Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Inline electrochemical reduction of NISTmAb for middle-up subunit liquid chromatography-mass spectrometry analysis†
Tomos E.
Morgan
*a,
Craig
Jakes
ab,
Hendrik-Jan
Brouwer
 c,
Silvia
Millán-Martín
a,
Jean-Pierre
Chervet
c,
Ken
Cook
d,
Sara
Carillo
a and
Jonathan
Bones
c,
Silvia
Millán-Martín
a,
Jean-Pierre
Chervet
c,
Ken
Cook
d,
Sara
Carillo
a and
Jonathan
Bones
 *ab
*ab
aCharacterisation and Comparability Laboratory, NIBRT – the National Institute for bioprocessing research and training, Foster Avenue, Mount Merrion, Blackrock, Co., Dublin, A94 X099, Ireland. E-mail: jonathan.bones@nibrt.ie
bSchool of Chemical Engineering and Bioprocessing, University College of Dublin, Belfield, Dublin 4, Ireland
cAntec Scientific, Industrieweg 12, 2382NV Zoeterwoude, The Netherlands
dThermo Fisher Scientific, Hemel Hempstead, Herts, HP2 7GE, UK
First published on 29th September 2021
Abstract
Disulfide bond reduction within antibody mass spectrometry workflows is typically carried out using chemical reducing agents to produce antibody subunits for middle-down and middle-up analysis. In this contribution we offer an online electrochemical reduction method for the reduction of antibodies coupled with liquid chromatography (LC) and mass spectrometry (MS), reducing the disulfide bonds present in the antibody without the need for chemical reducing agents. An electrochemical cell placed before the analytical column and mass spectrometer facilitated complete reduction of NISTmAb inter- and intrachain disulfide bonds. Reduction and analysis were carried out under optimal solvent conditions using a trapping column and switching valve to facilitate solvent exchange during analysis. The level of reduction was shown to be affected by electrochemical potential, temperature and solvent organic content, but with optimization, complete disulfide bond cleavage was achieved. The use of an inline electrochemical cell offers a simple, rapid, workflow solution for liquid chromatography mass spectrometry analysis of antibody subunits.
Introduction
Monoclonal antibodies (mAb) and their variants make up an increasingly large field within biotherapeutics. Due to their size (∼150 kDa) and heterogeneity deriving from expression and process related post-translational modifications (PTMs), analytical methods are being continuously developed for their characterization.1,2 MAb analysis is commonly broken down further into native, intact, subunits, top- and middle-down, and bottom-up workflows.3,4Intact mAb workflows are affective at confirming protein mass and possible PTMs via direct mass measurement mass spectrometry.5–7 Middle-down and middle-up techniques evaluate antibodies at the subunit level (e.g. light chain – Lc, heavy chain – Hc, crystallizable fragment – Fc, and antigen binding region – Fab). Modified subunits are more easily separated by chromatography than intact mAbs due to the reduction in molecular size, and high-resolution mass spectrometry methods can be utilized more effectively on smaller analytes. Localization of PTMs to an individual subunit is also beneficial for the assessment of critical quality attributes.8–10 Subunit production is often carried out by non-biological chemical reaction and/or enzyme digestion.10,11 Consistent subunit production is essential in middle-up and middle-down mass spectrometry workflows requiring effective reduction steps.
Both inter- and intra-chain disulfide bonds are common and important features of antibodies. Disulfide bonds create the 3D structure required for bioactivity and in antibodies, hold the light and heavy chains together.12 Mispairing of disulfide bonds may affect function and stability of the final drug, especially for mAbs with more complex disulfide bonds patterns, like IgG2, leading to the need for a thorough characterization. The presence of extensive intrachain disulfide bond formation within antibodies makes sequencing by tandem mass spectrometry (MS/MS) for top- and middle-down techniques much more challenging.13–15
Reduction of disulfide bonds is often carried out chemically by dithiothreitol (DTT)16 or tris(2-carboxyethyl)phosphine (TCEP)17 producing light chain (Lc) and heavy chain (Hc) units or further coupled with IdeS/IdeZ enzymes to produce Lc, Fc/2, and Fd subunits.18–20 Denaturing agents, such as guanidine hydrochloride, are often used to relax the tertiary structure of the antibody to allow for more complete reduction.21
Online chemical reduction has been successfully performed with the use of a trap column and DTT being introduced to the flow path to reduce the antibody.22 Online reduction methods with the use of DTT require the reaction to be performed under basic conditions which are not hydrogen–deuterium exchange (HDX) compatible. Disulfide bond reduction has also been carried out with the use of 254 nm radiation.23
Electrochemical reduction of disulfide bonds in proteins using a flow-through reactor cell prior to mass spectrometry analysis has been successfully applied as an alternative method previously. Disulfide bond cleavage using such an approach is achieved by inline electrochemical reduction using a titanium working electrode under acidic conditions with formic acid as an additive in the sample solution.24 It was postulated that the presence of formic acid under these conditions will result in the formation of carboxylic radical anions which act as mediator for disulfide reduction.25
Accessible disulfide bonds present during the reduction will be reduced to free thiol groups. Complete and partial electrochemical reduction of multiple proteins and peptides prior to mass spectrometry has been carried out by direct infusion through the electrochemical cell.24,26–30 Reduction of an intact antibody was also carried out through a direct infusion method, successfully producing Lc and Hc subunits but not completely reducing the intrachain disulfide bonds.31 A downside of the direct infusion methods are that the reduced products are not separated prior to analysis. The use of electrochemical reduction for HDX workflows has also been investigated with success as normal reductive conditions are not compatible with HDX workflows.32,33
This contribution presents an inline electrochemical reduction workflow that reduces both inter- and intra-chain disulfide bonds of NISTmAb. The fully reduced subunits produced were analyzed by reverse-phase separation. The method does not need the addition of chemical reducing agents or denaturing agents to produce complete reduction. A trapping column setup also allows the hyphenation with mass spectrometry using compatible solvents for liquid chromatography-mass spectrometry analysis. The system setup also allows switching between intact and middle up/down analysis with a single step.
