Open Access Article
Open Access ArticleIdentification of cellulose textile fibers†
Mikko
Mäkelä
 *a,
Marja
Rissanen
b and
Herbert
Sixta
*a,
Marja
Rissanen
b and
Herbert
Sixta
 b
b
aVTT Technical Research Centre of Finland Ltd, PO Box 1000, 02044 VTT Espoo, Finland. E-mail: mikko.makela@vtt.fi
bAalto University, School of Chemical Engineering, Department of Bioproducts and Biosystems, PO Box 16300, 00076 Aalto, Finland
First published on 3rd November 2021
Abstract
Distinguishing different textile fibers is important for recycling waste textiles. Most studies on non-destructive optical textile identification have focused on classifying different synthetic and natural fibers but chemical recycling requires more detailed information on fiber composition and polymer properties. Here, we report the use of near infrared imaging spectroscopy and chemometrics for classifying natural and regenerated cellulose fibers. Our classifiers trained on images of consumer textiles showed 100% true positive rates based on model cross-validation and correctly identified on average 8–9 out of 10 test set pixels using images of specifically made cotton, viscose and lyocell samples of known compositions. These results are significant as they indicate the possibility to monitor and control fiber dosing and subsequent dope viscosity during chemical recycling of cellulose fibers. Our results also suggested the possibility to identify fibers purely based on polymer chain length. This finding opens the possibility to indirectly estimate dope viscosity and creates entirely new hypotheses for combining imaging spectroscopy with classification and regression methods within the broader field of cellulose modification.
1. Introduction
Identification of cellulose fibers plays an important role in chemical recycling of textiles. Cotton and regenerated cellulose fibers differ in cellulose structure and polymer chain length, which governs their viscosity after chemical dissolution and the ease in which the dissolved fibers can be spun again into regenerated textile fibers. Textile recycling is a timely topic as the global fiber production for textiles has over doubled since the year 2000 to circa 120 million tons in 2019.1,2 Increasing textiles production and consumption are associated with decreasing average garment-use times,3 a trend which will increase the quantity of generated textiles waste. The need for textiles collection has been acknowledged by the European Commission, which requires European Union (EU) members states to organize separate collection of household textile waste by 2025.4 Chemical recycling of used textiles into regenerated man-made cellulose fibers enables converting waste materials into new fiber products with improved mechanical properties.5,6 Recycling of cellulose fibers therefore decreases our dependence on primary fiber production and contributes to bridging the cellulose gap generated by an increasing demand for regenerated textile fibers.7Here, we report the use of near infrared (NIR) imaging spectroscopy and chemometrics for classification of cotton, viscose and lyocell fibers. These three fibers are the main cellulose fibers for consumer applications and currently cover approximately 32% of annual textile fiber production.1 Used cotton, viscose and lyocell fibers are important raw materials for chemical recycling and are challenging to identify quickly and accurately. Most recent studies on non-destructive optical methods for textile identification have focused on classifying synthetic and natural fibers, such as polyester,8–11 cotton,8–11 viscose,9–11 and wool.8,9,12 These results are important for developing automated textile identification for efficient separation and sorting once the upcoming EU regulation on textile collection will be enforced. Chemical recycling of cellulose fibers, however, requires more detailed information on fiber composition and polymer properties.
Previous reports have shown 68% classification accuracy for cotton and viscose combined based on visible-NIR images9 and 76% accuracy for pure cotton using individual NIR spectra.10 The intrinsic viscosities of cotton and viscose fibers are in the range 150–2000 mg l−1,13 and improving classification accuracy could enable sorting fiber raw materials for controlling fiber dosing and subsequent dope viscosity for fiber spinning. Rashed et al.11 further compared two handheld NIR sensors for textile identification using random forests and obtained 94–96% classification accuracies for cotton and viscose based on a test set separated from 14 and 10 cotton and viscose samples, respectively. Saito et al.14 reported classification of natural and regenerated cellulose fibers. Their method, also covered by an international patent,15 was based on infrared spectroscopy followed by hierarchical modelling using regularized discriminant analysis. The authors reported 100% classification accuracy for cotton, lyocell, linen and other fibers, but were required to decrease spectral variation by mercerizing the fibers using aqueous sodium hydroxide.14 These recent results are encouraging for accurately identifying cotton and regenerated cellulose fibers for chemical recycling.
