Open Access Article
Open Access ArticleA new lateral flow assay to detect sIL-2R during T-cell mediated rejection after kidney transplantation†
Lisa K.
Seiler
*a,
Rebecca
Jonczyk
a,
Patrick
Lindner
a,
Ncog Linh
Phung
a,
Christine S.
Falk
b,
Jessica
Kaufeld
c,
Wilfried
Gwinner
c,
Irina
Scheffner
c,
Stephan
Immenschuh
d and
Cornelia
Blume
a
aInstitute of Technical Chemistry, Leibniz University Hannover, Callinstrasse 5, 30167 Hannover, Germany. E-mail: seiler@iftc.uni-hannover.de
bInstitute for Transplant Immunology, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany
cDepartment of Nephrology, Hannover Medical School, Carl-Neuberg-Straße 1, 30625 Hannover, Germany
dInstitute for Transfusion Medicine, Hannover Medical School, Germany
First published on 2nd August 2021
Abstract
Kidney is the most frequently transplanted among all solid organs worldwide. Kidney transplant recipients (KTRs) undergo regular follow-up examinations for the early detection of acute rejections. The gold standard for proving a T-cell mediated rejection (TCMR) is a biopsy of the renal graft often occurring as indication biopsy, in parallel to an increased serum creatinine that may indicate deterioration of renal transplant function. The goal of the current work was to establish a lateral flow assay (LFA) for diagnosing acute TCMR to avoid harmful, invasive biopsies. Soluble interleukin-2 (IL-2) receptor (sIl-2R) is a potential biomarker representing the α-subunit of the IL-2 receptor produced by activated T-cells, e.g., after allogen contact. To explore the diagnostic potential of sIL-2R as a biomarker for TCMR and borderline TCMR, plasma and urine samples were collected from three independent KTR cohorts with various distinct histopathological diagnostic findings according to BANFF (containing 112 rsp. 71 rsp. 61 KTRs). Samples were analyzed by a Luminex-based multiplex technique and cut off-ranges were determined. An LFA was established with two specific sIL-2R-antibodies immobilized on a nitrocellulose membrane. A significant association between TCMR, borderline TCMR and sIL-2R in plasma and between TCMR and sIL-2R in urine of KTRs was confirmed using the Mann–Whitney U test. The LFA was tested with sIL-2R-spiked buffer samples establishing a detection limit of 25 pM. The performance of the new LFA was confirmed by analyzing urine samples of the 2nd and 3rd patient cohort with 35 KTRs with biopsy proven TCMRs, 3 KTRs diagnosed with borderline TCMR, 1 mixed AMR/TCMR rsp. AMR/borderline TCMR and 13 control patients with a rejection-free kidney graft proven by protocol biopsies. The new point-of-care assay showed a specificity of 84.6% and sensitivity of 87.5%, and a superior estimated glomerular filtration rate (eGFR) at the time point of biopsy (specificity 30.8%, sensitivity 85%).
Introduction
Transplantations of kidneys largely outnumber those of other solid organs and according to the Global Observatory on Donation and Transplantation 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 kidneys were transplanted in 2018 worldwide.1 Furthermore, kidney transplant recipients (KTRs) require intensive immunosuppression even years after kidney transplantation (Ktx) especially when compared to liver recipients in whom a state of graft tolerance can develop.2–4 These treatments have severe side effects for the patients and it is therefore most desirable to decrease the intensity of the immunosuppressive regimen. Despite tremendous improvements in the quality and tolerability of immunosuppressive medications, numerous kidney grafts are rejected by the recipient's immune system and further graft function loss can occur.5 According to the U.S. Department of Health & Human Services, within the first year after transplantation, in 2.5% of the living donor organs and 6.8% of the deceased donor organs rejection episodes occur, most of which are acute T-cell mediated rejections (TCMRs). Although this form of rejection can be effectively treated e.g. by immediate high pulse corticoid therapy and increase of basic immunosuppression, the detection in due time is crucial for a complete remission. Ongoing rejection activity may favor later rejection episodes and facilitate late rejections, and there may be an association with the occurrence of antibody-mediated rejection (AMR) after frequent T-cell allogen recognition.6 In general, five years after transplantation, the probability of rejection increases up to 14.4% (living donations) and 25.6% (deceased donor organs). To detect and treat acute rejection as early as possible, regular follow-up examinations after Ktx are mandatory. Surveillance examinations after Ktx e.g. at the transplant centre of Hannover Medical School were usually performed two times weekly in the first month after transplantation, weekly in between month one and three and at least monthly until the end of the first year after Ktx, later on quarterly. The screening examinations in kidney transplanted children is even more frequent.7,8 An acute rejection can be assumed, when function parameters such as serum creatinine in KTRs rapidly increase, but this indicator is vague and may also indicate an unspecific function failure not triggered by the immune system. Actually, there is a wide range of specific (e.g. BKV nephritis, TMA, glomerulonephritis, pyelonephritis) and non-specific injury which can be reflected by kidney function loss. An acute TCMR or a T-cell driven borderline rejection (borderline TCMR) can be confirmed only by biopsy, categorized according to the BANFF classification.9 Therefore, a graft biopsy is the gold standard for diagnosing transplant rejection. However, this procedure is associated with potential severe clinical complications such as bleeding and infection.10–12 Moreover, such invasive procedures are time-consuming and costly, because they require hospitalization. Therefore, specific markers for kidney transplant rejection monitoring must be determined and it is crucial to develop a reliable and statistically validated non-invasive test that physicians and potentially even patients themselves can apply easily within minutes, which does not cause any pain.
664 kidneys were transplanted in 2018 worldwide.1 Furthermore, kidney transplant recipients (KTRs) require intensive immunosuppression even years after kidney transplantation (Ktx) especially when compared to liver recipients in whom a state of graft tolerance can develop.2–4 These treatments have severe side effects for the patients and it is therefore most desirable to decrease the intensity of the immunosuppressive regimen. Despite tremendous improvements in the quality and tolerability of immunosuppressive medications, numerous kidney grafts are rejected by the recipient's immune system and further graft function loss can occur.5 According to the U.S. Department of Health & Human Services, within the first year after transplantation, in 2.5% of the living donor organs and 6.8% of the deceased donor organs rejection episodes occur, most of which are acute T-cell mediated rejections (TCMRs). Although this form of rejection can be effectively treated e.g. by immediate high pulse corticoid therapy and increase of basic immunosuppression, the detection in due time is crucial for a complete remission. Ongoing rejection activity may favor later rejection episodes and facilitate late rejections, and there may be an association with the occurrence of antibody-mediated rejection (AMR) after frequent T-cell allogen recognition.6 In general, five years after transplantation, the probability of rejection increases up to 14.4% (living donations) and 25.6% (deceased donor organs). To detect and treat acute rejection as early as possible, regular follow-up examinations after Ktx are mandatory. Surveillance examinations after Ktx e.g. at the transplant centre of Hannover Medical School were usually performed two times weekly in the first month after transplantation, weekly in between month one and three and at least monthly until the end of the first year after Ktx, later on quarterly. The screening examinations in kidney transplanted children is even more frequent.7,8 An acute rejection can be assumed, when function parameters such as serum creatinine in KTRs rapidly increase, but this indicator is vague and may also indicate an unspecific function failure not triggered by the immune system. Actually, there is a wide range of specific (e.g. BKV nephritis, TMA, glomerulonephritis, pyelonephritis) and non-specific injury which can be reflected by kidney function loss. An acute TCMR or a T-cell driven borderline rejection (borderline TCMR) can be confirmed only by biopsy, categorized according to the BANFF classification.9 Therefore, a graft biopsy is the gold standard for diagnosing transplant rejection. However, this procedure is associated with potential severe clinical complications such as bleeding and infection.10–12 Moreover, such invasive procedures are time-consuming and costly, because they require hospitalization. Therefore, specific markers for kidney transplant rejection monitoring must be determined and it is crucial to develop a reliable and statistically validated non-invasive test that physicians and potentially even patients themselves can apply easily within minutes, which does not cause any pain.