Experimental
Antibody preparation
NISTmAb Reference Material 8671 acquired from The National Institute of Standards and Testing (NIST, Gaithersburg, Maryland) which contains high levels of lysine clipping and pyroglutamine modification was buffer exchanged into 1% formic acid (FA) and ultrapure water using centrifuge 10 kDa molecular weight cutoff filters (BioRad, Hercules, CA, USA) final concentration of NISTmAb was 1 mg mL−1. Experiments carried out under denaturing conditions were buffer exchanged into 1% FA and concentration adjusted to 20% acetonitrile 1% FA (v/v) at a concentration of 1 mg mL−1 of NISTmab. For FabRICATOR digestion product reduction: 100 μg of NISTmAb was digested with 100 units of FabRICATOR (Genovis AB) for 2 hours at 37 °C. Following digestion samples were reduced to dryness via vacuum centrifugation before being resuspended in 1% FA in water.Analytical instrumentation
All liquid chromatography-mass spectrometry (LC-MS) analyses were performed using a Vanquish™ Flex Duo UHPLC system (Thermo Scientific, Germering, Germany) and Q-Exactive™ Plus Hybrid Quadrupole Orbitrap MS with extended mass BioPharma Option utilising intact protein mode, equipped with an Ion Max source with HESI-II probe (Thermo Scientific, Bremen, Germany). All data was acquired using Thermo Scientific™ Chromeleon™ software 7.2.10.Mass spectrometry
The Q-Exactive orbitrap mass spectrometer remained in intact protein mode for the duration of the experiment. Full mass spectrometry conditions are contained within ESI Table S3.† The mass spectrometer was set to full positive polarity MS scans at 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolving power (at m/z 200, 512 ms transient length) for the Lc analysis before switching to 35
000 resolving power (at m/z 200, 512 ms transient length) for the Lc analysis before switching to 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolving power (at m/z 200, 128 ms transient length) for the heavy chain analysis, at 14 minutes. The mass spectrometer was held at 140
000 resolving power (at m/z 200, 128 ms transient length) for the heavy chain analysis, at 14 minutes. The mass spectrometer was held at 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolving power for the duration of the FabRICATOR sample experiment. The detected mass range was from m/z 600–4000, the AGC target value was 3 × 106. Tandem mass spectrometry was carried out at 140
000 resolving power for the duration of the FabRICATOR sample experiment. The detected mass range was from m/z 600–4000, the AGC target value was 3 × 106. Tandem mass spectrometry was carried out at 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolving power using higher energy collisional dissociation (HCD) with a collision energy of 12–18 V in 2 V steps, and the fragmentation coverage collated in the results. All mass spectrometry analysis was carried out using Thermo Scientific™ BioPharmaFinder™ 3.2. Hc deconvolution was carried out using the ReSpect algorithm and Lc, FabRICATOR and tandem mass spectrometry deconvolution was carried out with the Xtract algorithm.
000 resolving power using higher energy collisional dissociation (HCD) with a collision energy of 12–18 V in 2 V steps, and the fragmentation coverage collated in the results. All mass spectrometry analysis was carried out using Thermo Scientific™ BioPharmaFinder™ 3.2. Hc deconvolution was carried out using the ReSpect algorithm and Lc, FabRICATOR and tandem mass spectrometry deconvolution was carried out with the Xtract algorithm.
Electrochemical cell
A μ-PrepCell™ SS reactor cell in combination with the ROXY™ Exceed potentiostat (Antec Scientific, Leiden, Netherlands) was used for the online electrochemical reduction of NISTmAb. The potentiostat with integrated oven compartment was operated by instrument control in the Thermo Scientific™ Chromeleon™ Software, version 7.2.10. The μ-PrepCell SS is a dual electrode flow-through cell consisting of a replaceable platinum counter electrode (CE) and the titanium surface of the inlet block acting as the working electrode (WE) at which reduction takes place. PEEK tubing was used to connect the electrochemical cell into the flow path. The flow rate through the electrochemical cell was held at 50 μL min−1 for the duration of the experiment.The EC setup allows the control of both temperature and electrode potential within the EC using Thermo Scientific™ Chromeleon™ Software 7.2.10. The temperature was held at room temperature for the partially reduced experiments and 60 °C for the full reduced experiments. Solvent flow was fed directly from the autosampler switching valve through the electrochemical cell, where reduction occurred, and onto the trapping column. The electrochemical cell was operated in a pulse mode using a 2-step square-wave pulse with the following settings: E1 = 0–1 V (reduction), E2 = 0 volt, with a t1 = 1 s and t2 = 0.1 s. E2 is a short cleaning step with a duration of 100 ms. The electrode potential, (E1) was varied between 0 V to 1 V for comparison of reduction conditions in 0.2 V increments. All the experiments comparing reduction at different temperatures and organic solvent content used 1 V in the electrochemical cell.
