Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A fluorescent probe for STED microscopy to study NIP-specific B cells†
Selda Kabatas
Glowacki‡
ab,
Maria Angela
Gomes de Castro‡
 a,
Ka Man
Yip
c,
Ommolbanin
Asadpour
ab,
Matthias
Münchhalfen
d,
Niklas
Engels
*d and
Felipe
Opazo
a,
Ka Man
Yip
c,
Ommolbanin
Asadpour
ab,
Matthias
Münchhalfen
d,
Niklas
Engels
*d and
Felipe
Opazo
 *ab
*ab
aInstitute of Neuro- and Sensory Physiology, University Medical Center Göttingen, 37073, Göttingen, Germany
bCenter for Biostructural Imaging of Neurodegeneration (BIN), University Medical Center Göttingen, 37075, Göttingen, Germany
cDepartment of Structural Dynamics, MPI for Biophysical Chemistry, 37077, Göttingen, Germany
dInstitute for Cellular and Molecular Immunology, University Medical Center Göttingen, 37073, Göttingen, Germany. E-mail: fopazo@gwdg.de; nengels@gwdg.de
First published on 30th June 2021
Abstract
We have developed a series of monovalent fluorophore-conjugated affinity probes based on the hapten 3-nitro-4-hydroxy-5-iodophenylacetyl (NIP), which is widely used as a model antigen to study B lymphocytes and the functional principles of B cell antigen receptors (BCRs). We successfully used them in flow-cytometry, confocal and super-resolution microscopy techniques.
B lymphocytes are central components of the adaptive immune system in vertebrates. Activated B cells produce antibodies that promote the elimination of pathogens and furthermore have become invaluable reagents in research, diagnostics and clinical applications. In order to sense foreign substances (generally termed ‘antigens’), B lymphocytes express specialized protein complexes on their cell surface, designated B cell antigen receptors (BCRs). These BCRs have a unique antigen specificity in each individual B cell. A popular approach in the analysis of B cells is to generate and analyze cells that express BCRs with defined antigen specificity. Probably the most common antigen in these approaches is the small inorganic hapten 3-nitro-4-hydroxy-5-iodophenylacetyl (NIP). The amino acid sequence of NIP-reactive BCRs is known since the 1980s.1 When studying the biology of B cells, the functional interplay of cell surface receptors is becoming increasingly important. Therefore, NIP-binding BCRs are regularly used and very well characterized. This field of immunological research is developing hand in hand with the field of super-resolution microscopy.2–4
The majority of super-resolution techniques require small affinity probes with minimal size and monovalent binding.5,6 However, despite the great advances in super-resolution microscopy techniques,7 there is currently only a limited number of ideal affinity probes for the analysis of BCRs, which limits the progress in studying accurately their numbers and organization at the nano-scale. Therefore, there is an urgent need for the development of new tools that allow for precise and specific labeling of cell surface receptors in lymphocytes. NIP can be used to label BCRs by coupling it to carrier proteins such as bovine serum albumin that in addition are conjugated to fluorophores or biotin. The latter allows for the detection of NIP-binding cells by using e.g., fluorophore-coupled streptavidin as secondary reagent. However, these conventional approaches have their limitations especially when it comes to high-resolution imaging of cell surface BCRs.4,8,9 The most obvious constraints are the polyvalent nature and inaccuracy (ill-defined fluorophore to carrier ratio, batch to batch variations) of NIP-carrier reagents. Here, we developed a series of fluorescent monovalent affinity probes for NIP-reactive BCRs that proved suitable for flow cytometry and Stimulated Emission-Depletion (STED) super-resolution microscopy (Fig. 1).
To generate these NIP-based affinity probes, we chose different peptide spacers with good solubility in water yet varying net charge. Alternatively, we used electrically neutral, polyethylene glycol-(PEG) based spacers (Table 1). Furthermore, we conjugated each of the different spacers to one of two different fluorophores, AbberiorStar635P (Star635P) or Atto647N, which have similar spectral properties, yet different net charges (Table 1).