A variety of immunological biomarkers have been proposed that could be relevant as indicators of acute rejection, e.g. chemokines such as CXCL 9 or CXCL 10,13,14 growth factors such as PDGF-BB15 or cell receptors reflecting the state of immune-cell activity such as sIL-2R. Various candidate non-invasive biomarkers have been linked with the immunological status of the allograft and have shown potential in preliminary studies, but widespread translation of these biomarkers to the clinic will require robust validation studies, stringent standardization of assays and the availability of commercial assays.16
Levels of the interleukin-2 receptor (IL-2R) are increased on activated T-cells and may become detectable after cell shedding as soluble factors in human bodily fluids,17 therefore sIL-2R seemed suitable for the development of an LFA. Among three different subunits of this receptor (IL-2Rα, IL-2Rβ and IL-2Rγc), IL-2Rα is solely expressed by activated T-cells, has a unique structure and is specific for interleukin 2 (IL-2). When IL-2 binds to this cell-standing receptor, activation and proliferation of resting T-helper, T-suppressor and cytotoxic T-cells occur. These activated T-cells initiate and regulate immune reactions, also within pathogenic contexts.
IL-2R appears to be involved in auto-immune diseases that are characterized by a disturbed Treg/Teff balance. For example, elevated serum levels of sIL-2R correlate with disease activity in rheumatoid arthritis, sarcoidosis and complex regional pain syndrome18,19 and here increased serum sIL2R, with respect to IgG4, is accurate in monitoring disease activity and predicting the response of glucocorticoid therapy (91% vs. 41%). T-cell activity in myelofibrosis accompanied by hypersplenismus and anemia20 or within microinflammation of patients affected by diabetes mellitus type II21 was indicated by increased sIL-2R levels. A disturbed Treg/Teff balance also occurs after allogen contact within the context of cell transplantation. As a consequence, sIL-2R plasma levels were shown to be elevated within T-cell mediated graft-versus-host-disease after hematopoietic stem cell transplantation.22–24
sIL-2R levels have also been shown to be up-regulated early after organ transplantation.25 In cardiac graft recipients, “ISHLT rejection type 3” corresponding to a TCMR within the first year after heart transplantation correlated with increased sIL-2R plasma levels.26 Recently, sIL-2R was also shown to be increased in serum of children with an acute kidney graft rejection.27,28
The general problem with immunological markers after organ transplantation is, that their detection can be suppressed by immunosuppressive drug effects. In contrast, sIL-2R was already used as a marker of complications, e.g. after the introduction of the IL-2R antibody basiliximab into routine initial immunosuppressive therapy after renal transplantation at many transplant centers worldwide.29,30
Transcriptomics,31–33 modern digital polymerase chain reaction (dPCR),34–36 cell-free DNA37–39 and proteomics40–44 have already been examined as possible screening tools after Ktx. Due to the comprehensive analysis and low practicability within a short time interval at a transplant outpatient clinic, most of these diagnostic tools have not yet gained clinical acceptance, so a renal biopsy remains the tool of choice. Thus, antibody-dependent test assays such as an LFA and also an enzyme-linked immunosorbent assay (ELISA) appear to be suitable alternatives, but currently only some ELISAs for the detection of sIL-2R are available. Although ELISAs are sensitive methods for protein detection, they require expertise and additional equipment. Generally speaking, ELISAs do not represent point-of-care tests (POCT).
In contrast, an LFA can be read out by the naked eye within minutes. If the test is positive, the user can verify the result himself and turn to his physician for further evaluation of a suspected diagnosis. It is the aim of this study to develop an LFA as a POCT for KTRs to analyze urine samples in the case of a suggested acute TCMR.
Materials
Polyclonal goat anti-human sIL-2R antibody (AF-223-NA) was used as the capture antibody and polyclonal biotinylated goat anti-human sIL-2R antibody (BAF223) was used as the detection antibody. Both antibodies were purchased from R&D Systems (Minneapolis, USA). Rabbit anti-goat antibody (31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105) was obtained from Thermo Fisher Scientific (Rockford, USA) and served as the capture antibody in the control line of the LFA. sIL-2R was obtained from Abbexa Ltd (Cambridge, UK). Conjugate and absorbent pads (Grade 6615, Grade 222) were purchased from Åhlstrom-Munksjö (Helsinki, Finland). Nitrocellulose membrane (Sartorius Unisart® CN180 backed) was kindly provided by Sartorius Stedim Biotech (Goettingen, Germany) and gold nanoparticles (AuNPs) were kindly provided by Fassisi GmbH (Goettingen, Germany). Trehalose was obtained from Fluka BioChemika (Steinheim, Germany) and Tween20 was obtained from PanReac AppliChem (Darmstadt, Germany). As negative controls, C-reactive protein (CRP, Bio-Rad, Hercules, USA), human serum albumin (HSA, Sigma-Aldrich GmbH, Munich), as a further immunological parameter “monokine induced by gamma interferon” (MIG, also called CXCL9; Acris Antibodies GmbH, Herford, Germany) and immunoglobulin from human serum (I4506, Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) were used.
105) was obtained from Thermo Fisher Scientific (Rockford, USA) and served as the capture antibody in the control line of the LFA. sIL-2R was obtained from Abbexa Ltd (Cambridge, UK). Conjugate and absorbent pads (Grade 6615, Grade 222) were purchased from Åhlstrom-Munksjö (Helsinki, Finland). Nitrocellulose membrane (Sartorius Unisart® CN180 backed) was kindly provided by Sartorius Stedim Biotech (Goettingen, Germany) and gold nanoparticles (AuNPs) were kindly provided by Fassisi GmbH (Goettingen, Germany). Trehalose was obtained from Fluka BioChemika (Steinheim, Germany) and Tween20 was obtained from PanReac AppliChem (Darmstadt, Germany). As negative controls, C-reactive protein (CRP, Bio-Rad, Hercules, USA), human serum albumin (HSA, Sigma-Aldrich GmbH, Munich), as a further immunological parameter “monokine induced by gamma interferon” (MIG, also called CXCL9; Acris Antibodies GmbH, Herford, Germany) and immunoglobulin from human serum (I4506, Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) were used.
Phosphate buffered saline (PBS) was prepared with 137 mM sodium chloride, 2.7 mM calcium chloride, 10.1 mM disodium hydrogen phosphate and 1.8 mM calcium hydrogen phosphate (all purchased from Sigma-Aldrich Chemie GmbH). Solution pH was adjusted to 7.4. Sodium borate buffer was prepared with 2 mM sodium tetraborate (Sigma-Aldrich Chemie GmbH) and set to pH 9. Running buffer was prepared with 0.1% bovine serum albumin (BSA, Sigma-Aldrich Chemie GmbH) in PBS. Running buffer for patient samples was prepared with 0.2% bovine serum albumin (BSA, Sigma-Aldrich GmbH) in PBS. Tris buffer (100 mM, pH 8.0, Carl Roth) was prepared with 1% BSA and 0.05% Tween20. All solutions were prepared with distilled water and sterilized by filtration if not indicated otherwise. For the adjustment of pH of all solutions sodium hydroxide (1 M NaOH, Sigma-Aldrich Chemie GmbH) or hydrochloride acid (1 M HCl, Sigma-Aldrich Chemie GmbH) was used.