Intact electrochemical reduction analysis
With the switching valve initially held in the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 position for intact mAb analysis 5 μg of mAb sample was injected through the electrochemical cell at a flow rate of 50 μL min−1 fed from pump 1 and trapped onto a MAbPac™ RP column, 4 μm, 2.1 mm × 50 mm. Conditions were isocratic at 20% (v/v) acetonitrile, 1% (v/v) FA, in water, 1% (v/v) FA in water was used for the partial reduction experiments. After 5 minutes the switching valve moved to the 1
6 position for intact mAb analysis 5 μg of mAb sample was injected through the electrochemical cell at a flow rate of 50 μL min−1 fed from pump 1 and trapped onto a MAbPac™ RP column, 4 μm, 2.1 mm × 50 mm. Conditions were isocratic at 20% (v/v) acetonitrile, 1% (v/v) FA, in water, 1% (v/v) FA in water was used for the partial reduction experiments. After 5 minutes the switching valve moved to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 position. The mobile phases for the right pump consist of 0.1% FA in water (A) and 0.1% FA in acetonitrile (B). The flow was held for 5 minutes at 25% B at 0.3 mL min−1 to allow back flushing from the trap column onto the analytical column MAbPac™ RP column, 4 μm, 2.1 mm × 150 mm, at 10 minutes the gradient was immediately increased to 30% B and then increased to 40% over 10 minutes. At 20 minutes the mobile phase was adjusted to 80% B and held for 6 minutes before being dropped to 20% B from minutes 26–28 before being held at 20% B for a further 2 minutes for a total analytical analysis time of 30 minutes.
2 position. The mobile phases for the right pump consist of 0.1% FA in water (A) and 0.1% FA in acetonitrile (B). The flow was held for 5 minutes at 25% B at 0.3 mL min−1 to allow back flushing from the trap column onto the analytical column MAbPac™ RP column, 4 μm, 2.1 mm × 150 mm, at 10 minutes the gradient was immediately increased to 30% B and then increased to 40% over 10 minutes. At 20 minutes the mobile phase was adjusted to 80% B and held for 6 minutes before being dropped to 20% B from minutes 26–28 before being held at 20% B for a further 2 minutes for a total analytical analysis time of 30 minutes.
IdeS FabRICATOR® samples
With the switching valve in the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 position IdeS digested NISTmAb samples were loaded onto the trapping column using 20% acetonitrile and 1% FA in water. After five minutes the switching valve was moved to position 1
6 position IdeS digested NISTmAb samples were loaded onto the trapping column using 20% acetonitrile and 1% FA in water. After five minutes the switching valve was moved to position 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 at 25% B and held isocratically for a further 5 minutes. The mobile phase was increased to 32% B over 6 minutes and then up to 80% for a further 10 minutes, held at 80% B for 3 minutes before dropping the mobile phase down to 20% B for 2 minutes for a total length of 30 minutes.
2 at 25% B and held isocratically for a further 5 minutes. The mobile phase was increased to 32% B over 6 minutes and then up to 80% for a further 10 minutes, held at 80% B for 3 minutes before dropping the mobile phase down to 20% B for 2 minutes for a total length of 30 minutes.
Results and discussion
Trapping column and solvent switching
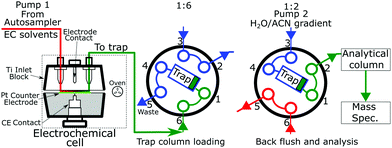
The key module of the Vanquish Flex Duo system is the dual pump that produced two discrete flows. One pump which feeds the electrochemical cell onto the trapping column. The second pump backflushed the trapping column and through the analytical column into the mass spectrometer. A switching valve setup (Fig. 1) is placed after the electrochemical cell and before the trapping and analytical columns. Fig. 1 presents the electrode and switching column used in the experimentation.Reduction potential testing
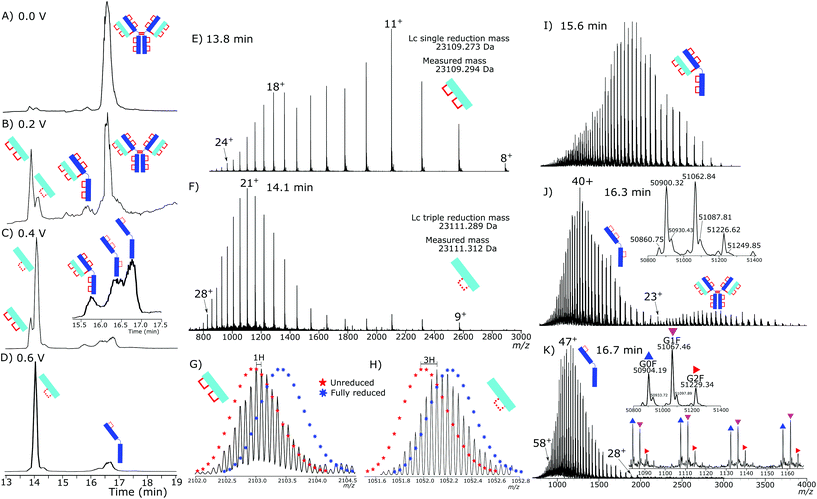
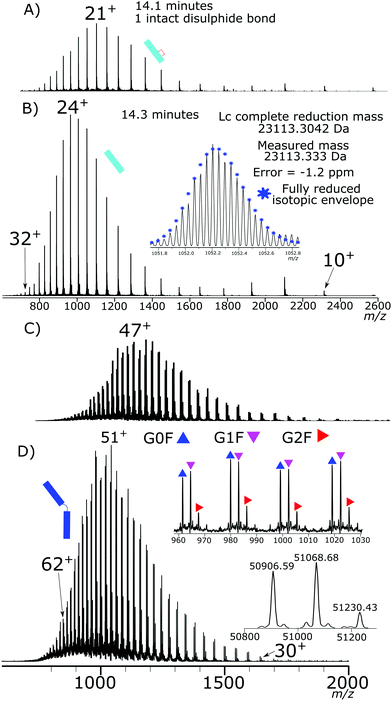