Our approach based on NIR imaging requires no sample pretreatment and can potentially be extended to automated fiber identification. We trained our classifiers on images of pure and mixed fiber consumer textiles and determined model performance on an independent pixel test set of specifically made pure cotton, viscose and lyocell fabrics of known compositions. Our results showed that 8–9 out of 10 test set pixels were on average classified correctly. These results enable estimating the potential of NIR imaging for cellulose fiber identification using only a limited number of pixel spectra, which is important for combining spectral imaging sensors with cameras operating in the visible range for faster image segmentation. The results also extend our current knowledge on the possibilities of NIR imaging in evaluating the properties of cellulose and cellulose textile fibers. Overall, this work continues our recent efforts16,17 on evaluating the potential of NIR imaging in estimating textile properties with the aim of developing machine vision tools for chemical recycling of textile fibers.
2. Experimental
2.1 Sampling and sample preparation
A total of 81 training samples of consumer textiles made from cotton and regenerated cellulose fibers were collected from an import and wholesale shop (Tekstiilipalvelu), local fabric shops (Lempala and Eurokangas), fabric sample providers (Finlayson and Lenzing) and Lahti University of Applied Sciences. The samples contained four pure fiber classes and their fiber blends as shown in Table 1. Further details on the samples are given in Table S1. in the ESI.† The textile samples were first cut into approximately 10 × 10 cm samples and were qualitatively evaluated using an optical microscope (Leica DMLAM, Leica Microsystems) to validate the fiber types given in the textile labels. Fibers from both warp and weft yarns were unraveled and placed on a microscope slide using a total magnification of 200× (objective lens 20×, eyepiece lens 10×). The fiber samples were then identified based on their visual longitudinal appearance. Examples of the microscopic images of the different fibers are illustrated in Fig. S1.†| Textile fiber | Number of samples | Viscosity (mg l−1)6,13 | Crystal structure14,18 |
|---|---|---|---|
| a Pre-consumer fibers >1500 mg l−1, post-consumer fibers 300–1200 mg l−1. b Virgin cotton: cellulose I, mercerized cotton: cellulose II. n.a. = not applicable. | |||
| Cotton | 22 | 300–2000a | Cellulose Ib |
| Viscose | 22 | 150–200 | Cellulose II |
| Lyocell | 12 | 320–450 | Cellulose II |
| Modal | 3 | 220–300 | Cellulose II |
| Fiber blends | 22 | n.a. | n.a. |
In addition to the training samples, three test set samples were prepared from pure cotton, viscose and lyocell fibers. Approximately 80 tex yarns were first plied and twisted from 20 tex commercial cotton, viscose and lyocell yarns provided by Orneule. The test set sample fabrics were then weft-knitted from the prepared yarns using a programmable knitting machine (Stoll CMS ADF 32 W, Karl Mayer Stoll GmbH).
2.2 Imaging spectroscopy
Reflectance images of the textile samples were recorded with a SWIR3.0 imaging sensor (Specim, Spectral Imaging Ltd). The camera operated in line-scanning mode and measured 384 spatial pixels on 288 spectral variables under halogen light illumination. The field of view was set to approximately 12 cm and the speed of the moving stage was adjusted to record square pixels of circa 0.3 × 0.3 mm2 dimensions. Integration time was adjusted so that the highest reflectance target signals were approximately 90% of the signal maximum. The raw images were median filtered to eliminate the effect of potential dead pixels in the camera detector and then converted into unitless reflectance using externally calibrated 2, 25, 50, 75 and 99% reflectance targets. Different reflectance transformations were determined as described in ref. 16. The final transformations were chosen to minimize the respective root mean squared errors determined based a 50% reflectance test set of approximately 2.6 × 106 voxels. Noisy variables on extreme wavelengths were excluded, which provided 271 wavelength variables within 1000–2500 nm.2.3 Fiber classification