Methods
Patient sample collection and ethic vote
The samples used in this study (plasma of 1st and 2nd patient cohort, urine of 2nd and 3rd patient cohort) were derived from three independent sample collections in the transplant department of the Hannover Medical School. The 120 KTRs of the 1st patient cohort – aged between 18 and 75 years – were transplanted between 1990 and 2012 (Ethic approval Medical school Hannover (MHH) no. 9682011 and 2765). The 2nd patient cohort contained 87 KTRs – aged between 4 and 81 years – which were transplanted between 1986 and 2017 (Ethical approval MHH no. 7370). The 3rd patient cohort contained 64 KTRs – aged between 26 and 75 – which were transplanted between 2010 and 2015. All experiments were performed with the guidelines according to the Helsinki Declaration and approved by the ethics committee at Hannover Medical School. Informed consent was obtained from human participants of this study. Clinical parameters were collected in a pseudonymized form after informed consent from clinical databases, from the patients’ clinical files and from a transplantation platform presented by https://www.eurotransplant.org. Biopsies were taken either as protocol or as for-cause biopsies (Table S1†) and analyzed using the BANFF classification including AMR, Borderline TCMR, TCMR and non-rejection, using the BANFF classification version valid at the time of biopsy.45 A borderline TCMR is a T-cell driven borderline rejection and non-rejection refers to samples of patients with a non-rejection biopsy finding. 8 data sets out of 120 patients in the 1st cohort and 16 data sets out of the 87 patients in the 2nd cohort were excluded from this analysis due to missing values or diagnoses other than rejection, which possibly interfere with the detection of sIL-2R (such as recurring glomerulopathies and others). Information on KTR characteristics, transplant characteristics and the immunosuppressive therapy was collected (Tables S2 and S3†). For sample collection, EDTA plasma was obtained by centrifugation of blood specimens at room temperature. Plasma and urine samples were collected at the time of the biopsy before possible anti-rejection therapies were given. Samples were processed within 3 hours after collection and stored in cryotubes (500–1000 μL) at −80 °C until utilization.Determination of assay relevant biomarker concentrations
Plasma and urine samples from the 1st and 2nd cohort were analyzed by multiplex testing (Bio-Plex MAGPIX Multiplex Reader, Bio-Rad Laboratories GmbH, Munich) to quantify the concentration of sIL-2R among up to 56 diverse immunological parameters (growth factors, cytokines and chemokines) in KTRs’ samples and determine the relevant protein concentrations for the here established LFA. According to the manufacturer's instructions, all samples were thawed on ice and immediately diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 with Bio-Rad sample diluent, before they underwent Luminex-based multiplex protein assay. Here, 65 markers out of the Bio-Plex Pro™ Human Cytokine Screening Panel and the Bio-Plex Pro™ Human Chemokine Assay were used and scored using a machine learning feature (see statistics) together with four clinical parameters (age at Ktx, time between biopsy and Ktx (in months), gender and serum creatinine at biopsy (μM L−1)). In the multiplex protein assay, plasma samples of 112 KTRs of the 1st cohort and urine and plasma samples of 71 KTRs of the 2nd cohort were used to determine the relevant concentration of sIL-2R. This assay is based on color-coded, magnetic microbeads coated with recombinant antibodies against the target protein. Target proteins in the patient sample bind to the antibodies and are detected by a fluorescent-labeled second antibody against the target protein. Both, the color-coding of microbeads as well as the fluorescence labeling of secondary antibodies, were detected and used for the calculation of the target protein concentration.
4 with Bio-Rad sample diluent, before they underwent Luminex-based multiplex protein assay. Here, 65 markers out of the Bio-Plex Pro™ Human Cytokine Screening Panel and the Bio-Plex Pro™ Human Chemokine Assay were used and scored using a machine learning feature (see statistics) together with four clinical parameters (age at Ktx, time between biopsy and Ktx (in months), gender and serum creatinine at biopsy (μM L−1)). In the multiplex protein assay, plasma samples of 112 KTRs of the 1st cohort and urine and plasma samples of 71 KTRs of the 2nd cohort were used to determine the relevant concentration of sIL-2R. This assay is based on color-coded, magnetic microbeads coated with recombinant antibodies against the target protein. Target proteins in the patient sample bind to the antibodies and are detected by a fluorescent-labeled second antibody against the target protein. Both, the color-coding of microbeads as well as the fluorescence labeling of secondary antibodies, were detected and used for the calculation of the target protein concentration.
Patient urine samples of the 3rd cohort were analyzed by enzyme-linked immunosorbent assay (ELISA; Human CXCL9/MIG DuoSetELISA, R&D Systems, Minneapolis, USA) to determine the sIL-2R concentration. 42 of these samples were applied on the here developed LFA. For standardization of sIL-2Rα-concentration of the samples used for the LFAs, 11 samples of the 2nd cohort (4 TCMRs, 2 borderline TCMRs, 2 mixed rejections with either AMR/TCMR or AMR/borderline TCMR and 3 non-rejection samples) were additionally analyzed by ELISA. The assay was performed according to the general ELISA protocol of the manufacturer.
Preparation of gold nanoparticle (AuNP)–antibody conjugates
The concentration of the nanoparticle solution was measured over 90 seconds by nanoparticle tracking analysis (Nanoparticle Analysis System, Firefly Camera, NanoSight, Modul LM10). The result was analyzed with NanoSight Nanoparticle Tracking Analysis, Version 2.3 Build 0033.
Flocculation test for AuNP–antibody conjugates
For the visualization of the tested biomarker, AuNP-labeled detection antibodies were part of the sandwich assay in the LFA. In order to find the lowest antibody concentration necessary to completely cover the gold nanoparticles and create stable particles, a flocculation test was performed.During the flocculation test, possible aggregations of gold nanoparticles due to the addition of NaCl as the electrolyte solution were formed. Aggregation generally occurs, if gold nanoparticles are not fully covered by antibodies, and can be photometrically visualized as a color change.46 Within the flocculation test, 20 μL of AuNPs and different volumes of the detection antibody were adjusted to a final volume of 23 μL by the addition of sodium borate buffer. Solutions were incubated in the dark for 20 min at 115 rpm at room temperature. The absorption spectrum was measured using NanoDrop (Peqlab ND-1000) from 300 to 745 nm.47,48
AuNP-detection antibody conjugation and characterization
Based on the highest observed particle stability, the AuNP-detection antibody conjugation was performed using the determined optimal antibody concentration (5.9 μg mL−1, see above). Therefore, AuNPs (54 pM, 500 μL, pH 9.5), detection antibody (90 μg mL−1, 37.5 μL) and sodium borate buffer (2 mM, 37.5 μL) were mixed and incubated for 20 min at 115 rpm at room temperature. After adding 10% BSA to the AuNP–antibody mixture, the solution was incubated for 20 minutes at 115 rpm at room temperature and subsequently centrifuged at 1300g for 30 min at 20 °C. The supernatant was removed and the pellet was resuspended in 290 μL of H2O to a final concentration of 70.4 pM.Lateral flow assay (LFA)
Assay procedure
For each strip, 100 μL of protein sample was prepared in running buffer using different concentrations of sIL-2R, analyte sample or negative control samples. As negative controls, HSA (750 μM), MIG (145 pM) and human IgG (67 mM) were used for pre-testing, while patient samples from KTRs with a kidney graft judged as non-rejection after biopsy served as negative controls for the final LFA testing. Patient samples were diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) in running buffer. In general, 53 patient urine samples were used, therefrom 35 were derived from patients with TCMR (4 from 2nd cohort, 31 from 3rd cohort), 3 from patients with borderline TCMR (2 from 2nd cohort, 1 from 1st cohort), 1 each from patients suffering from mixed rejection (AMR/TCMR or AMR/borderline TCMR; both 2nd cohort) and 13 samples from patients with a non-rejection biopsy finding (3 from 2nd cohort, 10 from 3rd cohort). Protein samples, controls and patient samples were provided in wells of a 96 well plate. The prepared strips were placed into the respective well to start the assay.
2) in running buffer. In general, 53 patient urine samples were used, therefrom 35 were derived from patients with TCMR (4 from 2nd cohort, 31 from 3rd cohort), 3 from patients with borderline TCMR (2 from 2nd cohort, 1 from 1st cohort), 1 each from patients suffering from mixed rejection (AMR/TCMR or AMR/borderline TCMR; both 2nd cohort) and 13 samples from patients with a non-rejection biopsy finding (3 from 2nd cohort, 10 from 3rd cohort). Protein samples, controls and patient samples were provided in wells of a 96 well plate. The prepared strips were placed into the respective well to start the assay.
