 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selective-sampling Raman imaging techniques for ex vivo assessment of surgical margins in cancer surgery
Maria Giovanna
Lizio†
 ,
Radu
Boitor†
and
Ioan
Notingher
*
,
Radu
Boitor†
and
Ioan
Notingher
*
School of Physics and Astonomy, University of Nottingham, Nottingham, Nottinghamshire, UK. E-mail: ioan.notingher@nottingham.ac.uk
First published on 12th May 2021
Abstract
One of the main challenges in cancer surgery is to ensure the complete excision of the tumour while sparing as much healthy tissue as possible. Histopathology, the gold-standard technique used to assess the surgical margins on the excised tissue, is often impractical for intra-operative use because of the time-consuming tissue cryo-sectioning and staining, and availability of histopathologists to assess stained tissue sections. Raman micro-spectroscopy is a powerful technique that can detect microscopic residual tumours on ex vivo tissue samples with accuracy, based entirely on intrinsic chemical differences. However, raster-scanning Raman micro-spectroscopy is a slow imaging technique that typically requires long data acquisition times wich are impractical for intra-operative use. Selective-sampling Raman imaging overcomes these limitations by using information regarding the spatial properties of the tissue to reduce the number of Raman spectra. This paper reviews the latest advances in selective-sampling Raman techniques and applications, mainly based on multimodal optical imaging. We also highlight the latest results of clinical integration of a prototype device for non-melanoma skin cancer. These promising results indicate the potential impact of Raman spectroscopy for providing fast and objective assessment of surgical margins, helping surgeons ensure the complete removal of tumour cells while sparing as much healthy tissue as possible.
Introduction
It is estimated that, worldwide, 24.5 million new cancer cases are diagnosed each year, with approximately 9.6 million cancer-related deaths.1 Surgery is the main treatment option for most cancer patients. The main challenge in cancer surgery is to remove the entire tumour while minimising the amount of healthy tissue being excised. The standard procedure to check the completeness of tumour removal is histology of paraffin embedded tissues.2 Using hematoxylin and eosin (H&E) stained sections, histology measures the distance from the tumour edge to the surface of the excised tissue (surgical margins), which provides an indication on whether all tumour had been successfully removed or not. However, histology using paraffin embedded tissue is time-consuming and is performed post-operatively only. The availability of accurate techniques for checking the completeness of tumour excision intra-operatively could improve the outcomes of cancer surgery, as surgeons could check the excision margins while the patient is still in the operative theatre and could therefore excise additional tissue if needed. For this purpose, techniques that have high sensitivities and specificities for detection of tumour as well as a high spatial resolution, are required.Intraoperative frozen section histology3,4 is a faster alternative to paraffin embedded histology, as tissue freezing, cryosectioning, staining and evaluation can be completed within 20–90 minutes.5 However, the quality of the frozen H&E sections is lower than for paraffin embedded staining, and sampling errors (typically < 1% of the excision surface) and freezing artifacts have limited its use.3,5 Currently, frozen section histology is adopted only in some specialised surgeries, such as Mohs surgery (evaluates 100% of excision surface),5,6 explorative surgeries7 or assessment of sentinel nodes.8
A wide range of optical techniques have been proposed for intraoperative assessment of surgical margins of the resected tissues, including optical coherence tomography,9–11 laser-induced fluorescence imaging,12,13 two-photon excited auto-fluorescence,14 second harmonic generation,15–17 Coherent anti-Stokes Raman scattering imaging, Fourier transform infrared (FTIR)18–20 and Fluorescence Life Time microscopy.21 Optical techniques have the potential to provide information at high spatial resolution (the diffraction limit of light is lower than the size of cells) and high contrast based on physical or chemical differences between normal tissue and cancerous tissue. Raman spectroscopy uses light to measure the molecular composition of tissue without requiring sectioning, staining or labelling.22 A key feature of Raman spectroscopy is the ability to develop multivariate classification models for quantitative and objective diagnosis of the samples.22–25 Raman microscopy can produce high-resolution diagnosis maps of fresh ex vivo tissue specimens, on which cancer cells are identified based on endogenous molecular contrast, without requiring subjective interpretation of morphological features, which are known to cause inter observer variability.26–28
During the last two decades, Raman spectroscopy has been used to discriminate between tumour and normal tissues on samples from various anatomical locations, including some related to the most common types of cancer: skin,29,30 breast,31,32 oesophagus33 and lung.34 Hand-held fibre optic Raman probes have been used to identify positive margins in breast, brain, oesophagus, cervical cancer and gastric dysplasia surgery,35–38 demonstrating the potential for intra-operative diagnosis. Nevertheless, for many cancer types, the spatial resolution required is difficult (or impossible) to achieve with hand-held Raman probes that measure single point spectra. Typical examples are Mohs micrographic surgery of non-melanoma skin cancers (residual infiltrative tumours as small as tens of μm) and residual tumour of ductal carcinoma in situ (DCIS, smaller than 1 mm), which is responsible for the higher rates of re-excision due to positive margins in breast cancer surgery.39