Sample backgrounds were removed from the images using principal components analysis (PCA). Each image was decomposed individually after mean centering and the first component successfully separated the background pixels from each sample. Comparable regions of interest (ROIs) of 200 × 200 pixels were then chosen from the center of each sample. The ROIs were used for extracting mean sample spectra for model training. The reflectance spectra were −log10 transformed into pseudo-absorbance and preprocessed using standard normal variate (SNV) transformation19 and mean centering. The differences in the training objects were first visualized using a PCA model.20 PCA model details including model residuals and confidence limits were calculated as described by Jackson.21 An example image ROI and the respective average spectrum are illustrated in Fig. 1.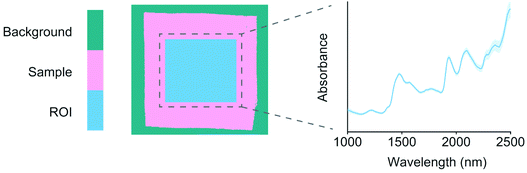 | ||
| Fig. 1 An example image ROI and the respective average spectrum of a random (lyocell) sample. The shaded areas around the spectrum show wavelength variation as mean ± standard deviation across the ROI pixels, which were SNV19 transformed. | ||
Cotton, viscose and lyocell fibers were then identified using linear classification models. The binary classifiers were trained separately for the cotton, viscose and lyocell classes using partial least squares discriminant analysis (PLS-DA)22 based on the SIMPLS algorithm.23 Each training object was given a class assignment which described whether the sample belonged to the class of interest (+1) or not (−1). Differences in class sizes were then adjusted for by determining the overall wavelength means for mean centering as an average of the class means (i.e., 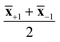 ).24 Predicted class assignments were determined based on the PLS predictions using a decision threshold. The decision threshold was first set to zero and was then updated for the pixel test set by maximizing the sensitivity and specificity of each model based on the training set. Classification errors were determined as an average of both classes and the results were used for estimating the appropriate number of latent variables based on model cross-validation. Cross-validation performance was determined by continuously dividing the training objects into five groups and using one group at a time (ca. 20% of objects) as a validation set. This procedure was repeated until all objects had been left out once. The final wavelengths used in the classifiers were selected using interval PLS.25 The wavelength range was first divided into 14 intervals where each interval contained 19 variables. Cross-validation errors were then determined based on all possible subset combinations and the relative frequency of each interval in the 10% of models with the lowest cross-validation errors was visualized as a function of the number of used intervals. The cross-validation and variable selection procedures are visualized in Fig. S2.† The final classifiers were validated on the pixel level using the specifically made test samples of known compositions. The test sample images in reflectance were −log10 and SNV transformed and the entire sample images without image backgrounds were used as a pixel test set.
).24 Predicted class assignments were determined based on the PLS predictions using a decision threshold. The decision threshold was first set to zero and was then updated for the pixel test set by maximizing the sensitivity and specificity of each model based on the training set. Classification errors were determined as an average of both classes and the results were used for estimating the appropriate number of latent variables based on model cross-validation. Cross-validation performance was determined by continuously dividing the training objects into five groups and using one group at a time (ca. 20% of objects) as a validation set. This procedure was repeated until all objects had been left out once. The final wavelengths used in the classifiers were selected using interval PLS.25 The wavelength range was first divided into 14 intervals where each interval contained 19 variables. Cross-validation errors were then determined based on all possible subset combinations and the relative frequency of each interval in the 10% of models with the lowest cross-validation errors was visualized as a function of the number of used intervals. The cross-validation and variable selection procedures are visualized in Fig. S2.† The final classifiers were validated on the pixel level using the specifically made test samples of known compositions. The test sample images in reflectance were −log10 and SNV transformed and the entire sample images without image backgrounds were used as a pixel test set.