| Fluorescent hapten | Charge at pH = 7.4 | Exitation/emissiona | ||||
|---|---|---|---|---|---|---|
| Name | Sequence | Spacer | Fluoroph. | Net | Max. Ex. | Max. Em. |
| a Values obtained from suppliers. | ||||||
| KM1 | ε-NIP-DEGDEK(Star635p)G-NH2 | −4 | −3 | −7 | 638 nm | 651 nm |
| KM4 | ε-NIP-PEG(12)-K(Star635p)-NH2 | 0 | −3 | −3 | 638 nm | 651 nm |
| KM5 | ε-NIP-PEG(12)-K(Atto647N)-NH2 | 0 | 1 | 1 | 646 nm | 664 nm |
| KM7 | ε-NIP-KSKGESAGC(Star635p)-NH2 | 1 | −3 | −2 | 638 nm | 651 nm |
| KM8 | ε-NIP-KSKGESAGC(Atto647N)-NH2 | 1 | 1 | 2 | 646 nm | 664 nm |
With super-resolution microscopy in sight, we designed all NIP probes as monovalent binders and with a defined NIP-to-fluorophore ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. For the synthesis of the different compounds, we first applied solid phase synthesis on a Sieber amide resin with orthogonal protection strategy to generate the spacer between NIP and fluorophore. Then, the amine reactive NIP-ε-aminocaproyl-OSu was coupled to a terminal amine group on resin, followed by acidic cleavage from a solid support and subsequent fluorophore conjugation in solution (for more details see ESI Scheme S1†). All precursor compounds and products were purified by HPLC and analyzed by ESI-MS. Their integrity was confirmed by HR-ESI-MS (ESI spectra S1–S8). To test the applicability of our affinity probes for specific labeling of NIP-reactive BCRs, we used a variant of the human B cell line Ramos that was made deficient for its endogenous BCR (kindly provided by M. Reth). We expressed a NIP-specific IgD-BCR in these cells by retroviral gene transfer. Flow cytometry using anti-human IgD primary antibody confirmed BCR expression on their cell surface (ESI Fig. S1†).
1. For the synthesis of the different compounds, we first applied solid phase synthesis on a Sieber amide resin with orthogonal protection strategy to generate the spacer between NIP and fluorophore. Then, the amine reactive NIP-ε-aminocaproyl-OSu was coupled to a terminal amine group on resin, followed by acidic cleavage from a solid support and subsequent fluorophore conjugation in solution (for more details see ESI Scheme S1†). All precursor compounds and products were purified by HPLC and analyzed by ESI-MS. Their integrity was confirmed by HR-ESI-MS (ESI spectra S1–S8). To test the applicability of our affinity probes for specific labeling of NIP-reactive BCRs, we used a variant of the human B cell line Ramos that was made deficient for its endogenous BCR (kindly provided by M. Reth). We expressed a NIP-specific IgD-BCR in these cells by retroviral gene transfer. Flow cytometry using anti-human IgD primary antibody confirmed BCR expression on their cell surface (ESI Fig. S1†).
We first tested whether the different NIP-containing probes were indeed specifically recognized by NIP-reactive BCRs. To this end, we incubated live IgDNIP-BCR-expressing cells (Ramos IgDNIP) with 250 nM of each NIP probe and after repeated washes, we measured their fluorescence intensities by flow cytometry (Fig. 2). As negative control, we used Ramos cells that did not express a NIP-specific BCR (Ramos). This way, we were able to calculate a signal-to-noise ratio for each compound using thousands of cells. Out of the five compounds tested, KM5 and in particular KM7 showed high signal-to-noise ratios, which were 11.4 and 19.6 respectively. However, NIP-reactive cells stained with KM7 were several times brighter than KM5 in this assay.
To corroborate that these differences in fluorescence were not due to possible changes in the excitation and emission of the fluorophores after conjugation to the differently designed NIP probes, we measured the dyes UV/visible fluorescence properties under the same physiological buffer condition before and after functionalization into the probe (ESI Fig. S2†). We observed minimal differences in their emission intensities, with KM7 being a little less bright than the pure fluorophore Star635P. Therefore, these measurements suggest that it is not the photophysical properties of the probes that are decisive for the differences in cellular fluorescence intensity, but rather the binding of the probes to the NIP-reactive BCRs. Next, we stained Ramos IgDNIP cells with various concentrations of the 5 different NIP-probes and imaged them live or after chemical fixation with 4% paraformaldehyde (ESI Fig. S3 and S4†). Under these conditions, the probes KM7 and KM8 showed the strongest fluorescence signals. The fluorescent signals of these two probes were also successfully retained after chemical fixation of the cells, however, KM7 provided the brightest and cleanest staining in both non-fixed and fixed cells. Fixation is generally necessary to obtain high quality super-resolution images. Therefore, KM1, KM7 and KM8 were designed with internal lysines (K) to enhance retention after fixation with aldehydes, however, KM1 failed to provide sufficient signal in non-fixed and fixed cells (ESI Fig. S3 and S4†). Although we have not investigated these differences between probes in a systematic manner, we can speculate that the net negative charge of −7 of KM1 at physiological pH (see Table 1) may impair its binding to the BCR. By contrast, KM7 and KM8, have the same amino acid spacer sequence (with a +1 charge, as shown in Table 1), but differ in their fluorophore type and final net charge. Thus, any difference between KM7 and KM8 must be caused by the fluorophore, reinforcing the notion that different fluorophores might have different characteristics when used in the context of biological membranes.10
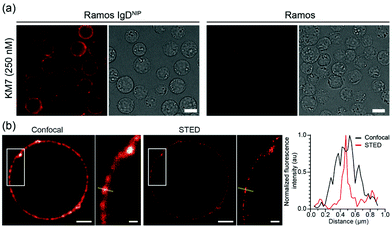
Finally, we confirmed the ability of KM7 to specifically detect NIP-specific BCRs using fluorescence microscopy (Fig. 3a), and furthermore demonstrated that KM7 is highly suitable to meet the high requirements that are needed for both confocal and STED imaging (Fig. 3b).
