysis no later than 2 hours after staining.
Distribution of 5TR1 aptamer binding between live and dead MCF-7 cells
Prepared 1 × 106 MCF-7 cell suspensions were centrifuged at 400 × g for 2 minutes and resuspended in 200 μl SPB containing 1 pmol, 10 pmol, 25 pmol, 50 pmol or 100 pmol additions of either Cy5-labelled 5TR1 aptamer or RND1 control sequences for 1 hour at 25 °C. Similarly, 1 μg of either the SM3 or IgG control antibodies were incubated with separate MCF-7 cell suspensions for 1 hour at 4 °C in 500 μl SPB. The primary antibody stained cells were washed three times with 1 ml SPB and incubated with 2.5 μg of the Dyl650-labelled goat anti-mouse IgG H&L secondary antibody for 1 hour at 4 °C in 500 μl SPB. Cell suspensions were washed three times with 1 ml SPB before data acquisition, whereby all cell events were collected until 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 live cells were counted.
000 live cells were counted.
Blocking efficacy of salmon sperm dsDNA on 5TR1 aptamer binding to MCF-7 cells
Salmon sperm double-stranded DNA (dsDNA, MilliporeSigma, USA) was ultrasonicated to within the oligonucleotide length of the MUC1-targeting aptamers,81 heat-denatured at 95 °C for 5 minutes and rapidly cooled to 4 °C to minimise re-annealing. The sheared salmon sperm dsDNA was prepared as 10 pmol, 100 pmol, 500 pmol, 1000 pmol, and 2000 pmol aliquots in 200 μl SPB. Each aliquot was incubated with 106 MCF-7 cells for 1 hour at 25 °C. After centrifugation at 400 × g to remove excess salmon sperm dsDNA, cells were incubated with 25 pmol of the Cy5-labelled 5TR1 or RND1 sequences for 1 hour at 25 °C in 200 μl SPB. After three 1 ml SPB wash steps, all cell events were collected until 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 live cells were counted.
000 live cells were counted.
Anti-MUC1 antibody and aptamer staining of live and dead cell fractions of MUC1-expressing cancer cell lines
After centrifugation at 400 × g, 106 cell suspensions were incubated with 250 pmol additions of Cy5-labelled 5TR1 aptamer or RND1 control, or FITC-labelled aptamers 5TR1, 5TR4, 5TRG2 or S22 for 1 hour at 25 °C in 200 μl SPB. Separate cell suspensions were stained by 1 μg of either the SM3 or IgG control antibody for 1 hour at 4 °C. The primary antibody stained cell samples were washed three times with SPB and incubated with 2.5 μg of the Dyl650-labelled goat anti-mouse IgG H&L secondary antibody for 1 hour at 4 °C. Under similar conditions, dead cell suspensions were used for MH-1 (1 μg) and anti-Armenian hamster IgG secondary control (2.5 μg) antibody staining. Following three 1 ml SPB wash steps and resuspension, data acquisition was separated into live and dead cell events. Except for the 4000 live cell events collected for SM3 and IgG control antibody stained cells, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 live cell events were collected for live cell analysis. For dead cells, all events were collected until 10
000 live cell events were collected for live cell analysis. For dead cells, all events were collected until 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dead cells were counted.
000 dead cells were counted.
Gating strategy to distinguish between live and dead cell subpopulations
Untreated (predominately live cells) and Triton X-100 permeabilised (dead cells) cell samples were stained with 1 μg ml−1 propidium iodide (PI) solution to distinguish between the live and dead cell subpopulations of each cancer cell line. Typically, the gate identifying live cells (PI-negative) was set to a fluorescent signal intensity of 1000 RFU; the dead cell gate (PI-positive) typically extended from 3000 RFU onwards. The FITC- and APC-positive gates were subsequently defined from the upper 95% percentile FITC and APC auto-fluorescence intensity of total live cell events from the untreated cell sample, determined using PI exclusion.To prevent potential PI intercalation with the ssDNA aptamers, PI was not used to co-stain cells with the MUC1-targeting aptamers or the RND1 control. Rather, live and dead cell subpopulations of aptamer or RND1 stained cells were identified from their distinct forward- and side-scattered light profiles inferred from the gates on untreated and Triton X-100 permeabilised cell samples stained with PI. The same procedure was used to define live and dead cell subpopulations of antibody stained cell samples. Following antibody, aptamer or RND1 control staining, the percentage of cells positive for FITC, Cy5 or Dyl650 fluorescence were determined from the number of live or dead cell events harbouring a fluorescence intensity greater than the above-defined FITC- and APC-positive gates. Separately, the median FITC, Cy5 or Dyl650 fluorescence intensity of antibody, aptamer or RND1 control stained live and dead cell subpopulations were determined from the total live and dead cell events respectively.
Statistical analysis
Statistical analysis was performed using Statistica 10 (Statsoft Inc, USA) and GraphPad Prism 8.4 (Graphpad Software Inc, USA) software. Where stated, experimental reproducibility is represented as the standard deviation from the mean and the median absolute deviation (where n = the number of independent measurements performed). Colocalization of DAPI and FITC fluorescence microscopy staining was quantified by Pearson's correlation coefficient, obtained by the Coloc 2 routine in ImageJ-Fiji.77 Pixel thresholds were auto-detected and validated according to Costes et al.82 Parametric Two-way ANOVA using Tukey–Kramer multiple comparison tests and nonparametric Kruskal–Wallis using Dunn's multiple comparison tests were used to evaluate significant differences (p < 0.05). The fluorescence intensities median, median absolute deviation and mean rank scores of Fig. 2 and 4 datasets are stated in Table S1 of the ESI.†Results and discussion
Nonspecific aptamer binding to extrinsic protein in ELONA
Among the published MUC1 aptamers, the sequences 5TR1, 5TR4, 5TRG2, and S22 were selected as each exhibit the highest reported binding affinity to the MUC1 peptide targets used in their respective SELEX enrichment.28,30 Despite reports of their broad ability to bind various glycoforms of MUC1, the selected aptamers were each only enriched to a single peptide analogue of the MUC1 VNTR.28–30 Reports detailing their subsequent use in aptasensor development against MUC1 also restrict their analysis to a single aptamer often with no consensus over the targeted MUC1 analogue.29,32,34,83–85In targeting MUC1-expressing cells, studies report high binding specificity by the selected aptamers and by association negligible nonspecific binding to extracellular or membrane-associated proteins. Yet, these findings are often cited without sufficient assessment relative to a nonbinding oligonucleotide or against peptides or whole proteins unrelated to the MUC1 VNTR.30,55,86
To cross-compare the published MUC1 aptamers, ELONA's using a nonglycosylated peptide analogue of the MUC1 VNTR as a target, and a milk powder blocking agent as a control were performed. The capability of the aptamers to bind endogenous MUC1 amongst unrelated cell-expressed protein was also investigated by southwestern blotting to denatured protein lysates of the cancer cell lines MCF-7, Hs578T, A549, SW480, and SW620. For both assays, the binding specificity of the MUC1-targeting aptamers was assessed relative to a randomized RND1 control sequence designed to share identical primer-binding sites but having no prior enrichment to MUC1. Fig. 1 summarises the colourimetric intensity of the assayed binding.