3. Results and discussion
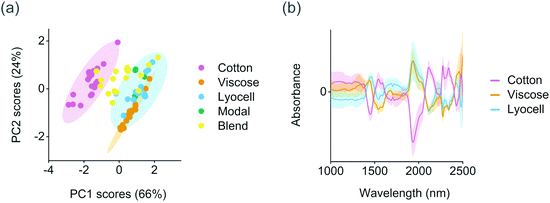
The training set samples were first microscopically evaluated to validate the fiber types reported in the textile labels. Fiber type can be identified with a microscope based on the physical structure of the individual fibers. Cotton fibers generally have a convoluted, ribbon-like structure whereas viscose fibers have strongly grooved, granulated structure (Fig. S1†). In comparison with viscose, modal fibers have a less grooved structure and lyocell fibers a smooth longitudinal appearance.26 Based on the results, 11 out of the 81 training samples contained fibers or fiber blends which were different from those reported in the textile labels. This information was used for assigning the final class assignments shown in Table 1. Differences within the training set were then determined using PCA. The first two principal components explained 91% of variation in the preprocessed and mean centered spectra and provided a clear separation between the natural and regenerated fibers. As illustrated in Fig. 2a, cotton fibers showed lower score values on the first principal component than the regenerated cellulose fibers. These lower score values were associated with higher absorbance especially at 1540–1590 nm and within the combination band region in the range 2100–2350 nm based on the average class spectra from the mean centered training set, Fig. 2b. Cotton fibers also showed comparatively lower absorbance at approximately 1440 and 1940 nm than the viscose and lyocell fibers. Four samples were excluded from the PCA as generic outliers due to high Hotelling T2 and Q residuals (Table S1†). Two of these four samples were partly composed of synthetic fibers based on microscopic evaluation. The remaining two samples contained a thick binder layer on the fabric surface.The crystalline structure of natural cellulose is readily converted from cellulose I to cellulose II during the production of regenerated fibers. For example, the effect of strong alkali irreversibly changes the arrangement of cellulose chains through the conformation of hydroxymethyl groups and chain polarity.27 Virgin cotton fibers are also known to have significantly higher molar mass than viscose and lyocell fibers due to differences in polymer chain length.13 These differences were reflected in the training set spectra. Crystalline cellulose I structures and the intramolecular hydrogen bonds in cellulose in wood have previously been reported to absorb at 1530–1590 nm,28 which suggested that the separation of cotton fibers based on PCA was in part due to differences in cellulose structure or polymer chain length (Fig. 2). The deformation and strain vibrations between mainly oxygen and hydrogen and carbon and hydrogen in cellulose have also been reported at 2110, 2277 and 2343 nm, which matched the positive absorbance peaks of cotton within the combination band region. The two negative peaks in the average cotton spectra in Fig. 2b at 1440 and 1940 nm were close to the well-known water-related vibrations in the NIR region. We did not find a clear separation between the regenerated viscose or lyocell fibers based on the score values of the first two, or the successive, principal components.
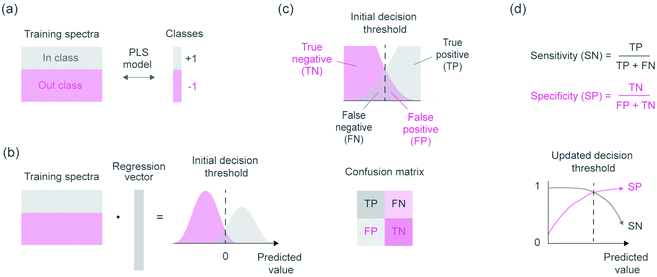
Supervised classifiers were then determined for identifying cotton, viscose and lyocell fibers. The binary classifiers were based on a one-versus-all procedure, where each of the cotton, viscose and lyocell fiber classes were modelled separately using an individual PLS-DA model. This required building three different models but enabled refining the models specifically for each fiber class. PLS is a multivariate regression method and applying it for classification tasks requires setting a decision threshold to separate the model predictions into discrete class assignments. This decision threshold can be chosen in different ways and becomes especially important with different class sizes.24,29 Our approach for updating the decision threshold is visualized in Fig. 3 and enabled considering the effect of unequal class sizes in model training.
The results of the individual classifiers are illustrated in Table 2. Variable selection reduced the number of used wavelengths by 37–44% for the three classifiers. The chosen variable subsets improved the estimation of model pseudorank based on cross-validation and decreased final classification errors. The final classifiers explained 67–86% of the variation in the given class assignments and led to average training set classification errors of 2–4% using 1–3 latent variables (Table 2). Four important wavelengths for separating the regenerated fibers based on the classifier regression vectors are shown in Fig. S3.† Although we did not find a clear band assignment for the difference in peaks at approximately 2147 nm, the lyocell class showed positive regression coefficients at approximately 1485, 1732 and 2368 nm. These wavelengths have been reported to absorb based on the intramolecular hydrogen bond vibrations in cellulose, the carbon and hydrogen bond vibrations of methyl groups in cellulose, and the oxygen and carbon or hydrogen and carbon deformation and strain vibrations in cellulose, respectively.28 As lyocell fibers are known to have a higher degree of polymerization than viscose fibers,13 our results on classifier performance suggested that the classifiers were successful in separating the regenerated cellulose fibers based on polymer chain length.