Here, we have developed and characterized an enhanced monovalent fluorescent affinity probe to study BCRs in living and fixed cells that can be used in flow cytometry and in conventional or STED super-resolution microscopy. The NIP-reactive BCR was cloned already in the early 1980s1 and since then has been used in countless studies. Its structure was solved in 199511 and it is still contributing to the understanding of B cell physiology.12,13 Although this BCR has been used extensively within the immunological community, there were no standard functionalization nor characterizations for its ligand, the hapten NIP. Typically, NIP has been used as an undefined polyvalent reagent that is presented to NIP-recognizing B cells. However, with the continual improvement of the optical resolution in light microscopy, major efforts have been directed to understand the numbers and spatial organization of BCRs in the B cell surface,4,8,9 where monovalent and well-functioning binders are needed.
The probe designated KM7, has a superb signal-to-noise ratio when used to label human B cells expressing NIP-binding BCRs on their cell surface. Due to its monovalent nature, it does not induce cross-linking of BCRs in the plasma membrane, thus avoiding unwanted BCR internalization and B cell activation. To this end, this newly characterized fluorescent and monovalent NIP probe provides a significant advantage to any existing NIP probe for the analysis of BCRs in their native environment, i.e., within the plasma membrane. It thus significantly expands the imaging toolbox and can furthermore be used in combination with additional affinity probes to study the B cell surface landscape with nanometer precision.
Author contributions
S. K. G., M. A. G. d. C., F. O. and N. E. conceived and planned the project and experiments. S. K. G., M. A. G. d. C., N. E., K. M. Y., O. A., and M. M. performed the experiments. S. K. G. and M. A. G. d. C., N. E. and F. O. performed data analysis and interpreted the data. F. O., S. K. G., M. A. G. d. C. wrote the manuscript. F. O. supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
N. E & F. O thank the Deutsche Forschungsgemeinschaft (DFG) for funding part of this work with the projects EN 834/5-1 and OP 261/3-1 respectively.Notes and references
- A. L. M. Bothwell, M. Paskind, M. Reth, T. Imanishi-Kari, K. Rajewsky and D. Baltimore, Cell, 1981, 24, 625–637 CrossRef CAS PubMed.
- M. R. Gold and M. G. Reth, Annu. Rev. Immunol., 2019, 37, 97–123 CrossRef CAS.
- T. Nerreter, S. Letschert, R. Götz, S. Doose, S. Danhof, H. Einsele, M. Sauer and M. Hudecek, Nat. Commun., 2019, 10, 1–11 CrossRef CAS.
- M. A. G. de Castro, H. Wildhagen, S. Sograte-Idrissi, C. Hitzing, M. Binder, M. Trepel, N. Engels and F. Opazo, Nat. Commun., 2019, 10, 820 CrossRef PubMed.
- S. Sograte-Idrissi, N. Oleksiievets, S. Isbaner, M. Eggert-Martinez, J. Enderlein, R. Tsukanov and F. Opazo, Cells, 2019, 8, 48 CrossRef CAS PubMed.
- M. Maidorn, S. O. Rizzoli and F. Opazo, Biochem. J., 2016, 473, 3385–3399 CrossRef CAS PubMed.
- G. Jacquemet, A. F. Carisey, H. Hamidi, R. Henriques and C. Leterrier, J. Cell Sci., 2020, 11, 133 Search PubMed.
- J. Lee, P. Sengupta, J. Brzostowski, J. Lippincott-Schwartz and S. K. Pierce, Mol. Biol. Cell, 2017, 28, 511–523 CrossRef CAS PubMed.
- P. C. Maity, A. Blount, H. Jumaa, O. Ronneberger, B. F. Lillemeier and M. Reth, Sci. Signaling, 2015, 8, ra93 CrossRef.
- L. D. Hughes, R. J. Rawle and S. G. Boxer, PLoS One, 2014, 9 DOI:10.1371/journal.pone.0087649.
- R. Mizutani, K. Miura, T. Nakayama, I. Shimada, Y. Arata and Y. Satow, J. Mol. Biol., 1995, 254, 208–222 CrossRef CAS PubMed.
- A. Nishiguchi, N. Numoto, N. Ito, T. Azuma and M. Oda, Mol. Immunol., 2019, 114, 545–552 CrossRef CAS PubMed.
- J. Jacob, R. Kassir and G. Kelsoe, J. Exp. Med., 1991, 173, 1165–1175 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting methods, figures, schemes and spectra. See DOI: 10.1039/d1an00601k |
| ‡ Both authors contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2021 |