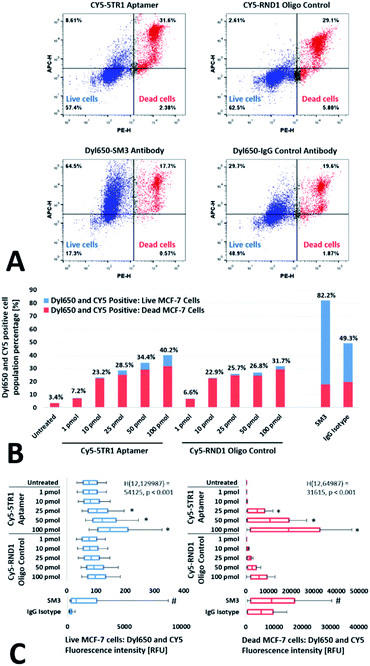
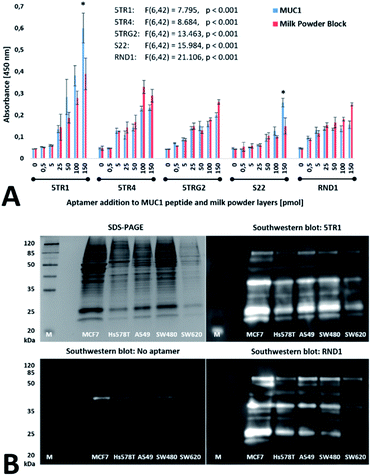 | ||
| Fig. 1 Aptamer and RND1 control binding to the MUC1 peptide and cell line lysates. (A) The ELONA response obtained for 5TR1, 5TR4, 5TRG2, S22 aptamer and RND1 control binding to MUC1 peptide (MUC1) and milk powder control (MP) layers. Significant binding by the biotin-labelled aptamers to wells containing the MUC1 peptide in comparison to the milk powder control layer only is denoted with an asterisk (*, p < 0.05). Error bars represent the standard deviation of the mean (n = 4). Two-way ANOVA tests were performed using the Tukey–Kramer post hoc test, with results from the two-way ANOVA annotated in the graph. (B) SDS-PAGE and corresponding southwestern blots of MCF-7, Hs578T, A549, SW480, and SW620 cell lysates using biotin-labelled 5TR1 aptamer, RND1 and no aptamer controls. Southwestern blots of cancer cell line lysates by the 5TR4, 5TRG2, and S22 aptamers shown in ESI as Fig. S1A–C.† | ||
As evaluated using ELONA, at 150 pmol of added aptamer the binding specificity (p < 0.001) of the 5TR1 (λ450 = 0.600 ± 0.069) and S22 (λ450 = 0.258 ± 0.019) aptamers to the MUC1 peptide was established relative to the milk powder control. Under the tested conditions, binding specificity to the MUC1 peptide was not observed for the aptamers 5TR4, 5TRG2, and the RND1 control sequence. All MUC1-targeting aptamers (and the RND1 control) bound to the milk powder coated surface in a concentration-dependent manner, denoted by a significant absorbance response above baseline (aptamer and RND1 additions ≥50 pmol, p < 0.001). These observations suggest the tested aptamers are capable of nonspecific binding to proteins other than MUC1. Furthermore, differences in MUC1 binding specificity between the tested aptamers was independent of their shared oligonucleotide length and primer binding sites, as was determined using the RND1 control.
When applied to cell-expressed protein, comparable binding between all aptamer sequences and the RND1 control was observed during southwestern blotting to each cancer cell line lysate (Fig. 1B and S1A–C†). Fewer luminescent bands were observed relative to the Coomassie-stained gel indicative of only a subset of the protein bands from each cell lysate stained by aptamer and RND1 blotting. Each resulted in multiple luminescent bands distinct from cell-expressed MUC1; the endogenous MUC1 protein harbouring a molecular weight >250 kDa for the mature glycoform (120–225 kDa nonglycosylated protein core).6,87–90 Native peroxidase activity of the cell lysates was accounted for by the aptamer- and RND1-free blot (Fig. 1B).
The propensity for extensive nonspecific binding by the MUC1-targeting aptamers to unrelated cell-expressed protein was evident by the high similarity in staining between southwestern blots of the aptamers 5TR1, 5TR4, 5TRG2, S22 and the RND1 control. This binding behaviour opposes previously reported studies detailing low cross-reactivity of the MUC1-targeting aptamers.66,91,92 However, the cell lysates used in southwestern blotting analysis contained both intracellular and cell membrane protein which poorly represents nonspecific binding sites accessible to aptamers targeting whole cells.
Distribution of 5TR1 aptamer binding to live and dead MCF-7 cells
To better resolve the capability of the MUC1-targeting aptamers to bind cell membrane expressed MUC1, flow cytometry was used to assess 5TR1 aptamer binding to MCF-7 cells relative to the RND1 control (Fig. 2A–C). Flow cytometry analysis incorporated PI as a cell viability stain to evaluate the distribution of 5TR1 aptamer binding between live (PI-negative) and dead (PI-positive) MCF-7 cells. The 5TR1 aptamer was selected based on findings of binding specificity from the MUC1 peptide ELONA (Fig. 1A).First, MUC1 expression by the MCF-7 cell line was confirmed by specific staining of the SM3 antibody in relation to an IgG antibody control (Fig. 2B and C). For the Cy5-labelled 5TR1 assay, standard additions of the aptamer resulted in a concentration-dependent increase in staining in the APC channel from 3.40% (0 pmol 5TR1) to 40.21% (100 pmol 5TR1) of the total MCF-7 cell population (live and dead cells). While approximately 70% of the total collected cell events consisted of live cells, most of the fluorescence response obtained after Cy5-labeled 5TR1 staining originated from the dead cell population. Specifically, dead MCF-7 cells which bound to the Cy5-labeled 5TR1 addition of 100 pmol comprised 31.60% of the total cell population with live MCF-7 cells the remaining 8.61% (Fig. 2A).