| Classifier | No. of outliers excluded from the training set | Chosen wavelengths (nm) | Latent variables | Average class error during model training and cross-validation | Updated decision threshold | Sensitivity and specificity | |
|---|---|---|---|---|---|---|---|
| Training set (object level) | Test set (pixel level) | ||||||
| Cotton | 4 | 1225–1537; 1648–2064; 2175–2274 | 1 | 3% | 0.31 | 100 and 96% | 100 and 93% |
| Viscose | 7 | 1331–1959; 2070–2379 | 3 | 2% | 0.20 | 100 and 98% | 83 and 90% |
| Lyocell | 6 | 1331–1537; 1648–2169; 2280–2379 | 3 | 4% | 0.35 | 100 and 94% | 91 and 67% |
An example of the variable selection results and model diagnostics for the lyocell class with the highest training and cross-validation errors are illustrated in Fig. 4. Wavelengths for the final classifiers were chosen based on the variable selection results. The lyocell model misclassified five training samples as false positives before adjusting the decision threshold for the pixel test set (Fig. 4). These false positives were cotton and lyocell blends where the share of lyocell fiber was 50–96% based on the information given in the textile labels. Updating the decision threshold based on model sensitivity and specificity excluded one sample containing 85% lyocell from the misclassified group. The cotton model with an updated decision threshold misclassified two cotton and lyocell blends as false positives. In these false positives the reported share of cotton fiber was 60–80%. The final viscose model misclassified one sample composed entirely of modal fiber as a false positive. Although we were not able to quantitatively verify the compositions of the fiber blends in the training set, the false positives of the cotton and lyocell blends show how the predictions of the regression models trained on discrete class assignments serve as indirect estimates of fiber composition. This information is useful in practice for diagnosing potential misclassifications and served as an additional tool for identifying cases where incorrect compositional information was given in the textile labels based on microscopic evaluation. All three classifiers showed 100% true positive rates by correctly classifying all training objects belonging to each class of interest based on cross-validation (Table 2).
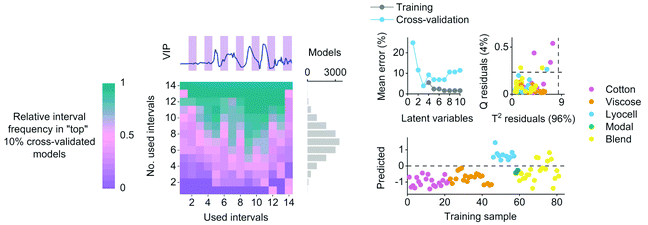 | ||
| Fig. 4 An example of relative interval frequency determined during variable selection, average classification errors as a function of latent variables, PLS model residuals and the predicted training sample values with the initial decision threshold for the lyocell class. The lyocell classifier showed the highest average training and cross-validation errors as illustrated in Table 2. | ||
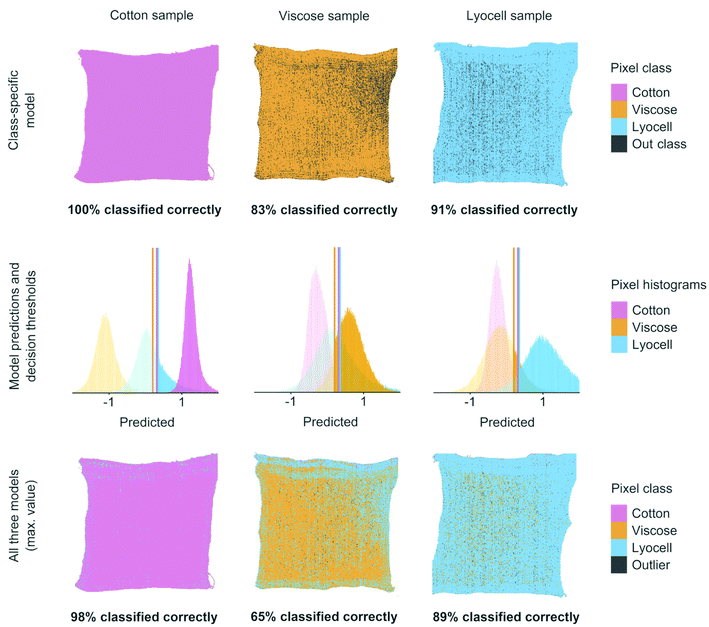
Classifier performance was finally validated using images of the prepared cotton, viscose and lyocell samples of known compositions, see Fig. 5. Using one binary classifier offered only two possible verdicts but combining several classifiers increased the number of verdicts and introduced a new class of outlier objects.30 With three classifiers there were a total of eight possible class combinations for each test set pixel. In three out of these eight cases one of the three models was able to identify a pixel correctly. One case was formed by the outlier class, where all three models failed in predicting a pixel value which was above the updated decision threshold. The remaining four were misclassifications or ambiguous cases where two or three models produced competing results which were above the respective decisions thresholds. We resolved the ambiguous class assignments by assigning the pixel to the class with the highest predicted value before using the decision threshold. As illustrated in Fig. 5, 8–9 out of 10 test set pixels were on average classified correctly when one or all three classifiers were combined on the same pixel test set.