Initial testing of different electrode potentials within the reduction step showed an increase in disulfide bond reduction at higher electrode potentials. The antibody was exposed to the reduction potentials in 0.2 V increments from 0 V to 1 V. Fig. 2(A)–(D) presents four total ion chromatograms (TICs) corresponding to the reduction of NISTmAb under the different electrode potential conditions.At 0 V, Fig. 2A, completely intact NISTmAb was observed, as expected, increasing the electrode potential to 0.2 V, Fig. 2B, showed formation of light chain (Lc) with both intrachain disulfide bonds still intact at 13.8 min, a Lc with one intrachain disulfide bond reduced at 14.1 min. A peak at 15.7 min was detected corresponding to a Lc/Hc pairing as well as significant intact antibody still detected. At 0.4 V, Fig. 2C, potential shifted the distribution significantly, the majority of Lc became partially reduced, only containing one intrachain disulfide bond, a series of Hc products were detected between 16 and 17 minutes each with different levels of intrachain disulfide bond reduction. At 0.6 V, Fig. 2D, two main products were seen, the partially reduced Lc and partially reduced Hc both containing one intrachain disulfide bond.
Reduction of the disulfide bonds increased the retention times of both the Lc and Hc subunits. The increase in retention time is not unexpected, the loss of native structure with disulfide bond reduction increased the antibody hydrodynamic volume and hydrophobicity.
The reduction of disulfide bonds resulted in two significant changes in the mass spectrum, firstly, the mass increased by 2 Da for each intrachain disulfide bond reduced. The increase in exposed surface area caused by the reduction of the disulfide bonds increased the net charge during electrospray ionization. At high resolving powers isotopically resolved Lc was observed and compared to theoretical isotopic envelopes. The two Lc distributions, Fig. 2E and F, show clearly the change caused by an increased reduction potential.
Fig. 2E presents the mass spectrum of the chromatographic peak eluting at 13.8 min. The distribution of charge states is bimodal, having maxima at +11 (m/z 2103.22) and +18 (m/z 1285.58) charges. The detectable charge state range was from +24 (m/z 964.69) to +8 (m/z 2891.68). Deconvoluting the spectrum gave a total mass of 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 109.294 Da accurately matching the predicted mass of the Lc with a single interchain disulfide bond reduced but with both intrachain disulfide bonds intact.
109.294 Da accurately matching the predicted mass of the Lc with a single interchain disulfide bond reduced but with both intrachain disulfide bonds intact.
The isotopic envelope, Fig. 2G, was shifted from the +11 charge state by m/z 0.091 producing a 1 Da shift after deconvolution. The shift after deconvolution of 1 Da is due to the addition of a single hydrogen after reduction of the interchain disulfide bond, producing the free Lc.
Fig. 2F presents the mass spectrum of the chromatographic peak eluting at 14.1 min. The distribution of Lc charge states for the peak at 14.1 min ranged from +28 (m/z 826.84), to +9 (m/z 2570.49) with the most abundant charge state +21 (m/z 1157.22). The charge state distribution was shifted significantly to higher charge states, demonstrating a relaxation of the 3D structure caused by increased reduction. Fig. 2H shows the isotopic shift corresponding to 3 Da due to reduction of the interchain disulfide bond and one of the intrachain disulfide bonds. The mass difference between the measured and theoretical masses aligns closely (−1 ppm). Only a single peak is observed for the partially reduced Lc species, eluting at 14.1 minutes, Fig. 2D. Partial reduction could have produced two products, intrachain disulfide bond reduction of the C23–C87 or C133–C193 disulfide bond positions. A single detected peak within the chromatogram for the partially reduced Lc suggests that either the chromatographic characteristics of the resultant Lc after either single intrachain disulfide bond reduction are the same, or that only a single position of intrachain disulfide bond is being reduced, and not a mixture of both reduced states.
Fig. 2I shows intact light- and heavy chain still bound, but with the hinge region disulfide bonds reduced. The intrachain disulfide bonds are still intact within the antibody structure. The partially reduced structure is only observed at low electrode potentials. Consistently across the experiments, the loss of interchain disulfide bonds occurs more readily than the intrachain disulfide bonds.