The significant difference in median Cy5 fluorescence intensity between 100 pmol 5TR1 stained live and dead cell populations further exemplified the disparity in 5TR1 aptamer binding to dead cells (p < 0.001, Fig. 2C). Similarly, more prominent binding to dead cells occurred throughout the tested 5TR1 aptamer concentrations at co-incubation conditions identical to the live MCF-7 cell fraction (Fig. 2A–C).
When compared to 5TR1, the 100 pmol addition of the Cy5-labeled RND1 control bound fewer live MCF-7 cells (2.61% total cell population) with near equivalent staining of the dead cell population (29.10% total cell population). The RND1 stained dead cells also showed a significantly greater median Cy5 fluorescence intensity than the RND1-bound live cells (p < 0.001, Fig. 2C). Disproportionate binding by the RND1 control indicates extensive nonspecific retention to dead cells independent of the MUC1 mucin. With high sequence identity to RND1, dead cell retention of the 5TR1 aptamer was thus attributed to nonspecific interactions.
The inclusion of salmon sperm dsDNA was subsequently assessed as a blocking agent to saturate nonspecific oligonucleotide binding sites.70,92,93 However, blocking with excess dsDNA did not prevent nonspecific binding by the 5TR1 aptamer or RND1 control to both live and dead MCF-7 cells (ESI, Fig. S2†). Sequence-dependent differences (at aptamer additions ≥25 pmol, p < 0.001) in dead cell retention were also noted between the 5TR1 aptamer and RND1 control. Given the lack of an effective blocking agent, this finding is problematic: without cell viability screening the sequence-dependent retention of aptamer sequences to dead cells may be misinterpreted as specific binding to the target even with the inclusion of a nonbinding oligonucleotide control within a study.
Nonspecific nuclear uptake of aptamers within dead cells
In flow cytometry, PI staining defined the dead cell population by a compromised plasma membrane integrity. While this increased membrane permeability allows access to intracellular DNA binding sites, the cause of extensive nonspecific aptamer binding to dead cells remains to be elucidated. To elaborate on dead cell retention, localization of MUC1-targeting aptamer binding was evaluated between live and dead cells of the cancer cell lines MCF-7, Hs578T, SW480, and SW620 by fluorescence microscopy (Fig. 3). Methanol fixation and NP-40 surfactant treatment were used to disrupt the plasma membrane integrity to yield dead permeable cells.94The FITC fluorescence of live (non-fixed) cells stained using the FITC-labelled 5TR1 aptamer was evenly distributed throughout each cell extending beyond the nuclear periphery. Staining each cancer cell line by the MUC1-specific SM3 antibody produced a similar fluorescence profile (ESI, Fig. S3E†). These observations support 5TR1 aptamer binding localised to the plasma membrane of live cells, attributable to complex formation with membrane-bound MUC1. In contrast, FITC-labeled 5TR1 aptamer binding to dead (MeOH fixed) cells was primarily localised to the DAPI-stained nuclei. When compared to live cell samples, an increase in the Pearson's correlation coefficient was attributed to the overlap of DAPI and FITC-aptamer staining within the nuclei of dead cells (Fig. 3C). As aptamer binding to live and dead cells was evaluated under identical conditions, aptamer diffusion through the perforated plasma membrane of dead cells resulted in internalization and subsequent nuclear uptake.
While distinct nuclear bodies were not resolved, aptamer localisation in dead cells appeared to concentrate within the optically-dense nucleoli (Fig. 3C and D, dead cells), additionally characterised by a low DAPI fluorescence intensity compared to the rest of the nucleus.95,96 Similar localisation was observed for the FITC-labelled aptamers 5TR4, 5TRG2, and S22 to each of the live and dead populations of the cancer cell lines MCF-7, Hs578T, SW420, and SW620 (ESI, Fig. S3A–D†). This corresponds to previous fluorescence microscopy studies which noted the uptake of short sequences of ssDNA (<100 nucleotide length) into the nuclei of fixed cells.68 However, these findings are not applied in the assessment of ssDNA aptamer binding to cell-expressed targets.
Nuclei localisation of FITC-labelled SM3 and IgG control antibodies to dead cell samples was not observed (ESI, Fig. S3E†). Consequently, the increased nuclei fluorescence evident in dead cell samples following aptamer addition was not attributed to previously reported artefacts of methanol fixation, elevated autofluorescence of dead cells, nor the hydrophobic association of the FITC fluorophore to intracellular structures.94,97 Hence, aptamer localisation to the nuclei of dead cells is attributed to distinct nonspecific interactions between the nucleic acid of the aptamers and nuclear structures within the cell, particularly the nucleolus, occurring across all tested cancer cell lines.