For optimization, assays were performed after a pre-incubation of the protein samples with the AuNP-labeled detection antibody solution. Here, 2 μL of the AuNP-labeled detection antibody (70.4 pM) were added to protein samples, negative controls or patient samples in the 96 well plate and incubated under exclusion of light for 15 minutes at room temperature before the LFA assay was started.
Analysis of the lateral flow assay (LFA)
The strips were analyzed by semi-quantitative visual analysis of scans (Epson Perfection V370) after 20 and 60 minutes of running time. The LFA scans were analyzed according to the histogram of intensities of the red lines using ImageJ. Here, the analysis window was adjusted to 126 × 738 pixels. During optimization, each LFA was repeated three times. When urine patient samples were applied, LFAs were run once. To compare the different strips of the pretests of different runs with each other, for each run the mean of the control line of 0 pM sIL-2R was set to 100%.Statistical methods
To identify the key immunological biomarkers for categorization into ‘rejection’ or ‘non-rejection’ and subsequently for acute TCMR, two different statistic approaches were used. First, a feature selection (FS) process was performed with neural net analysis. This analysis was based on 112 patient's files of the 1st cohort with a total of 69 features (65 biomarkers and 4 clinical features such as age at Ktx, time between biopsy and Ktx (in months), gender and serum creatinine at biopsy (μM L−1)). A set of training samples contained input values together with the desired output. In combination with an error function, the training set was used to adjust the internal parameters of the network and mapped the values of the immunological parameters to ‘rejection’ or ‘non-rejection’. For the evaluation of the method, two-layer feed-forward networks with 5–20 neurons in the hidden layer were used. For each neuron, 250 FS runs were carried out. A ranking order for the various markers was calculated by the evaluation of score values and the 15 candidates with the highest score were elected (ESI Table S1†). Second, the Wilcoxon test (Mann–Whitney U test, suitable for a non-parametric value distribution) was used for subgroup analysis of multiplex testing results according to BANFF scores (borderline TCMR, TCMR or AMR versus non-rejection samples), and asterisk indicates the level of statistical significance reached (* p < 0.05, ** p < 0.01; *** p < 0.001).Sensitivity and specificity were defined according to a binary classification test for sIL-2R in the LFA and the estimated glomerular filtration rate (eGFR) at the time of biopsy. For eGFR, 25 mL min−1 was set as the cut off. The eGFR was calculated using the chronic kidney disease epidemiology collaboration (CKD-EPI-) equation:49
MS Excel XLSTAT 2020 (Addinsoft Inc., New York, USA) was used to plot receiver operating characteristics (ROC) curves and to calculate the area under curve (AUC); for the determination of the eGFR, the serum creatinine values at the time of graft biopsy were used and GFR estimates according to CKD-EPI, whereas for the LFA the intensity of the test line was used.
Results and discussion
Determination of assay relevant biomarker concentrations
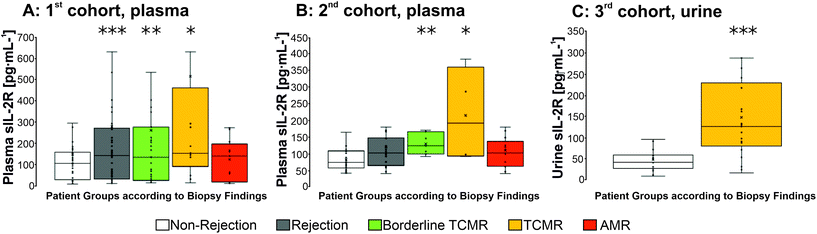
To investigate whether sIL-2R is a biomarker for TCMR, plasma and urine samples from three independent KTR cohorts were included in the current study. In the applied multiplex assay, up to 65 different immunological parameters and additional clinical parameters had been analyzed in parallel, 15 best candidates were chosen by scoring (ESI Table S1†) and validated in a 2nd patient cohort. Among those, sIL-2R was found to be increased significantly in plasma and urine of KTRs with acute TCMR and also in the plasma of KTRs with borderline TCMR (Fig. 1). In detail, according to the BANFF classification, biopsies of the 1st and 2nd patient cohort (recruited between 2011 and 2012 rsp. recruited from 2017 and 2018) were categorized as “biopsy proven rejection-free sample” – termed “non-rejection sample” in the following borderline TCMR, acute TCMR and AMR (for details see ESI Tables S1–S3†). The term ‘rejection’ comprises the above-mentioned rejection categories.The sIL-2R-selected results of a multiplex protein assay as well as ELISA for sIL-2R are shown in Fig. 1. In the 1st patient cohort, sIL-2R concentrations in samples of the KTR group with all kinds of reception (‘rejection’) and the KTR group with ‘TCMR’ or ‘borderline TCMR’ were significantly higher (‘rejection’: 282.5 ± 498.5 pg mL−1; ‘TCMR’: 595.6 ± 817.4 pg mL−1, ‘borderline TCMR’ 264 ± 400 pg mL−1, ‘AMR’ n. s. with 126.4 ± 95.5 pg mL−1) compared to samples of patients with a non-rejection biopsy finding (149.7 ± 324.6 pg mL−1). In the 2nd patient cohort, sIL-2R concentrations were significantly higher in plasma samples of patients with TCMR (plasma: ‘TCMR’: 217.61 ± 124.9 pg mL−1, N = 7; ‘borderline TCMR’ with 132 ± 29.6 pg mL−1; N = 12) compared to those of non-rejecters (plasma: non-rejecters: 76.5 ± 35.4 pg mL−1, N = 37). To answer the question, whether increased sIL-2R levels are also detectable in the urine of KTRs with acute TCMR rejection, urine samples from KTRs out of a 3rd cohort were tested with 35 urine samples being categorized as TCMRs, 26 as non-rejection samples (Fig. 1C and ESI Table S4† for details according the KTRs, data documenting a homogeneous distribution of different immunosuppressive regimens in these groups). ELISA results showed an sIL-2R mean concentration of 124.3 ± 103.3 pg mL−1 for the urine samples of KTRs with TCMR. The mean concentration of sIL-2R in patients with non-rejection as determined by histopathological classification of graft biopsies was significantly lower (48.9 ± 35.5 pg mL−1).
The significantly elevated sIL-2R concentration in acute TCMR after Ktx corresponds to the findings of others.28,50,51 For example, Rasool et al. showed that up-regulation of sIL-2R levels occur at an earlier time than that of serum creatinine52 indicating that sIL-2R might be an early immunological specific marker for TCMR.
Thus, it has been successfully demonstrated in all three patient cohorts that sIL-2R is an indicator of TCMR and – according to plasma levels – possibly also in borderline TCMR after Ktx. Furthermore, it was successfully shown that the biomarker sIL-2R is not only statistically reliably detectable in plasma, but also in urine (from the 3rd patient cohort), and the medium preferably used for commonly utilized POC testing due to its easy availability for the patient and lower concentration of other proteins may be competitive in antibody-binding.