The Hc mass spectrum shown in Fig. 2J presents the chromatographic peak at 16.3 minutes from Fig. 2C. The deconvoluted mass spectrum gives a mass at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 Da for the G0F Hc, 6 Da less than a completely reduced Hc, Table S4† contains comparisons of masses. The mass difference aligns with three internal disulfide bonds still being intact. The charge distribution of the Hc had a maximum at +40 (m/z 1273.49) and the highest detected charge was +51 (m/z 999.06) and minimum detected charge was +23 (m/z 2213.97). Further reduction at higher electrode potentials, Fig. 2K, showed a shift to higher charge states with the highest detected charge state +57 (m/z 893.99), the maximum intensity charge state was +47 (m/z 1084.01) with a minimum charge state of +28 (m/z 1819.07).
900 Da for the G0F Hc, 6 Da less than a completely reduced Hc, Table S4† contains comparisons of masses. The mass difference aligns with three internal disulfide bonds still being intact. The charge distribution of the Hc had a maximum at +40 (m/z 1273.49) and the highest detected charge was +51 (m/z 999.06) and minimum detected charge was +23 (m/z 2213.97). Further reduction at higher electrode potentials, Fig. 2K, showed a shift to higher charge states with the highest detected charge state +57 (m/z 893.99), the maximum intensity charge state was +47 (m/z 1084.01) with a minimum charge state of +28 (m/z 1819.07).
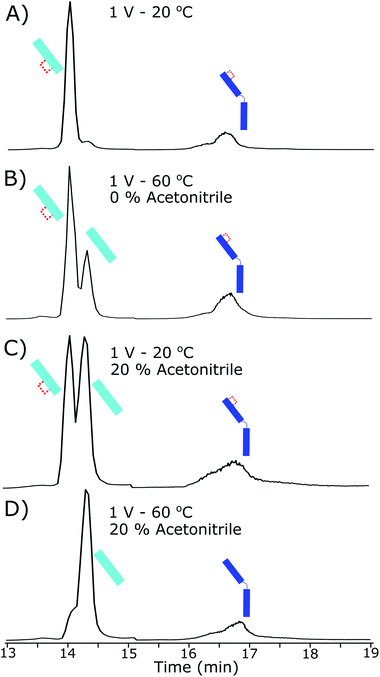
Lack of complete intrachain disulfide bond reduction but complete reduction of the interchain disulfide bond is evidence that, although the electrode potential is high enough for disulfide bond reduction to occur, an external factor was limiting further reduction. Higher electrode potentials above 0.6 V did not increase reduction further, evidence for the constrained intrachain disulfide bonds limiting the progress of the reaction. As the intrachain disulfide bonds were restricted it is likely this is an effect of the electrochemical reaction kinetics. Fig. 3 shows the increased reduction that occurs when the antibody is further denatured. By increasing the temperature inside the electrochemical cell to 60 °C the presence of a fully reduced Lc was observed in the subsequent chromatographic separation. At 60 °C the ratio of partially reduced to fully reduced Lc was approximately 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The increase in temperature to 60 °C will have increased the flexibility of the structure allowing reduction to occur.
1. The increase in temperature to 60 °C will have increased the flexibility of the structure allowing reduction to occur.
Adjusting the solvent used for electrochemical reduction to 20% acetonitrile, but maintaining the cell at room temperature, resulted in approximately 50% of the Lc reducing fully. The current within the electrochemical cell was reduced slightly by the increase in organic content (by approximately 5%) although reduction increased in total. The addition of acetonitrile and increase in reduction is strong evidence to the loss of 3D structure causing the increase in reduction, as acetonitrile will not be an effective charge carrier for the reduction to occur but acted as a denaturing agent at these concentrations.
By coupling both an increase in temperature and increased organic portion of the mobile phase the Lc was completely reduced, as shown in Fig. 3D. Similar results were observed for the Hc reduction but were harder to quantitate due to a lack of separation of the partially reduced species. At the highest denaturing conditions, the majority of the Hc product was fully reduced.
Fig. 4 shows the corresponding mass spectrum to both the Lc, Fig. 4B, and Hc, Fig. 4D, from the completely reduced sample where the Lc charge distribution has shifted further to higher charge states. Above them, Fig. 4A, and C, from Fig. 2F and K, are the corresponding partially reduced states for reference. The corresponding isotopic distribution now aligns with that of a fully reduced Lc. The deconvoluted mass now aligning to the addition of 5 hydrogen atoms in total represented the reduction of the interchain and two intrachain disulfide bonds.
 | ||
| Fig. 4 (A) Mass spectrum of partially reduced Lc from Fig. 2F compared to (B) mass spectrum of fully reduced Lc showing charge state shift as well as accurate alignment of mass and isotopic envelope of the +22 charge state with the theoretical isotope distribution of the fully reduced Lc. (C) Partially reduced Hc distribution from Fig. 2K compared to (D) fully reduced Hc showing mass agreement with the fully reduced species of the three main glycoforms, as well as significant charge state shift of the fully reduced Hc. | ||
The Lc charge distribution significantly shifted from the partially reduced Lc again. Shifting to higher charge states is indicative of further denaturing and structure loss. The detected charge state range was +32 (m/z 723.67) to +10 (m/z 2313.54), with a peak maximum at +24 (m/z 964.65).
The Hc charge state shift from +63 (m/z 808.75) to +30 (m/z 1697.80) with a charge state maximum at +51 (m/z 1043.24). The Hc mass aligns with that of fully reduced Hc, Table S4.† Overall, complete reduction of the Hc and Lc was achieved under increased denaturing conditions from the intact antibody. The electrochemical reduction is effective without the use of specific denaturing agents or reducing agents. The reduction of the antibody just prior to analysis allows reduction of the disulfide bonds without evidence of disulfide bond reformation on the analysis timescale. The electrochemical reduction also showed no evidence of antibody modification further than disulfide bond reduction.