Aptamer binding to cancer cell lines by dead cell exclusion
With the necessary displacement of adherent cells for flow cytometry analysis, disruption of the cell membrane integrity is largely unavoidable. Consequently, nonspecific uptake of the MUC1-targeting aptamers in dead cells of a compromised membrane integrity effectively masks their binding affinity to cell-expressed MUC1. To counter such shortcomings, flow cytometry utilised cell viability and membrane integrity screening by PI staining to collect and analyse MUC1-targeting aptamer binding to live cell events separate from dead cells and cellular debris (Fig. 4).The expression of MUC1 by the cancer cell lines MCF-7, Hs578T, SW480, and SW620 was confirmed by FITC-labeled MH-1 antibody staining of the MUC1 C-terminal, independent of the MUC1 VNTR O-glycosylation state. Staining by the MH-1 antibody was apparent by the significant increase in median FITC fluorescence of MH-1 antibody-bound cells as compared to those only stained with the secondary antibody control (p < 0.001, Fig. 4D). High-level MUC1 expression by the cell lines MCF-7 and SW620 are in accordance with the literature,55,88,98–103 although mixed consensus exists relating immunochemical and qRT-PCR mRNA detection of MUC1 in Hs578T and SW480 cells with reports stating either negative or low-level MUC1 expression.100,104–107
In SM3 antibody staining of the MUC1 VNTR, a significant increase in median Dyl650 fluorescence intensity relative to the IgG isotype control was only observed for live MCF-7 and SW620 cells (p < 0.001, Fig. 4C). With MUC1 expression sufficient for MH-1 antibody detection, no apparent SM3 antibody binding specificity to Hs578T and SW480 cells was attributed to a MUC1 VNTR O-glycosylation state which disrupts SM3 antibody recognition.21,108,109 Accordingly, the differential binding response between the antibodies SM3 and MH-1 reflects the VNTR O-glycosylation complexity of the MUC1 expressed between the tested cancer cell lines.20,21,38
The exclusion of dead cells, those identified as having a compromised plasma membrane integrity through PI staining, allowed for the resolution of Cy5-labelled 5TR1 aptamer binding specificity to live MCF-7, Hs578T, and SW480 cells. Binding specificity was apparent by the significant increase in median Cy5 fluorescence response of Cy5-labelled 5TR1 binding in relation to equivalent additions of the Cy5-labelled RND1 control (p < 0.001, Fig. 4A). In contrast, a comparable binding response was observed between the Cy5-labelled 5TR1 and RND1 control to live SW620 cells. With MUC1 expression confirmed by MH-1 and SM3 antibody staining to SW620 cells, the fluorescence response was attributed to peptide epitope availability and localised steric or conformational constraints exerted on 5TR1-MUC1 complex formation resulting from the MUC1 VNTR O-glycan profile of the SW620 cell line.20,21,55,98,110,111
Similarly, sensitivity to O-glycan related constraints may explain the binding affinity differences observed between the FITC-labeled aptamers to cell-expressed MUC1. When compared to the FITC-labelled 5TR1 aptamer, equivalent additions of the FITC-labelled 5TR4 aptamer produced a significantly greater median FITC fluorescence response in live cancer cells (p < 0.001, Fig. 4B). Separately, the median FITC fluorescence response following binding by the FITC-labelled aptamers 5TRG2 and S22 to live cells of the cancer cell lines was significantly lower than the FITC-labelled aptamers 5TR1 and 5TR4 (p < 0.001, Fig. 4B).
While the complexity of MUC1 O-glycosylation sets inherent limitations on the identification of specific O-glycan configurations responsible for aptamer binding to each cancer cell line,112–114 key differences in the binding response between the MUC1-targeting aptamers were resolved by accounting for nonspecific binding to dead cells and cellular debris. The observed binding response allows for further insights into the mechanism of aptamer-MUC1 recognition, and the impact of post-translational modification on aptamer binding performance to cancer-associated MUC1.
Conclusions
Intracellular accumulation of the MUC1-targeting aptamers to membrane-compromised dead cells was shown to far exceed aptamer binding to the cell plasma membrane of intact live cells. When compared to an equivalent number of live cells, flow cytometry analysis of dead cells showed a ∼120-fold greater Cy5 fluorescence response following 100 pmol Cy5-labeled 5TR1 aptamer binding to MCF-7 cells. Extensive nonspecific adherence to dead cells is consistent with cell-SELEX studies and occurred despite the high MUC1 binding affinity reported for each tested aptamer.28,30,69,70Elaborating on previous cell-SELEX studies, most nonspecific binding by the MUC1-targeting aptamers to dead cells was found to accumulate in the cell nucleus and related nuclear bodies, particularly to the nucleolus. As SM3 antibody staining confirmed no localisation of the MUC1 VNTR within the nucleus, aptamer uptake within the cell interior was indicative of nonspecific aptamer complexation to nuclear protein, RNA and/or DNA.39,115 While no distinction was made between nuclear, intracellular or membrane protein during southwestern blotting, multiple band staining identical to the RND1 control confirmed nonspecific aptamer binding to numerous proteins distinct from the MUC1 mucin.
Though preservation of the cell membrane is necessary to prevent access to intracellular nonspecific binding sites, disruption of the cell membrane integrity is considered unavoidable during adherent cell detachment and isolation. Even without detachment and following aptamer co-incubation directly to the tissue culture flask, nonspecific uptake would persist due to necrotic cells embedded within the adherent cell monolayer.70 Lacking an effective blocking agent, the inclusion of cell viability monitoring during FACS analysis was, therefore, necessary to evaluate aptamer binding events to cancer-associated MUC1 on live cells separate from nonspecific binding to dead cells and cellular debris. Additionally, nonspecific aptamer binding along the plasma membrane of live cells further necessitated the design and inclusion of the RND1 control sequence to identify MUC1-specific aptamer binding.
By incorporating the above findings, the MUC1 binding specificity of the 5TR1 aptamer in relation to the RND1 control was demonstrated towards live cell populations of the cancer cell lines MCF-7, Hs578T, and SW480. Constraints on MUC1 recognition were only apparent after accounting for nonspecific 5TR1 adherence to the plasma membrane of live cells, and the exclusion of dead cell uptake. Furthermore, aberrant VNTR O-glycosylation expressed MUC1 was potentially identified by the lack of binding specificity of 5TR1 to the SW620 cell line.20,21,98,110,111,116
In fluorescence microscopy analysis, the endocytic mechanisms responsible for MUC1 trafficking were halted during methanol fixation. Coupled to the loss of plasma membrane integrity of methanol-fixed cells, aptamer staining of the cell nucleus noted in this study and other literature31,44,46,51,53,54,86,117 is instead attributed to passive diffusion and intracellular nonspecific accumulation44,51,72,73,86,117 as opposed to reports of aptamer-MUC1 complex formation and endocytic cycling between the cell plasma membrane and Golgi.29,30,47,48,53–56,118–123 While no nuclear localisation of the MUC1 aptamers was observed in live cells, indiscriminate association to extrinsic transmembrane protein may still allow for intracellular uptake of the aptamers through endocytic pathways analogous to MUC1.124–128 Consequently, intracellular uptake of the tested aptamers provides no indication of their binding specificity to MUC1 without the inclusion of a nonbinding control sequence, ssDNA pool, or a MUC1 negative control cell line.31,45,51,86,129
Overall, extensive nonspecific binding to extrinsic cellular protein distinct from cancer-associated MUC1 and associated uptake to dead cells and cellular debris are important limitations in the targeting capabilities of the MUC1 aptamers. The identified shortfalls of the MUC1-targeting aptamers illustrate the importance of more comprehensive aptamer selection methodology. In particular, the consideration of dead cell uptake as a principal contributor of nonspecific aptamer binding towards cell-expressed targets. Accounting for the influence of cell viability on aptamer-analyte recognition may improve the physiological relevance of existing and novel aptamer sequences for diagnostic and therapeutic application.