Sadeghi et al. showed that elevated sIL-2R plasma levels within a TCMR were accompanied by low CD8+ lymphocyte plasma counts53 presumably because peripheral activated T-cells in rejecting patients infiltrate the allograft.54 These Th1 cells normally used to lead to an increased cell-mediated response against external antigens oppose graft specific allogens in transplantation settings.55 Furthermore, in Sadeghi's study with mainly cyclosporine-treated KTRs, sIL-2R-increase was part of a Th2-cytokine pattern with increased IL-5 and IL-13 – levels. The latter result was not confirmed by our results (no elevated IL-5 and IL-13 levels, ESI Table S1†). This may be due to a higher percentage of samples deriving from tacrolimus- and mycophenolate-treated KTRs in the study presented here, in which Th2 immune responses are suppressed by higher mycophenolate-levels overtime under treatment with tacrolimus.56
Characterization and preparation of AuNP–antibody conjugates
Determination of antibody concentrations
A critical step in manufacturing an LFA is to prepare conjugates consisting of detection antibodies bound to AuNPs. These conjugates are used for the visualization of the target biomarker on the test line as well as to confirm the functionality of the test on the control line. As described in the Methods section, the flocculation test was used to optimize the theoretical ratio of antibody concentration to nanoparticles when antibodies completely cover the surface of the AuNPs. This test relies on the fact that optimized conjugates reach the highest possible stability without aggregation. Since antibodies contribute to the colloidal state of AuNPs, aggregation occurs within a salty milieu, when the surface of the AuNPs is not fully covered by these micromolecules.58 The appearance of aggregates was recognized photometrically within the flocculation test (ESI Fig. S3†) using non-antibody-conjugated AuNPs as the control.Antibody-loaded AuNPs were considered stable, when the AuNP-concentration curve converged with the curve of AuNPs without antibodies and an antibody concentration of 1.17 μg mL−1 did not stabilize the gold nanoparticles (ESI Fig. S3†). Beginning with an antibody concentration of 3.13 μg mL−1, stable conjugates were detected and the curve converged with the curve of AuNPs without antibodies. Gold nanoparticles reached maximum stability at an antibody concentration of 5.87 μg mL−1 (no color change due to agglomeration after the addition of NaCl), which was used for all further AuNP preparations. Adding even higher antibody concentrations did not improve the stability of AuNP-conjugates.
Lateral flow assay
An LFA is made up of at least four different components: sample pad, nitrocellulose membrane, absorbent pad and backing card. The sample fluid is placed on the sample pad and migrates driven by capillary forces to the particle-bound detection antibodies directed against the analyte. AuNPs were applied here for visualization of the target. The migrating sample couples the complex of analyte and AuNP-labeled detection antibody and this complex passes the test line first and the control line second. On the test line, capture antibodies specific for the analyte are immobilized, whereas capture antibodies specific for the detection antibody are immobilized on the control line (Fig. 2). If the analyte is present in the applied sample, a sandwich consisting of capture antibody–analyte–AuNP-labeled detection antibody is formed and a red line appears as recognized by the naked eye. In this study, lines were additionally and objectively quantified with a scanner and ImageJ for pixel estimation. In each LFA, the control line indicates whether the test performance is appropriate by proper complex formation of immobilized antibodies with the detection antibody bound to AuNPs (Fig. 2B). If the protein of interest is not present in the sample, only the control line is visible (Fig. 2C). The unbound sample is collected in the absorbent pad.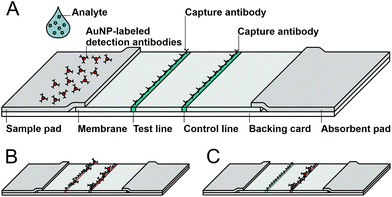 | ||
| Fig. 2 Schematic illustration of a lateral flow assay. (A) Test composition, (B) positive test (two red lines) and (C) negative test (one red line). | ||
In this work, the sIL-2R molecules (analyte) present in the sample were bound to the AuNP-labeled detection antibody and detected at the test line. The remaining AuNP-labeled detection antibodies were bound by the capture antibody at the control line directed against the detection antibody.
Assay procedure and sensitivity
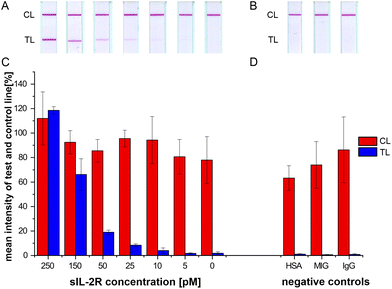
Various parameters are critical for the test performance, sensitivity and specificity of an LFA. Membrane properties corresponding to capillary flow are relevant for sensitivity. To increase the formation of antigen conjugates with the AuNP-labeled detection antibody, a nitrocellulose membrane with a low capillary speed was chosen (Sartorius Stedim Biotech; Unisart® CN180 backed), on which pure water exhibited a capillary speed of 135 to 175 seconds to move along 40 mm.59 The slow sample mobility on the membrane favors the sustainable formation of sIL-2R-AuNP-detection antibody-complexes; the test line therefore gains intensity. Furthermore, the right choice of antibodies is important. Three different monoclonal antibodies and one polyclonal antibody were tested as capture antibodies. Additionally, two polyclonal antibodies where tested as detection antibodies (ESI Fig. S4†). Of these, only the polyclonal capture antibody showed a remarkable result on the test line. For all further experiments, the polyclonal antibody was chosen as the capture antibody.To test the here developed LFA system, different concentrations of sIL-2R in running buffer (0 to 250 pM) were applied on the strip. The introduction of a 15 minute-pre-incubation of conjugates and target led to a stronger intensity of the test line (ESI Fig. S5†).
To determine the optimal test performance time, the LFA strips were analyzed qualitatively by visual observation of the color change of the test line after 20 and 60 minutes. After 60 minutes, sIL-2R concentrations as low as 25 pM and even 10 pM were visible (Fig. 3A). The control line which indicates a reliable test performance was detectable on all strips after two minutes.
Since polyclonal antibodies may exhibit unspecific binding to other proteins, three proteins with a certain probability to occur in patients’ urine after Ktx were tested competitively as negative controls: human serum albumin (HSA), CXCL9 and human immunoglobulin G (IgG). MIG is a chemokine and has been described as an indicator for acute rejections in transplanted organs.60 HSA is the most abundant protein in serum and is excreted in urine within proteinuria in the case of a damaged glomerular filtration barrier of the kidney graft.61 The LFA system revealed negative test lines and positive control lines for these negative control proteins (Fig. 2C, 3B & D) indicating specificity for the detection of sIL-2R.
In addition to visual control, scans of the red bands of the test lines were additionally quantified with ImageJ. The initial scan after 20 minutes was not distinct and clear whereas the second scan after 60 minutes exhibited accurately formed lines. Color intensities of test lines appeared to decrease with lower analyte concentrations, which was proportional to the sIL-2R concentrations (ESI Fig. S6†). Using the scan quantification method, the subjective results by visual estimation were confirmed and sIL-2R-levels as low as 10 pM were detectable.
Determination of sIL-2R in patient samples
This LFA developed as POCT for patients in the monitoring interval after Ktx represents a qualitative test with a binary diagnostic answer. If a red test line is visible, the patient should receive a consultation of a transplant physician.After running the newly developed LFA firstly with running buffer containing defined concentrations of sIL-2R (so called spiked samples), plasma and urine samples of KTRs with acute TCMR, borderline TCMR or mixed AMR/borderline TCMR or mixed AMR/TCMR or non-rejection were applied (Fig. 4).
As the application of plasma samples only exhibit a faint control line on a yellowish membrane, the LFA appeared not to be suitable for plasma samples. The low test performance here might be due to the fact, that plasma contains a huge amount of competing binding proteins for the antibodies used in the LFA and specific binding of sIL-2R is therefore decreased.