Electrochemical reduction of IdeS digested products
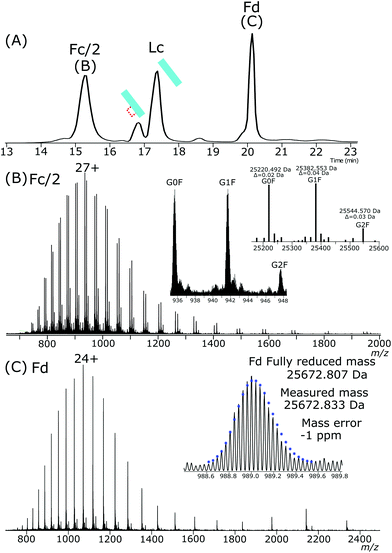
NISTmAb was treated with IdeS enzyme to produce Fc and Fab subunits. Completely reduced Fc/2, Fd and Lc subunits were present after exposure of the IdeS generated Fab and Fc subunits to the same electrochemical conditions used for intact NISTmAb. The resulting subunits, the chromatogram of which is shown in Fig. 5A, Fc/2, Fd and Lc have highly abundant completely reduced forms. Fc/2 had a retention time of 15.2 minutes, partially reduced Lc at 16.8 minutes, fully reduced Lc at 17.3 minutes and the Fd peak at 20.1 minutes. Comparing the abundances of partially reduced to fully reduced Lc gives a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.
5.
The Fc/2, Fig. 5B, is completely reduced and has a charge state maximum at +27. The mass agreements between the measured deconvoluted masses and theoretical masses are less than 2 ppm for the three main glycoforms.
The Fd, Fig. 5C, was completely reduced with a +24 charge maximum. The isotopic envelope closely aligns with that of a completely reduced theoretical isotopic distribution.
The chromatography conditions for the separation of the IdeS produced subunits separated the partially reduced Lc from the fully reduced Lc. The presence of a single peak for the partially reduced Lc indicates either a single intrachain disulfide bond was selectively reduced across the ensemble or that chromatographic properties are not significantly affected by reduction of one disulfide bond compared to the other.
Tandem mass spectrometry analysis
Tandem mass spectrometry analysis of the fully reduced and partially reduced Lc gave insights into the selectivity of the intrachain disulfide bond reduction that was occurring. Evenly distributed disulfide bond reduction would have produced fragments across the Lc, even in the partially reduced species, as the ensemble average of partially reduced Lc should give fragmentation from both intrachain disulfide bonds. 2 Da shifted fragments were also used to ascertain the position of intact intrachain disulfide bonds. By chromatography, the partially reduced peak was intact, leading to two potential conclusions; that the reduction of one disulfide bond compared to the other did not affect chromatographic behavior significantly, or that only a single intrachain disulfide bond was being reduced in the NISTmAb sample.HCD fragmentation was carried out at low energies as to not maximize fragmentation coverage, but to minimize the chance of internal or secondary fragmentation to accurately compare the reduction of intrachain disulfide bonds across the partially and fully reduced Lc.
At the low fragmentation energies used, although exhibiting low coverage, significant differences between the fragmentation coverage were observed.
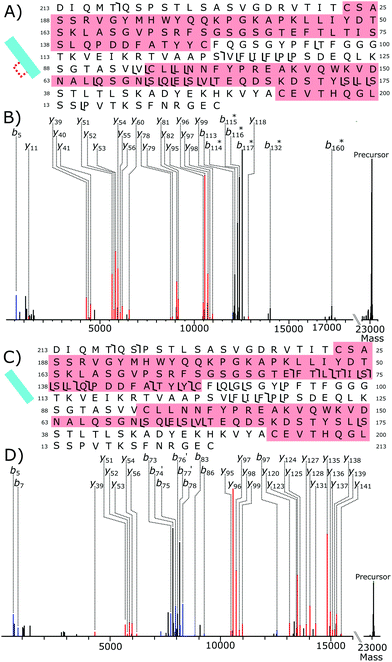
Fig. 6A shows the fragmentation coverage of the partially reduced Lc, detected at 14.1 minutes in the chromatogram, Fig. 3A. Within the partially reduced structure no fragmentation occurs in the Lc from C23 to C87 suggesting that, even across the ensemble average, the disulfide bond within this region is still intact. Fragmentation within the second intrachain disulfide bond, C133–C193, with y-fragments present from y51–y56 and from y78–y82. Detected fragments outside of the C23–C87 intrachain disulfide bond region but offset by 2 Da is further evidence of selective disulfide bond reduction occurring. Fragments b114–b117 are all offset by 2 Da, as well as b132 and b170. The lack of any 2 Da shifted y fragments gives further evidence that, even across the ensemble average, most of the reduction has occurred to the C133–C193 disulfide bond. Overall, the tandem mass spectrometry data of the partially reduced species gives evidence that for NISTmAb reduction of the intrachain disulfide bonds has not occurred equally under partial reduction conditions.
Within the fully reduced sample, fragments y141 to y120 show that fragmentation has occurred in the C23–C87 disulfide bond region. Fragments y51–y53 show the second, C133–C193, disulfide bond region has also been reduced. There are no fragment peaks that correspond to the difference of 2 Da caused by an intact disulfide bond showing again that reduction has occurred to the disulfide bonds exclusively.