Author contributions
Shane Patrick Flanagan: Conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft and writing – review & editing. Ronen Fogel: Conceptualization, formal analysis, methodology, project administration, supervision, writing – review & editing. Adrienne Lesley Edkins: Formal analysis, methodology, supervision, writing – review and editing. Lance St. John Ho: Formal analysis, writing – review and editing. Janice Limson: Conceptualization, funding acquisition, project administration, supervision, writing – review and editing.Conflicts of interest
All authors declare that they have no conflicts of interest. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.Acknowledgements
This work was supported wholly or in part by funding from the Department of Science and Innovation/Mintek Nanotechnology Innovation Centre and by the National Research Foundation of South Africa (grant number 95319 and 98566).Notes and references
- M. A. Cheever, J. P. Allison, A. S. Ferris, O. J. Finn, B. M. Hastings, T. T. Hecht, I. Mellman, S. A. Prindiville, J. L. Viner, L. M. Weiner and L. M. Matrisian, Clin. Cancer Res., 2009, 15, 5323–5337 CrossRef.
- F. Levitin, O. Stern, M. Weiss, C. Gil-Henn, R. Ziv, Z. Prokocimer, N. I. Smorodinsky, D. B. Rubinstein and D. H. Wreschner, J. Biol. Chem., 2005, 280, 33374–33386 CrossRef CAS.
- B. Macao, D. G. a Johansson, G. C. Hansson and T. Härd, Nat. Struct. Mol. Biol., 2006, 13, 71–76 CrossRef CAS.
- S. Parry, F. G. Hanisch, S. H. Leir, M. Sutton-Smith, H. R. Morris, A. Dell and A. Harris, Glycobiology, 2006, 16, 623–634 CrossRef CAS.
- D. J. Gill, H. Clausen and F. Bard, Trends Cell Biol., 2011, 21, 149–158 CrossRef CAS.
- Y. Altschuler, C. L. Kinlough, P. A. Poland, J. B. Bruns, G. Apodaca, O. A. Weisz and R. P. Hughey, Mol. Biol. Cell, 2000, 11, 819–831 CrossRef CAS.
- X. Liu, Z. Yuan and M. Chung, Biochem. Biophys. Res. Commun., 2008, 376, 688–693 CrossRef CAS.
- D. W. Kufe, Cancer Biol. Ther., 2009, 8, 1197–1203 CrossRef CAS.
- M. A. Hollingsworth and B. J. Swanson, Nat. Rev. Cancer, 2004, 4, 45–60 CrossRef CAS.
- S. Cascio and O. Finn, Biomolecules, 2016, 6, 39 CrossRef.
- N. Yamada, Y. Nishida, H. Tsutsumida, T. Hamada, M. Goto, M. Higashi, M. Nomoto and S. Yonezawa, Cancer Res., 2008, 68, 2708–2716 CrossRef CAS.
- P. Mukhopadhyay, S. Chakraborty, M. P. Ponnusamy, I. Lakshmanan, M. Jain and S. K. Batra, Biochim. Biophys. Acta, Rev. Cancer, 2011, 1815, 224–240 CrossRef CAS.
- S. Joshi, S. Kumar, A. Choudhury, M. P. Ponnusamy and S. K. Batra, Oncotarget, 2014, 5, 7272–7284 CrossRef.
- O. J. Finn, K. R. Jerome, R. A. Henderson, G. Pecher, N. Domenech, J. Magarian-Blander and S. M. Barratt-Boyes, Immunol. Rev., 1995, 145, 61–89 CrossRef CAS.
- J. D. Fontenot, S. V Mariappan, P. Catasti, N. Domenech, O. J. Finn, G. Gupta, S. V. Santhana Mariappan, P. Catasti, N. Domenech, O. J. Finn and G. Gupta, J. Biomol. Struct. Dyn., 1995, 13, 245–260 CrossRef CAS.
- U. Karsten, C. Diotel, G. Klich, H. Paulsen, S. Goletz, S. Müller and F. G. Hanisch, Cancer Res., 1998, 58, 2541–2549 CAS.
- E. Petrakou, A. Murray and M. R. Price, Tumor Biol., 1998, 19, 21–29 CrossRef CAS.
- M. R. Price, P. D. Rye, E. Petrakou, A. Murray, K. Brady, S. Imai, S. Haga, Y. Kiyozuka, D. Schol, M. F. A. Meulenbroek, F. G. M. Snijdewint, S. von Mensdorff-Pouilly, R. A. Verstraeten, K. Nil, A. Blockzjil, N. Nil, O. Nilsson, R. Nil, M. R. Suresh, K. Nil, S. Fortier, B. Nil, A. Berg, M. B. Longenecker, H. Nil, M. Boer, K. Nil, I. F. C. McKenzie, G. Nil, L. A. Simeoni, A. G. Ter-Grigoryan, I. M. Belyanchikov, N. V. Bovin, Y. Cao, U. Karsten, J. Dai, W. J. Allard, G. Davis, K. K. Yeung, F.-G. Hanisch, K. O. Lloyd, V. Kudryashov, R. Sikut, A. Sikut, K. Zhang, D. Baeckström, G. C. Hansson, C. A. Reis, H. Hassan, E. P. Bennett, H. Claussen, L. Norum, T. Varaas, B. Kierulf, K. Nustad, P. Ciborowski, W. M. Konitzki, J. Magarian- Blander, O. J. Finn and J. Hilgers, Tumor Biol., 1998, 19, 1–20 CrossRef.
- J. S. Grinstead, J. T. Schuman and A. P. Campbell, Biochemistry, 2003, 42, 14293–14305 CrossRef CAS.
- U. Karsten, Glycobiology, 2004, 14, 681–692 CrossRef CAS.
- S. Rangappa, G. Artigas, R. Miyoshi, Y. Yokoi, S. Hayakawa, F. Garcia-Martin, H. Hinou and S.-I. Nishimura, Medchemcomm, 2016, 7, 1102–1122 RSC.
- U. Karsten, S. von Mensdorff-Pouilly and S. Goletz, Tumour Immunobiol., 2005, 26, 217–220 CrossRef CAS.
- S. Rachagani, M. P. Torres, N. Moniaux and S. K. Batra, BioFactors, 2009, 35, 509–527 CrossRef CAS.
- O. Blixt, D. Bueti, B. Burford, D. Allen, S. Julien, M. Hollingsworth, A. Gammerman, I. Fentiman, J. Taylor-Papadimitriou and J. M. Burchell, Breast Cancer Res., 2011, 13, R25 CrossRef CAS.