Raman spectroscopy can be implemented in microscopes with scanning modalities to generate Raman maps of tissues. However, one main limitation of Raman micro-spectroscopy is the relatively low Raman scattering cross-section for most of the relevant molecules found in tissues. While this is not a significant issue in conventional single-point Raman measurements, acquiring Raman maps with a spatial resolution of 50 μm would require acquisition times of several hours, which is not compatible with intraoperative timescale of 20–30 minutes.30
To overcome this limitation, different modalities of Raman spectroscopy have been reported with the aim to reduce the acquisition time while retaining high spatial resolution. For example, surface–enhanced Raman spectroscopy (SERS)40,41 uses functionalized gold nanoparticles that can enhance the Raman signals by several orders of magnitude. The potential of such approaches for assessing surgical margins have been shown recently for breast cancer.42 Coherent anti-stoke Raman Scattering (CARS) microscopy has been used for brain cancer margin assessment and a miniaturized CARS endoscope was used for intraoperative imaging of prostate cancer.43,44 CARS has also been used in combination with two-photon excited fluorescence for in vivo imaging during brain surgery.45 Another strategy for reducing the acquisition time in Raman mapping is to reduce the number of Raman spectra by optimising the location of the sampling points to target the regions of the tissue that are most likely to contain tumour. This can be achieved by either using adaptive sampling algorithms to calculate locations of new sampling points in real-time, or by using a faster screening technique that can obtain a map of the tissue surface, which can be used to direct the Raman spectroscopy measurements. In this set-up, Raman spectra can be recorded from fewer locations without sacrificing the diagnosis accuracy. This approach (selective-sampling Raman spectroscopy) can drastically reduce the time required to assess the surgical margins of large tissue specimens when compared to raster scanning Raman microscopy. This review describes the latest developments in selective-sampling Raman spectroscopy and its applications on ex vivo tumour resections for the intraoperative assessment of surgical margins.
Multi-modal selective sampling Raman spectroscopy
Raman mapping by raster scanning has been widely used for acquiring high-resolution maps of tissues. Investigation of surgical margins has shown that tumours tend to be co-localised in clusters within the tissue specimens.46–48 Due to this high level of spatial correlation, it is not necessary to maintain the same level of spatial resolution over the entire surface of the excised tissue. For example, the sampling density for Raman spectroscopy can be reduced in large areas of normal tissue in favour of higher sampling density in the regions with higher likelihood of containing residual tumour. High resolution images of the tissue surface obtained by other optical modalities can be used to adapt the sampling density and target the Raman spectroscopy measurements towards regions that have a high probability of containing residual cancer cells. Rowlands et al. showed that selective-sampling Raman spectroscopy can approximate raster scans when new Raman spectra were acquired in areas selected by an algorithm that used interpolation of information obtained from previously measured Raman spectra (Fig. 1a).49–51 By calculating the difference between two interpolating surfaces (a cubic spline and a Kriging interpolation), the regions of the sample with the highest spectral variations were selected for further Raman spectroscopy measurements so that the sampling density was higher in the areas of the sample with higher spectral heterogeneity. Another approach for selective sampling was described by Zhang et al. which obtained Raman spectral images of bisulfate mixtures comparable to raster scan images (error of less than 0.5%) using only ∼16% of the spectra required for a raster scan.52 For this method, the locations of the new sampling positions (following an initial seed of measurements) were directed towards pixel locations that maximized the expected reduction in distortion, with each spectrum being classified by a multivariate classification model (LDA-SVM). While such selective sampling methods can reduce the number of required Raman spectra, they also have some limitations. For instance, in order to retain the improvements in measurement speed, small tissue structures may be missed or under-sampled. The algorithms can be optimised to increase the likelihood of capturing smaller spatial features by increasing the number of spectra, however, to adequately detect features that are in the order of tens of μm, the increase in measurement speed over raster scanning is minimal.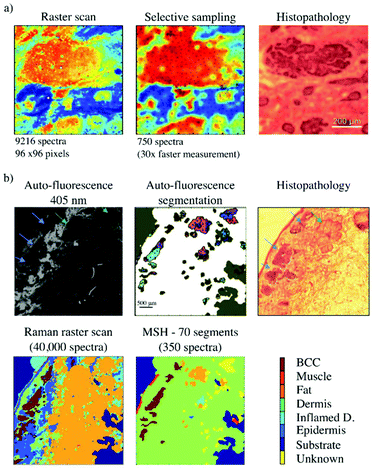 | ||
| Fig. 1 Methods for selective sampling: (a) adaptive selective sampling based on real time estimation of sampling points (reproduced from ref. 49 with permission from Journal of Biophotonics, copyright 2012). Basal cell carcinoma is depicted in red in the Raman maps, which is confirmed by histopathology. The 30× reduction in the number of acquired spectra resulted in minimal changes to the morphology of BCC between the two maps; (b) stratified sampling based on multi-modal approach (reproduced from ref. 46 with permission from PNAS, copyright 2013). Arrowheads point towards regions of low AF intensity which are further sampled via Raman spectroscopy (blue – BCC; green – non-BCC). The multimodal spectral histopathology (MSH) map shows the same relevant information as the raster scan and the histopathology section. | ||