The results in Fig. 5 show two different practical scenarios for using the classifiers. The class-specific model results show correctly classified test set pixels when only one model is used for identifying each specific fiber of interest. Such a situation could occur when only one fiber type is considered as suitable raw material for chemical recycling. The results on all three models in Fig. 5 illustrate how the three classifiers can be combined for identifying each of the three fiber types. These results are important for situations where all three fibers need to be identified correctly for controlling fiber dosing for chemical recycling.
4. Conclusions
We have shown how different virgin and regenerated cellulose textile fibers were identified using NIR imaging spectroscopy and chemometrics. Our results from the three classifiers indicated 100% true positive rates based on model cross-validation and correctly classified on average 8–9 out of 10 test set pixels using images of separate cotton, viscose and lyocell samples of known compositions. These results are significant as they indicate the possibility to reliably monitor and control fiber dosing and subsequent dope viscosity for fiber spinning during chemical recycling. Spinning processes to produce regenerated fibers are sensitive to polymer chain length which, together with the molecular mass distribution of the cellulose substrates, controls the viscoelastic properties of the spinning dope and its spinnability and regeneration behavior. It is therefore important to reliably distinguish different cellulose fibers to control the logistics and performance of chemical recycling and to ultimately increase the use of textile waste for fiber regeneration. The regression vectors of the viscose and lyocell classifiers also suggested the possibility to separate fibers purely based on polymer chain length. This finding opens the possibility to indirectly estimate dope viscosity and creates entirely new hypotheses for combining imaging spectroscopy with classification and regression methods within the broader field of cellulose modification.Author contributions
All authors conceptualized the study, H. S. and M. M. contributed to acquiring the funding. M. M. and M. R. acquired the study resources, devised the methodology, performed the investigation, and curated the data. M. M. developed the required software for formal analysis of the data, validated the results, carried out the visualization and wrote the original draft. All authors critically revised and edited the draft.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the contribution of Kirsti Cura from Lahti University of Applied Sciences for providing some of the training samples and Anna Leinonen, a knitting workshop master at Aalto University, for preparing the test set fabrics. This work was financially supported by the Strategic Research Council of the Academy of Finland under grant agreement no. 327296 – the FINIX project (finix.aalto.fi).References
- The Fiber Year 2020, The Fiber Year GmbH, 2020.
- C. Palacios-Mateo, Y. van der Meer and G. Seide, Environ. Sci. Eur., 2021, 33, 2 CrossRef CAS PubMed.
- K. Niinimäki, G. Peters, H. Dahlbo, P. Perry, T. Rissanen and A. Gwilt, Nat. Rev. Earth Environ., 2020, 1, 189–200 CrossRef.
- European Council, 2018. Waste management and recycling: council adopts new rules, press release. Available at: http://www.consilium.europa.eu/en/press/press-releases/2018/05/22/waste-management-and-recycling-council-adopts-new-rules/ [accessed August 4th, 2021].
- S. Asaadi, M. Hummel, S. Hellsten, T. Härkäsalmi, Y. Ma, A. Michud and H. Sixta, ChemSusChem, 2016, 9, 3250–3258 CrossRef CAS PubMed.