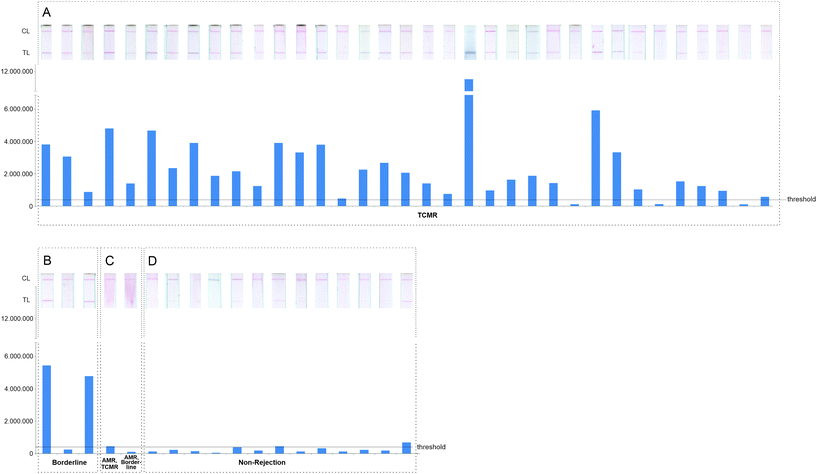
53 exemplary patients’ urine samples (deriving form KTRs with either TCMR, borderline TCMR or mixed rejections or non-rejection samples) were diluted with running buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), pre-incubated for 15 minutes with conjugates of AuNP-labeled detection antibodies as described above and applied on the LFA (for details of patient samples see ESI Table S6†). The resulting LFA strips were scanned after 20 and 60 minutes. Scans taken after 20 minutes were more distinct and clearer and therefore chosen for analysis. Most of these LFAs showed an intensive, defined red color at the test and at the control line, indicating that the strip is functional and detects sIL-2R. It is plausible, that a KTR with a borderline TCMR (which represents a limited form of a TCMR according to BANFF classification) is also identified by this LFA test.
2), pre-incubated for 15 minutes with conjugates of AuNP-labeled detection antibodies as described above and applied on the LFA (for details of patient samples see ESI Table S6†). The resulting LFA strips were scanned after 20 and 60 minutes. Scans taken after 20 minutes were more distinct and clearer and therefore chosen for analysis. Most of these LFAs showed an intensive, defined red color at the test and at the control line, indicating that the strip is functional and detects sIL-2R. It is plausible, that a KTR with a borderline TCMR (which represents a limited form of a TCMR according to BANFF classification) is also identified by this LFA test.
Furthermore, we tested samples of kidney grafts classified as mixed rejections (AMR/TCMR and AMR/borderline TCMR). The threshold between rejection and non-rejection samples was set to 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A.U. of the scan taken 20 min after starting the LFA due to a distinct discrimination of the test line development. Five LFAs out of 40 show a very weak test line and are therefore below this threshold (Fig. 4A–C), although the urine samples derived from patients with a biopsy proven rejection.
000 A.U. of the scan taken 20 min after starting the LFA due to a distinct discrimination of the test line development. Five LFAs out of 40 show a very weak test line and are therefore below this threshold (Fig. 4A–C), although the urine samples derived from patients with a biopsy proven rejection.
To verify that the test line is specific for sIL-2R, 13 samples of patients with a non-rejection biopsy finding (for details of patient samples see ESI Table S6†) were also tested on the LFAs (Fig. 4D). Here, a control line was formed but mostly just a very weak test line. Only two out of 13 LFAs showed a test line higher than 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A. U.
000 A. U.
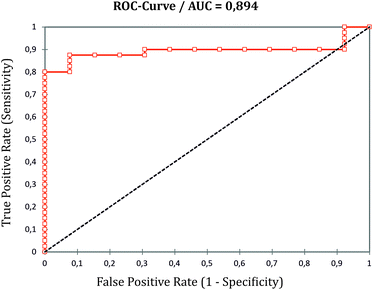
As to the correct classification of KTR samples according to the graft biopsy (TCMR yes or no), a specificity of 84.6% and a sensitivity of the LFA of 87.5% were determined. In comparison, serum creatinine values at the time point of biopsy of the 3rd cohort showed a sensitivity of 85% and a specificity of 30.8% for TCMR and an AUC of 0.565 in receiver operating curve analysis (ESI Fig. S7†). Comparison of sIL-2R-LFA ROC curves with eGFR at the day of biopsy shows that the applied LFA is clearly more specific and even slightly more sensitive than the eGFR on the day of diagnosing TCMR per biopsy (Fig. 5 and ESI Fig. S7†). It cannot be excluded that a significant decrease of eGFR within a distinct time interval before graft biopsy would be more specific and indicative of TCMR, but patients’ clinical files used did not comprise all historical laboratory data to judge the alteration of kidney graft function over time which frequently occurs in clinical practice.
As LFAs are qualitative assays and target-binding may be influenced by the individual sample consistency, sIL-2R concentrations do not necessarily correspond with the intensity of the test lines. In our study, the irregular width and intensity of the test and control lines may also be due to the (not industry standardized) use of a printing device. This device sets drops not creating defined lines. It may also be due to the variable content of the individual urine sample that may contain relevant amounts of other proteins cross reacting, some of which were ruled out by competing binding proteins (Fig. 3B and D). Hence, our results show that sIL-2R is a suitable indicator for TCMR and the newly developed antibody-based LFA is able to categorize urine samples of KTRs with a biopsy proven TCMR, borderline TCMR or a non-rejection biopsy finding. It cannot be ruled out that sIL-2R is detected in the urine of KTRs with a polyomavirus infection62 or an unspecific bacterial urogenital infection or another state of inflammatory or autoimmune disease. Furthermore, the number of urine samples of KTRs with a borderline TCMR did not suffice to evaluate the test's potency for this specificity and ambiguous biopsy diagnosis, in which the need-to-treat is still a matter of current debate among transplant physicians. These questions have to be evaluated in prospective clinical studies using an even broader variety of patients’ samples.
Conclusions
In this work, a novel LFA was established for detecting increased sIL-2R levels with high specificity (84.6%) and sensitivity (87.5%) in TCMR. In a first step, samples from patients with a biopsy proven acute TCMR and other biopsy proven forms of rejection (borderline TCMR or AMR according to the BANFF classification) were examined by the Luminex-based multiplex technique. It was successfully shown, that the biomarker sIL-2R is an indicator of TCMR and statistically reliably detectable in plasma and urine of patients with TCMR. Based on these studies, the relevant mean concentration of sIL-2R was determined in patient samples during TCMR. In the next step, an antibody-based LFA for sIL-2R in running buffer was established. After optimization of the test by introducing a pre-incubation of 15 minutes of the AuNP-labeled detection antibody with the antigen, the LFA reached a limit of detection of 10 pM. Quantification of the LFA test lines also corroborated these results. Finally, 53 samples of KTRs were tested on the LFA. Specificity and sensitivity were calculated for discrimination between TCMR, borderline TCMR, mixed rejections (AMR/TCMR and AMR/borderline TCMR) and non-rejection samples (negative).The newly developed LFA for the detection of sIL-2R is highly sensitive and specific for TCMR and easy to use. Moreover, it was also developed with low material resources and represents a rapid and reproducible test method. The results of the test are unambiguous and can be interpreted by non-experts. The LFA has the limitations – however – that it affords a relatively long pre-incubation time of 15 minutes and that the maximum level of the marker lines can be read out as late as after 20 minutes. Of note, the LFA can be performed as a POCT method by the KTRs at home using urine samples. The KTRs can check specifically, if a TCMR rejection might have occurred, and then turn to an expert specialized transplant physician. We expect KTRs with an increased serum creatinine specific for e.g. TCMR can check the plausibility of this specific rejection condition using the test. We suggest a higher TCMR-specificity for the test than for serum creatinine, and this may help to identify KTRs with the urgent need for a graft biopsy.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank H. Doehring, G. Pfeiffer and A. Sarti-Jacobi for excellent help with patient care and sample collection of the 1st and 2nd cohort and Kerstin Daemen and Jana Keil (Institute for Transplant, Immunology at the Medical School Hannover, MHH) for excellent technical assistance. We further thank Yvonne Mergel for excellent assistance with microarray analyses to test the antibodies used in this work. Finally, we thank Fassisi GmbH (Goettingen, Germany) for the friendly provision of gold nanoparticles and excellent counseling in test application forms to be used in this work and Sartorius-Stedim GmbH for support and supply of materials.The work addressing the 1st cohort was funded by the start-up project SU02 and the Core Facility Diagnostic Centre of the IFB-Tx, ref. no. 01EO0802, the DFG SFB738-B8 projects and the DZIF TTU-IICH07.801. The continued biomarker evaluation was funded by the European Regional Development Fund (ERDF), subproject A (ref. no. 85006385). All further parts of this work have been carried out within the framework of the SMART-BIOTECS alliance between the Technical University Braunschweig and the Leibniz University Hannover. This initiative is supported by the Ministry of Science and Culture (MWK) of Lower Saxony, Germany. We further thank Prof. Dr Stephan Immenschuh for his critical proofreading of the manuscript.