- N. Jonckheere, N. Skrypek and I. Van Seuningen, Biochim. Biophys. Acta, 2014, 1846(1), 142–151 CAS.
- V. Ghai, K. Sharma, K. K. S. Abbi and H. A. Harvey, Open Breast Cancer J., 2011, 3, 29–30 CrossRef CAS.
- G. S. Baird, Am. J. Clin. Pathol., 2010, 134, 529–531 CrossRef.
- C. S. M. Ferreira, C. S. Matthews and S. Missailidis, Tumor Biol., 2006, 27, 289–301 CrossRef CAS.
- C. S. M. Ferreira, K. Papamichael, G. Guilbault, T. Schwarzacher, J. Gariepy and S. Missailidis, Anal. Bioanal. Chem., 2008, 390, 1039–1050 CrossRef CAS.
- C. S. M. Ferreira, M. C. Cheung, S. Missailidis, S. Bisland and J. Gariépy, Nucleic Acids Res., 2009, 37, 866–876 CrossRef CAS.
- Y. Hu, J. Duan, Q. Zhan, F. Wang, X. Lu and X. Yang, PLoS One, 2012, 7, e31970 CrossRef CAS.
- Y. He, Y. Lin, H. Tang and D. Pang, Nanoscale, 2012, 4, 2054 RSC.
- L. Tan, K. G. Neoh, E.-T. Kang, W.-S. Choe and X. Su, Anal. Biochem., 2012, 421, 725–731 CrossRef CAS.
- J. Zhao, X. He, B. Bo, X. Liu, Y. Yin and G. Li, Biosens. Bioelectron., 2012, 34, 249–252 CrossRef CAS.
- J. Tian, T. Huang and J. Lu, Anal. Methods, 2016, 8, 2375–2382 RSC.
- S. von Mensdorff-Pouilly, E. Petrakou, P. Kenemans, K. van Uffelen, A. A. Verstraeten, F. G. Snijdewint, G. J. van Kamp, D. J. Schol, C. A. Reis, M. R. Price, P. O. Livingston and J. Hilgers, Int. J. Cancer, 2000, 86, 702–712 CrossRef CAS.
- J. Schuman, a P. Campbell, R. R. Koganty and B. M. Longenecker, J. Pept. Res., 2003, 61, 91–108 CrossRef CAS.
- U. Westerlind, H. Schröder, A. Hobel, N. Gaidzik, A. Kaiser, C. M. Niemeyer, E. Schmitt, H. Waldmann and H. Kunz, Angew. Chem., Int. Ed., 2009, 48, 8263–8267 CrossRef CAS.
- O. Martínez, E. Bellard, M. Golzio, S. Mechiche-Alami, M.-P. Rols, J. Teissié, V. Ecochard and L. Paquereau, Nucleic Acid Ther., 2014, 24, 217–225 CrossRef.
- T. Matsushita, W. Takada, K. Igarashi, K. Naruchi, R. Miyoshi, F. Garcia-Martin, M. Amano, H. Hinou and S. I. Nishimura, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 1105–1116 CrossRef CAS.
- M. Movahedin, T. M. Brooks, N. T. Supekar, N. Gokanapudi, G.-J. Boons and C. L. Brooks, Glycobiology, 2017, 27, 677–687 CAS.
- N. Li, J. N. Ebright, G. M. Stovall, X. Chen, H. H. Nguyen, A. Singh, A. Syrett and A. D. Ellington, J. Proteome Res., 2009, 8, 2438–2448 CrossRef CAS.
- T. Li, Q. Fan, T. Liu, X. Zhu, J. Zhao and G. Li, Biosens. Bioelectron., 2010, 25, 2686–2689 CrossRef CAS.
- M. Chang, C.-S. Yang and D.-M. Huang, ACS Nano, 2011, 5, 6156–6163 CrossRef CAS.
- R. Savla, O. Taratula, O. Garbuzenko and T. Minko, J. Controlled Release, 2011, 153, 16–22 CrossRef CAS.
- C. Da Pieve, E. Blackshaw, S. Missailidis and A. C. Perkins, Bioconjugate Chem., 2012, 23, 1377–1381 CrossRef CAS.
- L. Cai, Z.-Z. Chen, M.-Y. Chen, H.-W. Tang and D.-W. Pang, Biomaterials, 2013, 34, 371–381 CrossRef CAS.
- S. H. Jalalian, S. M. Taghdisi, N. Shahidi Hamedani, S. A. M. Kalat, P. Lavaee, M. ZandKarimi, N. Ghows, M. R. Jaafari, S. Naghibi, N. M. Danesh, M. Ramezani and K. Abnous, Eur. J. Pharm. Sci., 2013, 50, 191–197 CrossRef CAS.
- H. Chen, J. Zhao, M. Zhang, H. Yang, Y. Ma and Y. Gu, Mol. Imaging Biol., 2015, 17, 38–48 CrossRef CAS.
- R.-C. Huang, W.-J. Chiu, I. Po-Jung Lai and C.-C. Huang, Sci. Rep., 2015, 5, 10292 CrossRef CAS.
- S. Taghavi, A. HashemNia, F. Mosaffa, S. Askarian, K. Abnous and M. Ramezani, Colloids Surf., B, 2016, 140, 28–39 CrossRef CAS.
- R. Yazdian-Robati, M. Ramezani, S. H. Jalalian, K. Abnous and S. M. Taghdisi, Pharm. Res., 2016, 33, 2289–2297 CrossRef CAS.
- M. J. Piña, A. Girotti, M. Santos, J. C. Rodríguez-Cabello and F. J. Arias, Mol. Pharm., 2016, 13, 795–808 CrossRef.
- C. Yu, Y. Hu, J. Duan, W. Yuan, C. Wang, H. Xu and X.-D. Yang, PLoS One, 2011, 6, e24077 CrossRef CAS.
- T. Kurosaki, N. Higuchi, S. Kawakami, Y. Higuchi, T. Nakamura, T. Kitahara, M. Hashida and H. Sasaki, Gene, 2012, 491, 205–209 CrossRef CAS.
- L. Civit, S. M. Taghdisi, A. Jonczyk, S. K. Haßel, C. Gröber, M. Blank, H. J. Stunden, M. Beyer, J. Schultze, E. Latz and G. Mayer, Biochimie, 2018, 145, 53–62 CrossRef CAS.