Conclusions
Electrochemical reduction is an effective method for the reduction of protein disulfide bonds. Complete electrochemical reduction of an antibody can be carried out inline with LC-MS analysis without the need for reducing agents and denaturing agents, save for common organic solvents. The use of a trapping column enabling solvent switching from electrochemical compatible solvents to mass spectrometry compatible solvents and produced fully reduced light and heavy chain subunits from an intact antibody without the need for prior sample workup.A kinetic limit was reached for the electrochemical reduction of the antibody requiring higher temperatures and addition of organic solvent to increase denaturation of the antibody. Selectivity of the disulfide bond electrochemical reduction was also shown, further investigation into the control of electrochemical reduction of antibodies via top-down analysis, could yield useful information into the formation of non-uniform disulfide bonding structures within antibodies.
The electrochemical cell flowing into a trapping column presented here could be used for a wide range of samples by simply adjusting the trapping column stationary phase. Consistent acidic conditions allow the use of HDX with electrochemical reduction as well as minimizing possible analytical artefacts. The introduction of an effective means of reducing proteins without the need for chemical reducing agents, while still allowing standard LC-MS methods to be used, will hopefully prove beneficial to the wider community.
Author contributions
Tomos E. Morgan experimental, authorship, conception, and data analysis. Craig Jakes experimental, authorship, conception and design. Hendrik-Jan Brouwer experimental, data analysis and inter-pretation, conception and design. Silvia Millán Martín, data analysis and interpretation. Jean-Pierre Chervet, conception and design. Ken Cook, conception, design, and data interpretation. Sara Carillo, conception, design and data interpretation. Jonathan Bones conception, design and data interpretation. All authors were involved in drafting and revision and have given approval to the final version of the manuscript.Conflicts of interest
H.-J. Brouwer and J. P. Chervet are employees of Antec Scientific, manufacturers of the Roxy™ Exceed Electrochemical cell device. K. Cook is an employee of Thermo Fisher Scientific, the corporation that develops and produces Vanquish UHPLC systems and Orbitrap mass spectrometers. Beyond this, the authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this article.Acknowledgements
Thanks are extended to Dr Florian Füssel, Dr Josh Smith, Dr Lisa Strasser, and Dr Kai Scheffler for useful discussions. The authors gratefully acknowledge funding from Thermo Fisher Scientific as a collaborative research agreement with NIBRT.Notes and references
- K. Sandra, I. Vandenheede and P. Sandra, J. Chromatogr. A, 2014, 1335, 81–103 CrossRef CAS PubMed.
- A. Beck, E. Wagner-Rousset, D. Ayoub, A. Van Dorsselaer and S. Sanglier-Cianferani, Anal. Chem., 2013, 85, 715–736 CrossRef CAS PubMed.
- F. Lermyte, Y. O. Tsybin, P. B. O'Connor and J. A. Loo, J. Am. Soc. Mass Spectrom., 2019, 30, 1149–1157 CrossRef CAS PubMed.
- K. Srzentic, K. O. Nagornov, L. Fornelli, A. A. Lobas, D. Ayoub, A. N. Kozhinov, N. Gasilova, L. Menin, A. Beck, M. V. Gorshkov, K. Aizikov and Y. O. Tsybin, Anal. Chem., 2018, 90, 12527–12535 CrossRef CAS PubMed.
- D. P. Donnelly, C. M. Rawlins, C. J. DeHart, L. Fornelli, L. F. Schachner, Z. Lin, J. L. Lippens, K. C. Aluri, R. Sarin, B. Chen, C. Lantz, W. Jung, K. R. Johnson, A. Koller, J. J. Wolff, I. D. G. Campuzano, J. R. Auclair, A. R. Ivanov, J. P. Whitelegge, L. Pasa-Tolic, J. Chamot-Rooke, P. O. Danis, L. M. Smith, Y. O. Tsybin, J. A. Loo, Y. Ge, N. L. Kelleher and J. N. Agar, Nat. Methods, 2019, 16, 587–594 CrossRef CAS PubMed.
- J. Zhang, H. Liu and V. Katta, J. Mass Spectrom., 2010, 45, 112–120 CAS.
- N. J. Thompson, S. Rosati, R. J. Rose and A. J. Heck, Chem. Commun., 2013, 49, 538–548 RSC.
- R. D. Melani, K. Srzentic, V. R. Gerbasi, J. P. McGee, R. Huguet, L. Fornelli and N. L. Kelleher, mAbs, 2019, 11, 1351–1357 CrossRef CAS PubMed.
- C. Blochl, C. Regl, C. G. Huber, P. Winter, R. Weiss and T. Wohlschlager, Sci. Rep., 2020, 10, 18080 CrossRef PubMed.
- P. Liu, X. Zhu, W. Wu, R. Ludwig, H. Song, R. Li, J. Zhou, L. Tao and A. M. Leone, Rapid Commun. Mass Spectrom., 2019, 33, 31–40 CrossRef CAS PubMed.
- A. Farrell, S. Carillo, K. Scheffler, K. Cook and J. Bones, Anal. Methods, 2018, 10, 3100–3109 RSC.
- H. Liu and K. May, mAbs, 2012, 4, 17–23 CrossRef PubMed.