References
- Summary from Global Observatory on Donation and Transplantation. http://www.transplant-observatory.org/summary/, (last accessed July 2019).
- I. N. Crispe, Hepatology, 2014, 60, 2109–2117 CrossRef CAS PubMed.
- H. Dai, Y. Zheng, A. W. Thomson and N. M. Rogers, Front. Immunol., 2020, 11, 1044 CrossRef CAS PubMed.
- T. E. Nevins, P. W. Nickerson and M. A. Dew, J. Am. Soc. Nephrol., 2017, 28, 2290–2301 CrossRef PubMed.
- H. Neuwirt, M. Rudnicki, P. Schratzberger, M. Pirklbauer, A. Kronbichler and G. Mayer, Memo, 2019, 12, 216–221 CrossRef.
- P. W. Nickerson and D. N. Rush, Am. J. Transplant., 2013, 13, 2239–2240 CrossRef CAS PubMed.
- B. L. Kasiske, M. A. Vazquez, W. E. Harmon, R. S. Brown, G. M. Danovitch, R. S. Gaston, D. Roth, J. D. Scandling and G. G. Singer, J. Am. Soc. Nephrol., 2000, 11(Suppl 15), 1–86 CrossRef.
- R. Mehta, W. Cherikh, P. Sood and S. Hariharan, Clin. Transplant., 2017, 31(5) DOI:10.1111/ctr.12945.
- C. Roufosse, N. Simmonds, M. Clahsen-van Groningen, M. Haas, K. J. Henriksen, C. Horsfield, A. Loupy, M. Mengel, A. Perkowska-Ptasińska, M. Rabant, L. C. Racusen, K. Solez and J. U. Becker, Transplantation, 2018, 102, 1795–1814 CrossRef PubMed.
- D. Serón, F. Anaya, R. Marcén, R. G. del Moral, E. V. Martul, A. Alarcón, A. Andrés, D. Burgos, L. Capdevila, M. G. Molina, C. Jiménez, J. M. Morales, F. Oppenheimer, L. Pallardó and A. S. Fructuoso, Nefrología, 2008, 28, 385–396 Search PubMed.
- A. Schwarz, W. Gwinner, M. Hiss, J. Radermacher, M. Mengel and H. Haller, Am. J. Transplant., 2005, 5, 1992–1996 CrossRef PubMed.
- H. E. Wilczek, Transplantation, 1990, 50, 790–797 CrossRef CAS PubMed.
- H. Hu, J. Kwun, B. D. Aizenstein and S. J. Knechtle, Transplantation, 2009, 87, 1814–1820 CrossRef CAS PubMed.
- H. S. Ciftci, T. Tefik, M. K. Savran, E. Demir, Y. Caliskan, Y. D. Ogret, T. Oktar, O. Sanlı, T. Kocak, Y. Ozluk, F. S. Oguz, I. Kilicaslan, F. Aydın, A. Turkmen and I. Nane, Int. J. Organ Transplant. Med., 2019, 10, 53–63 CAS.
- J. Savikko, E. A. Kallio, E. Taskinen and E. von Willebrand, Transplantation, 2002, 73, 506–511 CrossRef CAS PubMed.
- D. J. Lo, B. Kaplan and A. D. Kirk, Nat. Rev. Nephrol., 2014, 10, 215–225 CrossRef CAS PubMed.
- M. E. Hemler, M. B. Brenner, J. M. McLean and J. L. Strominger, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 2172–2175 CrossRef CAS PubMed.
- J. Tang, S. Cai, C. Ye and L. Dong, Semin. Arthritis Rheum., 2020, 50, 354–359 CrossRef CAS PubMed.
- R. M. M. Vanmaris and G. T. Rijkers, Sarcoidosis Vasc. Diffuse Lung Dis., 2017, 34, 122–129 Search PubMed.
- M. V. Barabanshikova, I. A. Dubina, S. V. Lapin, E. V. Morozova, J. J. Vlasova, M. O. Ivanova, I. S. Moiseev and B. V. Afanasyev, Oncol. Res. Treat., 2017, 40, 574–578 CrossRef PubMed.
- B. Cai, J. Zhang, M. Zhang, L. Li, W. Feng, Z. An and L. Wang, Exp. Clin. Endocrinol. Diabetes, 2013, 121, 214–219 CrossRef CAS PubMed.
- C. Kitko, D. Couriel, J. E. Levine, P. Martin, M. E. Flowers, J. A. Hansen, L. Chang, T. Braun, D. Fox, M. Conlon, B. Fiema, R. Morgan, A. Gomez, P. Pongtornpipat, K. Lamiman, B. Storer, J. L. M. Ferrara, S. Lee and S. Paczesny, Conference Paper, 2013, 48.
- M. Shaiegan, M. Iravani, G. R. Babaee and A. Ghavamzadeh, Transplant Immunol., 2006, 15, 223–227 CrossRef CAS PubMed.
- Y. Nakamura, Y. Tanaka, M. Tanaka, A. Sugiyama, K. Yamamoto, Y. Tokunaga, T. Yujiri and Y. Tanizawa, Int. J. Hematol., 2016, 103, 436–443 CrossRef CAS PubMed.
- G. C. Zucchelli, A. Clerico, R. de Maria, M. Carmellini, R. Di Stefano, S. Masini, A. Pilo and L. Donato, Clin. Chem., 1990, 36, 2106–2109 CrossRef CAS.
- M. C. Deng, M. Erren, L. Kammerling, F. Gunther, S. Kerber, A. Fahrenkamp, G. Assmann, G. Breithardt and H. H. Scheld, Transplantation, 1995, 60, 1118–1124 CrossRef CAS PubMed.
- A. M. Hagras, D. M. Salah, D. H. Ahmed, O. K. Abd Elaal, H. A. F. Elghobary and F. I. Fadel, Nephron, 2018, 139, 30–38 CrossRef CAS PubMed.
- P. García-Roca, Y. A. Vargas, Y. Fuentes, A. M. Hernández, L. Ortiz, S. Valverde, L. Velásquez-Jones, B. Romero-Navarro, G. Ramón-Garcia and M. Medeiros, Pediatr. Transplant., 2012, 16, 274–279 CrossRef PubMed.
- A. Hamza, K. Fischer, H. Loertzer and P. Fornara, Urol. Int., 2006, 77, 232–239 CrossRef CAS PubMed.
- Z. Fang, Z. Zhang and C. Huang, Chin. J. New Drugs Clin. Rem., 2005, 50–53 Search PubMed.
- M. Kölling, G. Haddad, U. Wegmann, A. Kistler, A. Bosakova, H. Seeger, K. Hübel, H. Haller, T. Mueller, R. P. Wüthrich and J. M. Lorenzen, Clin. Chem., 2019, 65, 1287–1294 CrossRef PubMed.
- W. Zhang, Z. Yi, K. L. Keung, H. Shang, C. Wei, P. Cravedi, Z. Sun, C. Xi, C. Woytovich, S. Farouk, W. Huang, K. Banu, L. Gallon, C. N. Magee, N. Najafian, M. Samaniego, A. Djamali, S. I. Alexander, I. A. Rosales, R. N. Smith, J. Xiang, E. Lerut, D. Kuypers, M. Naesens, P. J. O'Connell, R. Colvin, M. C. Menon and B. Murphy, J. Am. Soc. Nephrol., 2019, 30, 1481–1494 CrossRef CAS PubMed.