- S. E. Wilner, B. Wengerter, K. Maier, M. de Lourdes Borba Magalhães, D. S. Del Amo, S. Pai, F. Opazo, S. O. Rizzoli, A. Yan and M. Levy, Mol. Ther.--Nucleic Acids, 2012, 1, e21 CrossRef.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS.
- A. Díaz-Fernández, R. Miranda-Castro, N. De-los-Santos-Álvarez and M. J. Lobo-Castañón, Anal. Bioanal. Chem., 2018, 410, 2059–2065 CrossRef.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS.
- L. Cerchia and V. de Franciscis, Trends Biotechnol., 2010, 28, 517–525 CrossRef CAS.
- P. Dua, S. Kim and D. Lee, Methods, 2011, 54, 215–225 CrossRef CAS.
- C. Pestourie, L. Cerchia, K. Gombert, Y. Aissouni, J. Boulay, V. De Franciscis, D. Libri, B. Tavitian and F. Ducongé, Oligonucleotides, 2006, 16, 323–335 CrossRef CAS.
- Y. Liu, C.-T. Kuan, J. Mi, X. Zhang, B. M. Clary, D. D. Bigner and B. A. Sullenger, Biol. Chem., 2009, 390, 137–144 CAS.
- P. Ray, B. A. Sullenger and R. R. White, Nucleic Acid Ther., 2013, 23, 435–442 CrossRef CAS.
- S. C. Simmons, H. Jämsä, D. Silva, C. M. Cortez, E. A. McKenzie, C. C. Bitu, S. Salo, S. Nurmenniemi, P. Nyberg, J. Risteli, C. E. B. DeAlmeida, P. E. C. Brenchley, T. Salo and S. Missailidis, PLoS One, 2014, 9, e96846 CrossRef.
- P. Ray and R. R. White, J. Nucleic Acids, 2017, 2017, 1–9 CrossRef.
- Q. Zhao, S. Matson, C. J. Herrera, E. Fisher, H. Yu and A. M. Krieg, Antisense Res. Dev., 1993, 3, 53–66 CrossRef CAS.
- M. L. Raddatz, A. Dolf, E. Endl, P. Knolle, M. Famulok and G. Mayer, Angew. Chem., Int. Ed., 2008, 47, 5190–5193 CrossRef CAS.
- M. Avci-Adali, M. Metzger, N. Perle, G. Ziemer and H. P. Wendel, Oligonucleotides, 2010, 20, 317–323 CrossRef CAS.
- G. Mayer, M.-S. L. Ahmed, A. Dolf, E. Endl, P. A. Knolle and M. Famulok, Nat. Protoc., 2010, 5, 1993–2004 CrossRef CAS.
- U. Schnell, F. Dijk, K. A. Sjollema and B. N. G. Giepmans, Nat. Methods, 2012, 9, 152–158 CrossRef CAS.
- A. N. Kuzmin, A. Pliss and P. N. Prasad, Anal. Chem., 2014, 86, 10909–10916 CrossRef CAS.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS.
- L. Young, J. Sung, G. Stacey and J. R. Masters, Nat. Protoc., 2010, 5, 929–934 CrossRef CAS.
- K.-A. Frith, R. Fogel, J. P. D. Goldring, R. G. E. Krause, M. Khati, H. Hoppe, M. E. Cromhout, M. Jiwaji and J. L. Limson, Malar. J., 2018, 17, 191 CrossRef.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS.
- P. O. Krutzik and G. P. Nolan, Cytometry, Part A, 2003, 55, 61–70 CrossRef.
- P. O. Krutzik, J. M. Irish, G. P. Nolan and O. D. Perez, Clin. Immunol., 2004, 110, 206–221 CrossRef CAS.
- M. Kaur and L. Esau, Biotechniques, 2015, 59(3), 119–126 CrossRef CAS.
- M. A. Gomes de Castro, C. Höbartner and F. Opazo, Bio-Protoc., 2017, 7, 1–13 Search PubMed.
- S. V. Costes, D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis and S. Lockett, Biophys. J., 2004, 86, 3993–4003 CrossRef CAS.
- A. K. H. Cheng, H. Su, Y. A. Wang and H.-Z. Yu, Anal. Chem., 2009, 81, 6130–6139 CrossRef CAS.
- F. Ma, C. Ho, A. K. H. Cheng and H.-Z. Yu, Electrochim. Acta, 2013, 110, 139–145 CrossRef CAS.
- S. Shin, H. Y. Nam, E. J. Lee, W. Jung and S. S. Hah, Bioorg. Med. Chem. Lett., 2012, 22, 6081–6084 CrossRef CAS.
- N. Liu, C. Zhou, J. Zhao and Y. Chen, Cancer Invest., 2012, 30, 577–582 CrossRef CAS.
- S. J. Gendler and A. P. Spicer, Annu. Rev. Physiol., 1995, 57, 607–634 CrossRef CAS.
- S. Müller and F.-G. Hanisch, J. Biol. Chem., 2002, 277, 26103–26112 CrossRef.
- S. Müller, K. Alving, J. Peter-Katalinic, N. Zachara, A. A. Gooley and F. G. Hanisch, J. Biol. Chem., 1999, 274, 18165–18172 CrossRef.
- C. M. Klinge, B. N. Radde, Y. Imbert-Fernandez, Y. Teng, M. M. Ivanova, S. M. Abner and A. L. Martin, Mol. Cancer Ther., 2011, 10, 2062–2071 CrossRef CAS.
- C. M. Cortez, D. Silva, C. M. C. Silva and S. Missailidis, Spectrochim. Acta, Part A, 2012, 95, 270–275 CrossRef CAS.
- E. F. Silva, E. B. Ramos and M. S. Rocha, J. Phys. Chem. B, 2013, 117, 7292–7296 CrossRef CAS.
- X. Wu, H. Liang, Y. Tan, C. Yuan, S. Li, X. Li, G. Li, Y. Shi and X. Zhang, PLoS One, 2014, 9, e90752 CrossRef.
- A. J. Hobro and N. I. Smith, Vib. Spectrosc., 2017, 91, 31–45 CrossRef CAS.
- P. Shaw and J. Doonan, Cell Cycle, 2005, 4, 102–105 CrossRef CAS.
- Z. A. Steelman, W. J. Eldridge and A. Wax, J. Biophotonics, 2018, 11, e201800091 CrossRef.
- M. C. O'Brien and W. E. Bolton, Cytometry, 1995, 19, 243–255 CrossRef.