- E. J. Larson, Y. Zhu, Z. Wu, B. Chen, Z. Zhang, S. Zhou, L. Han, Q. Zhang and Y. Ge, Anal. Chem., 2020, 92, 15096–15103 CrossRef CAS PubMed.
- V. C. Cotham and J. S. Brodbelt, Anal. Chem., 2016, 88, 4004–4013 CrossRef CAS PubMed.
- K. Srzentic, L. Fornelli, Y. O. Tsybin, J. A. Loo, H. Seckler, J. N. Agar, L. C. Anderson, D. L. Bai, A. Beck, J. S. Brodbelt, Y. E. M. van der Burgt, J. Chamot-Rooke, S. Chatterjee, Y. Chen, D. J. Clarke, P. O. Danis, J. K. Diedrich, R. A. D'Ippolito, M. Dupre, N. Gasilova, Y. Ge, Y. A. Goo, D. R. Goodlett, S. Greer, K. F. Haselmann, L. He, C. L. Hendrickson, J. D. Hinkle, M. V. Holt, S. Hughes, D. F. Hunt, N. L. Kelleher, A. N. Kozhinov, Z. Lin, C. Malosse, A. G. Marshall, L. Menin, R. J. Millikin, K. O. Nagornov, S. Nicolardi, L. Pasa-Tolic, S. Pengelley, N. R. Quebbemann, A. Resemann, W. Sandoval, R. Sarin, N. D. Schmitt, J. Shabanowitz, J. B. Shaw, M. R. Shortreed, L. M. Smith, F. Sobott, D. Suckau, T. Toby, C. R. Weisbrod, N. C. Wildburger, J. R. Yates 3rd, S. H. Yoon, N. L. Young and M. Zhou, J. Am. Soc. Mass Spectrom., 2020, 31, 1783–1802 CrossRef CAS PubMed.
- M. M. C. Sun, K. S. Beam, C. G. Cerveny, K. J. Hamblett, R. S. Blackmore, M. Y. Torgov, F. G. M. Handley, N. C. Ihle, P. D. Senter and S. C. Alley, Bioconjugate Chem., 2005, 16, 1282–1290 CrossRef CAS PubMed.
- H.-M. Zhang, S. M. McLoughlin, S. D. Frausto, H. Tang, M. R. Emmett and A. G. Marshall, Anal. Chem., 2010, 82, 1450–1454 CrossRef CAS PubMed.
- V. Faid, Y. Leblanc, N. Bihoreau and G. Chevreux, J. Pharm. Biomed. Anal., 2018, 149, 541–546 CrossRef CAS PubMed.
- J. Sjogren, F. Olsson and A. Beck, Analyst, 2016, 141, 3114–3125 RSC.
- B. P. Johansson, O. Shannon and L. Bjorck, PLoS One, 2008, 3, e1692 CrossRef PubMed.
- Z. Lai, J. McCulloch, H. A. Lashuel and J. W. Kelly, Biochemistry, 1997, 36, 10230–10239 CrossRef CAS PubMed.
- L. Verscheure, M. Oosterlynck, A. Cerdobbel, P. Sandra, F. Lynen and K. Sandra, J. Chromatogr. A, 2021, 1637, 461808 CrossRef CAS PubMed.
- X. Yang and Y. Xia, J. Am. Soc. Mass Spectrom., 2021, 32, 307–314 CrossRef CAS PubMed.
- A. Kraj, H. J. Brouwer, N. Reinhoud and J. P. Chervet, Anal. Bioanal. Chem., 2013, 405, 9311–9320 CrossRef CAS PubMed.
- B. B. Stocks and J. E. Melanson, Anal. Bioanal. Chem., 2019, 411, 4729–4737 CrossRef CAS PubMed.
- S. Nicolardi, M. Giera, P. Kooijman, A. Kraj, J. P. Chervet, A. M. Deelder and Y. E. van der Burgt, J. Am. Soc. Mass Spectrom., 2013, 24, 1980–1987 CrossRef CAS PubMed.
- C. N. Cramer, K. F. Haselmann, J. V. Olsen and P. K. Nielsen, Anal. Chem., 2016, 88, 1585–1592 CrossRef CAS PubMed.
- L. Buter, L. M. Frensemeier, M. Vogel and U. Karst, J. Chromatogr. A, 2017, 1479, 153–160 CrossRef PubMed.
- L. Switzar, S. Nicolardi, J. W. Rutten, S. A. Oberstein, A. Aartsma-Rus and Y. E. van der Burgt, J. Am. Soc. Mass Spectrom., 2016, 27, 50–58 CrossRef CAS PubMed.
- Q. Zheng, H. Zhang and H. Chen, Int. J. Mass Spectrom., 2013, 353, 84–92 CrossRef CAS PubMed.
- S. Nicolardi, A. M. Deelder, M. Palmblad and Y. E. van der Burgt, Anal. Chem., 2014, 86, 5376–5382 CrossRef CAS PubMed.
- S. Mysling, R. Salbo, M. Ploug and T. J. Jorgensen, Anal. Chem., 2014, 86, 340–345 CrossRef CAS PubMed.
- E. Trabjerg, R. U. Jakobsen, S. Mysling, S. Christensen, T. J. Jorgensen and K. D. Rand, Anal. Chem., 2015, 87, 8880–8888 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01184g |
| This journal is © The Royal Society of Chemistry 2021 |