- S. Haslinger, Y. Wang, M. Rissanen, M. B. Lossa, M. Tanttu, E. Ilen, M. Määttänen, A. Harlin, M. Hummel and H. Sixta, Green Chem., 2019, 21, 5598 RSC.
- C. Felgueiras, N. G. Azoia, C. Gonçalves, M. Gama and F. Dourado, Front. Bioeng. Biotechnol., 2021, 9, 608826 CrossRef PubMed.
- J. Li, X. Meng, W. Wang and B. Xin, Text. Res. J., 2019, 89, 3752–3767 CrossRef CAS.
- C. Blanch-Perez-del-Notario, W. Saeys and A. Lambrechts, J. Spectr. Imaging, 2019, 8, a17 CrossRef.
- K. Cura, N. Rintala, T. Kamppuri, E. Saarimäki and P. Heikkilä, Recycling, 2021, 6, 11 CrossRef.
- H. S. Rashed, P. Mishra, A. Nordon, D. S. Palmer and M. J. Baker, Vib. Spectrosc., 2021, 113, 103205 CrossRef CAS.
- C. Tan, H. Chen, Z. Lin and T. Wu, Vib. Spectrosc., 2019, 100, 71–78 CrossRef CAS.
- H. Wedin, M. Lopes, H. Sixta and M. Hummel, Text. Res. J., 2019, 89, 5067–5075 CrossRef CAS.
- K. Saito, T. Yamagata, M. Kanno, N. Yoshimura and M. Takayanagi, Spectrochim. Acta, Part A, 2021, 257, 119772 CrossRef CAS PubMed.
- M. Takayanagi, N. Yoshimura, K. Saito, T. Ando and M. Kanno, Fiber differentiation method. International patent application, WO2018074002, 2018.
- M. Mäkelä, P. Geladi, M. Rissanen, L. Rautkari and H. Sixta, Anal. Chim. Acta, 2020, 1105, 56–63 CrossRef PubMed.
- M. Mäkelä, M. Rissanen and H. Sixta, Resour., Conserv. Recycl., 2020, 161, 105007 CrossRef.
- T. Röder, J. Moosbauer, M. Fasching, A. Bohn, H.-P. Fink, T. Baldinger and H. Sixta, Lenzinger Ber., 2006, 86, 132–136 Search PubMed.
- T. Fearn, C. Riccioli, A. Garrido-Varo and J. E. Guerrero-Ginel, Chemom. Intell. Lab. Syst., 2009, 96, 22–26 CrossRef CAS.
- P. Geladi, Spectrochim. Acta, Part B, 2003, 58, 767–782 CrossRef.
- J. E. Jackson, A user's guide to principal components, John Wiley & Sons, Inc., United States, 1991 Search PubMed.
- C. L. M. Morais, K. M. G. Lima, M. Singh and F. L. Martin, Nat. Protoc., 2020, 15, 2143–2162 CrossRef CAS PubMed.
- S. de Jong, Chemom. Intell. Lab. Syst., 1993, 18, 251–263 CrossRef CAS.
- R. G. Brereton and G. R. Lloyd, J. Chemom., 2014, 28, 213–225 CrossRef CAS.
- L. Nørgaard, A. Saudland, J. Wagner, J. P. Nielsen, L. Munck and S. B. Engelsen, Appl. Spectrosc., 2000, 54, 413–419 CrossRef.
- T. Röder, J. Moosbauer, G. Kliba, S. Schlader, G. Zuckerstätter and H. Sixta, Lenzinger Ber., 2009, 87, 98–105 Search PubMed.
- M. Makarem, C. H. Lee, K. Kafle, S. Huang, I. Chae, H. Yang, J. D. Kubicki and S. H. Kim, Cellulose, 2019, 26, 35–79 CrossRef CAS.
- M. Schwanninger, J. C. Rodrigues and K. Fackler, J. Near Infrared Spectrosc., 2011, 19, 287–308 CrossRef CAS.
- N. F. Pérez, J. Ferré and R. Boqué, Chemom. Intell. Lab. Syst., 2009, 95, 122–128 CrossRef.
- R. G. Brereton, J. Chemom., 2011, 25, 225–246 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01794b |
| This journal is © The Royal Society of Chemistry 2021 |