- J. H. L. Chik, J. Zhou, E. S. X. Moh, R. Christopherson, S. J. Clarke, M. P. Molloy and N. H. Packer, J. Proteomics, 2014, 108, 146–162 CrossRef CAS.
- Q. Shen, J. J. Rahn, J. Zhang, N. Gunasekera, X. Sun, A. R. E. Shaw, M. J. Hendzel, P. Hoffman, A. Bernier and J. C. Hugh, Mol. Cancer Res., 2008, 6, 555–567 CrossRef CAS.
- J. M. David, D. H. Hamilton and C. Palena, Oncoimmunology, 2016, 5, e1117738 CrossRef.
- P. de Cremoux, J. M. Extra, M. G. Denis, J. Y. Pierga, E. Bourstyn, C. Nos, K. B. Clough, E. Boudou, E. C. Martin, A. Müller, P. Pouillart and H. Magdelénat, Clin. Cancer Res., 2000, 6, 3117–3122 CAS.
- X.-Q. Liu, H.-X. Ren and Z.-P. Wu, Chin. J. Cancer, 2008, 27, 1267–1270 CAS.
- M. a Houston, L. H. Augenlicht and B. G. Heerdt, PLoS One, 2011, 6, e25207 CrossRef.
- J. Ren, N. Agata, D. Chen, Y. Li, W. Yu, L. Huang, D. Raina, W. Chen, S. Kharbanda and D. Kufe, Cancer Cell, 2004, 5, 163–175 CrossRef CAS.
- U. Schumacher and E. Adam, J. Histochem. Cytochem., 1998, 46, 127–134 CrossRef CAS.
- S. Ogata, H. Uehara, A. Chen and S. H. Itzkowitz, Cancer Res., 1992, 52, 5971–5978 CAS.
- J. Kao, K. Salari, M. Bocanegra, Y.-L. Choi, L. Girard, J. Gandhi, K. a Kwei, T. Hernandez-Boussard, P. Wang, A. F. Gazdar, J. D. Minna and J. R. Pollack, PLoS One, 2009, 4, e6146 CrossRef.
- H. Möller, N. Serttas, H. Paulsen, J. M. Burchell, J. Taylor-Papadimitriou and B. Meyer, Eur. J. Biochem., 2002, 269, 1444–1455 CrossRef.
- H. Coelho, T. Matsushita, G. Artigas, H. Hinou, F. J. Canada, R. Lo-Man, C. Leclerc, E. J. Cabrita, J. Jimenez-Barbero, S. I. Nishimura, F. Garcia-Martin and F. Marcelo, J. Am. Chem. Soc., 2015, 137, 12438–12441 CrossRef CAS.
- F. Leccia, A. Nardone, S. Corvigno, L. Del Vecchio, S. De Placido, F. Salvatore and B. M. Veneziani, Cytometry, Part A, 2012, 81, 960–972 CrossRef.
- T. Piyush, J. M. Rhodes and L.-G. Yu, Cell Death Discovery, 2017, 3, 17044 CrossRef CAS.
- W. Song, E. S. Delyria, J. Chen, W. Huang, J. S. Lee, E. A. Mittendorf, N. Ibrahim, L. G. Radvanyi, Y. Li, H. Lu, H. Xu, Y. Shi, L. X. Wang, J. A. Ross, S. P. Rodrigues, I. C. Almeida, X. Yang, J. Qu, N. S. Schocker, K. Michael and D. Zhou, Int. J. Oncol., 2012, 41, 1977–1984 CrossRef CAS.
- M. a Wolfert and G.-J. Boons, Nat. Chem. Biol., 2013, 9, 776–784 CrossRef.
- S. R. Stowell, T. Ju and R. D. Cummings, Annu. Rev. Pathol.: Mech. Dis., 2015, 10, 473–510 CrossRef CAS.
- K. K. H. Stanlis and J. R. McIntosh, J. Histochem. Cytochem., 2003, 51, 797–808 CrossRef CAS.
- H. Takahashi, C. Jin, H. Rajabi, S. Pitroda, M. Alam, R. Ahmad, D. Raina, M. Hasegawa, Y. Suzuki, A. Tagde, R. T. Bronson, R. Weichselbaum and D. Kufe, Oncogene, 2015, 34, 5187–5197 CrossRef CAS.
- F. Dai, Y. Zhang, X. Zhu, N. Shan and Y. Chen, Target. Oncol., 2012, 7, 217–225 CrossRef.
- P. Wu, Y. Gao, H. Zhang and C. Cai, Anal. Chem., 2012, 84, 7692–7699 CrossRef CAS.
- L. Tan, K. G. Neoh, E.-T. Kang, W. S. Choe and X. Su, Macromol. Biosci., 2011, 11, 1331–1335 CrossRef CAS.
- B. Dai, Y. Hu, J. Duan and X.-D. Yang, Oncotarget, 2016, 7(25), 38257–38269 CrossRef.
- S. Taghavi, M. Ramezani, M. Alibolandi, K. Abnous and S. M. Taghdisi, Cancer Lett., 2017, 400, 1–8 CrossRef CAS.
- M. Alibolandi, F. Hoseini, M. Mohammadi, P. Ramezani, E. Einafshar, S. M. Taghdisi, M. Ramezani and K. Abnous, Int. J. Pharm., 2018, 549, 67–75 CrossRef CAS.
- A. Bahreyni, M. Alibolandi, M. Ramezani, A. Sarafan Sadeghi, K. Abnous and S. M. Taghdisi, Colloids Surf., B, 2019, 175, 231–238 CrossRef CAS.
- A. Wells and U. Marti, Nat. Rev. Mol. Cell Biol., 2002, 3, 697–702 CrossRef CAS.
- L. Rajendran, H.-J. Knölker and K. Simons, Nat. Rev. Drug Discovery, 2010, 9, 29–42 CrossRef CAS.
- Y.-N. Wang, H. Yamaguchi, J.-M. Hsu and M.-C. Hung, Oncogene, 2010, 29, 3997–4006 CrossRef CAS.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537–550 CrossRef CAS.
- S. Xu, B. Z. Olenyuk, C. T. Okamoto and S. F. Hamm-Alvarez, Adv. Drug Delivery Rev., 2013, 65, 121–138 CrossRef CAS.
- X. Zhu, J. Yang, M. Liu, Y. Wu, Z. Shen and G. Li, Anal. Chim. Acta, 2013, 764, 59–63 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ay01878c |
| This journal is © The Royal Society of Chemistry 2021 |