- M. L. Lubetzky, T. Salinas, J. E. Schwartz and M. Suthanthiran, Clin. J. Am. Soc. Nephrol., 2021, 16 DOI:10.2215/CJN.14010820.
- L. Cao, X. Cui, J. Hu, Z. Li, J. R. Choi, Q. Yang, M. Lin, Y. H. Li and F. Xu, Biosens. Bioelectron., 2017, 90, 459–474 CrossRef CAS PubMed.
- S. K. Goh, V. Muralidharan, C. Christophi, H. Do and A. Dobrovic, Clin. Chem., 2017, 63, 742–750 CrossRef CAS PubMed.
- M. Waterhouse, D. Pfeifer, M. Follo, J. Duyster, H. Schäfer, H. Bertz and J. Finke, Clin. Chem. Lab. Med., 2017, 55, 1115–1121 CAS.
- R. D. Bloom, J. S. Bromberg, E. D. Poggio, S. Bunnapradist, A. J. Langone, P. Sood, A. J. Matas, S. Mehta, R. B. Mannon, A. Sharfuddin, B. Fischbach, M. Narayanan, S. C. Jordan, D. Cohen, M. R. Weir, D. Hiller, P. Prasad, R. N. Woodward, M. Grskovic, J. J. Sninsky, J. P. Yee and D. C. Brennan, J. Am. Soc. Nephrol., 2017, 28, 2221–2232 CrossRef CAS PubMed.
- P. Celec, B. Vlková, L. Lauková, J. Bábíčková and P. Boor, Expert reviews in molecular medicine, 2018, 20, e1 CrossRef PubMed.
- S. C. Jordan, S. Bunnapradist, J. S. Bromberg, A. J. Langone, D. Hiller, J. P. Yee, J. J. Sninsky, R. N. Woodward and A. J. Matas, Transplant. Direct, 2018, 4, e379 CrossRef CAS PubMed.
- T. K. Sigdel, Y. Gao, J. He, A. Wang, C. D. Nicora, T. L. Fillmore, T. Shi, B.-J. Webb-Robertson, R. D. Smith, W.-J. Qian, O. Salvatierra, D. G. Camp and M. M. Sarwal, Kidney Int., 2016, 89, 1244–1252 CrossRef CAS PubMed.
- J. Mühlbacher, K. Doberer, N. Kozakowski, H. Regele, S. Camovic, S. Haindl, G. Bond, H. Haslacher, F. Eskandary, J. Reeve, G. A. Böhmig and M. Wahrmann, Front. Med., 2020, 7, 114 CrossRef PubMed.
- L. Krupickova, M. Fialova, M. Novotny, V. Svachova, K. Mezerova, E. Cecrdlova, O. Viklicky and I. Striz, Mediators Inflammation, 2021, 2021, 5513690 Search PubMed.
- T. K. Sigdel and M. M. Sarwal, Front. Med., 2017, 4, 80 CrossRef PubMed.
- I. Mertens, H. Willems, E. van Loon, K. Schildermans, K. Boonen, G. Baggerman, D. Valkenborg, W. Gwinner, D. Anglicheau, M. Essig, P. Marquet and M. Naesens, Kidney Int. Rep., 2020, 5, 1448–1458 CrossRef PubMed.
- A. Loupy, M. Haas, C. Roufosse, M. Naesens, B. Adam, M. Afrouzian, E. Akalin, N. Alachkar, S. Bagnasco, J. U. Becker, L. D. Cornell, M. C. Clahsen-van Groningen, A. J. Demetris, D. Dragun, J.-P. van Duong Huyen, A. B. Farris, A. B. Fogo, I. W. Gibson, D. Glotz, J. Gueguen, Z. Kikic, N. Kozakowski, E. Kraus, C. Lefaucheur, H. Liapis, R. B. Mannon, R. A. Montgomery, B. J. Nankivell, V. Nickeleit, P. Nickerson, M. Rabant, L. Racusen, P. Randhawa, B. Robin, I. A. Rosales, R. Sapir-Pichhadze, C. A. Schinstock, D. Seron, H. K. Singh, R. N. Smith, M. D. Stegall, A. Zeevi, K. Solez, R. B. Colvin and M. Mengel, Am. J. Transplant., 2020, 20, 2318–2331 CrossRef CAS PubMed.
- V. G. Panferov, I. V. Safenkova, Y. A. Varitsev, N. V. Drenova, K. P. Kornev, A. V. Zherdev and B. B. Dzantiev, Talanta, 2016, 152, 521–530 CrossRef CAS PubMed.
- H. Stückmann, Entwicklung immunchemischer Schnelltestverfahren, Hannover, 2007 Search PubMed.
- D. S. Kim, Y. T. Kim, S. B. Hong, J. Kim, N. S. Huh, M.-K. Lee, S. J. Lee, B. I. Kim, I. S. Kim, Y. S. Huh and B. G. Choi, Sensors, 2016, 16(12) DOI:10.3390/s16122154.
- A. S. Levey, L. A. Stevens, C. H. Schmid, Y. L. Zhang, A. F. Castro, H. I. Feldman, J. W. Kusek, P. Eggers, F. van Lente, T. Greene and J. Coresh, Ann. Intern. Med., 2009, 150, 604–612 CrossRef PubMed.
- F. W. R. Vondran, K. Timrott, S. Kollrich, A.-K. Steinhoff, A. Kaltenborn, H. Schrem, J. Klempnauer, F. Lehner and R. Schwinzer, Clin. Transplant., 2014, 28, 968–979 CrossRef CAS PubMed.
- R. B. Colvin, T. C. Fuller, L. MacKeen, P. C. Kung, S. H. Ip and A. B. Cosimi, Clin. Immunol. Immunopathol., 1987, 43, 273–276 CrossRef CAS PubMed.
- R. Rasool, Q. Yousuf, K. Z. Masoodi, I. A. Bhat, Z. A. Shah, I. A. Wani and M. S. Wani, Int. J. Organ Transplant. Med., 2015, 6, 8–13 CAS.
- M. Sadeghi, V. Daniel, R. Weimer, M. Wiesel, O. Hergesell and G. Opelz, Clin. Transplant., 2003, 17, 151–157 CrossRef CAS PubMed.
- A. Bachetoni, P. Lionetti, P. Cinti, P. Alò, E. R. Molajoni, U. Di Tondo, V. Barnaba, D. Alfani and R. Cortesini, Clin. Transplant., 1995, 9, 433–437 CAS.
- K. J. Wood and R. Goto, Transplantation, 2012, 93, 1–10 CrossRef PubMed.
- D. R. Kuypers, K. Claes, P. Evenepoel, B. Maes, W. Coosemans, J. Pirenne and Y. Vanrenterghem, J. Clin. Pharmacol., 2003, 43, 866–880 CrossRef CAS PubMed.
- W. Haiss, N. T. K. Thanh, J. Aveyard and D. G. Fernig, Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- H. Jans, X. Liu, L. Austin, G. Maes and Q. Huo, Anal. Chem., 2009, 81, 9425–9432 CrossRef CAS PubMed.
- Sartorius. https://www.sartorius.de/sartoriusDE/de/EUR/membranes-for-ivd-microarray-oem/unisart-lateral-flow, (last accessed December 2019).
- C. N. Naresh, A. Hayen, A. Weening, J. C. Craig and S. J. Chadban, Am. J. Kidney Dis., 2013, 62, 1095–1101 CrossRef CAS PubMed.
- G. Fanali, A. Di Masi, V. Trezza, M. Marino, M. Fasano and P. Ascenzi, Mol. Aspects Med., 2012, 33, 209–290 CrossRef CAS PubMed.
- V. Bröcker, A. Schwarz and J. U. Becker, Pathologe, 2011, 32, 399–405 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01001h |
| This journal is © The Royal Society of Chemistry 2021 |