Another option, to target the Raman sampling points more effectively to tissue structures of interest (such as tumours), is to measure the spatial properties of the sample, at the required spatial resolution, prior to acquiring the Raman spectra. For this purpose, alternative imaging techniques can be used, as long as they are fast and have high sensitivity to the required features of interest. Such optical techniques include confocal fluorescence microscopy,46,47,53 high-wavenumber Raman spectroscopy,54 coherent anti-Stokes Raman-scattering, second harmonic generation55,56 and quantitative phase microscopy.57 These techniques can be used to identify regions of interest in the samples and screen out regions that can be confidently identified as normal tissue. For surgical margin assessment in cancer surgery, the imaging techniques must have high sensitivity for detecting tumour but not necessarily high specificity (Fig. 1b). The decision on whether a particular region, depicted as a segment, contains cancer or not is made after the acquisition of Raman spectra and application of a multivariate spectral classification model. As these imaging techniques are much faster than confocal Raman microscopy, a large proportion of the resection margin can be investigated in several minutes. Furthermore, if the generated segments are homogeneous, only a small number of Raman spectra need to be acquired within each segment for an accurate detection, regardless of actual segment size. This multimodal approach can result in the investigation of centimetre sized tissue specimens in less than an hour.24,46,58,59 This measurement procedure is called multimodal spectral histopathology (MSH).
Assessment of surgical margins in Mohs micrographic surgery of basal cell carcinoma (BCC)
Basal cell carcinoma (BCC) is the most common type of cancer in the world, with ∼5.7 million cases diagnosed worldwide 1. Mohs micrographic surgery is the treatment with the lowest rate of tumour recurrence and is mostly used to treat lesions occurring on the head-and-neck, large (>2 cm) and recurrent BCCs.60–62 In Mohs surgery, sequential thin layers of skin are removed and analysed by frozen section histology to check for residual tumour.5 While effective at removing the tumour with minimal damage to healthy tissue, Mohs surgery is expensive due to its reliance on frozen section histopathology and availability of specialist surgeons trained to interpret histopathology sections.63 Selective-sampling Raman spectroscopy that uses auto-fluorescence (AF) imaging to direct the Raman measurements is investigated for intra-operative assessment of surgical margins in Mohs surgery, as a potential replacement to frozen section histology.Firstly, Na et al. reported the use of a 370 nm laser excitation to acquire the auto-fluorescence emission spectra of BCC and of the surrounding normal tissue from 21 lesions, to determine the differences between the two tissue types.64 The emission intensity of the peaks in the spectral region between 420 nm and 500 nm was observed to be 53% lower for BCC compared to healthy tissue. The emission spectral band in this range corresponds mainly to collagen (a structural component of normal dermis), with small contributions from nicotinamide adenine dinucleotide (NADH) and keratin.65,66 Therefore, auto-fluorescence spectroscopy has a high sensitivity of detecting BCC, but not sufficient specificity, as regions of BCC could be confused with other normal tissue structures that have low collagen composition, such as epidermis, hair follicles, fat, sebaceous glands, etc (Fig. 1b).66
Kong et al. tested the use of wide-field auto-fluorescence images, recorded using 292 nm and 357 nm excitation, as well as confocal auto-fluorescence images acquired with a 457.9 nm excitation as possible screening techniques for selective-sampling Raman spectroscopy.46 While both techniques were able to highlight the location of BCC, auto-fluorescence confocal imaging was preferred as it produced more detailed images with a higher contrast for thick tissue specimens. Auto–fluorescence confocal imaging was able to record images of 2 cm2 tissue surfaces in less than 2 minutes and correctly classified typically more than 50% of the healthy tissue surface. Segmentation algorithms were developed to group the remaining tissue surface into regions of similar auto-fluorescence intensity.67 Sampling points were generated within the generated segments and the recorded Raman spectra (785 nm excitation) were used to produce the final diagnosis with the use of a multivariate classification model.
Kong et al. investigated frozen tissue specimen sections from 75 patients to develop a fully automated measurement and analysis protocol that was implemented to allow user-independent analysis of new skin samples.46 The measurements were performed with a microscope-based instrument, for which the only manual operation was switching between the two microscope objectives required for AF imaging and Raman spectroscopy. The Raman spectral classification model had a sensitivity of 95% and a specificity of 94% for the detection of BCC. BCC was identified based on the relative changes in intensity of Raman bands corresponding to DNA and proteins (O–P–O symmetric stretching 788 cm−1, PO2- stretching at 1098 cm−1, 851 and 950 cm−1 bands assigned to proline and hydroxyproline, etc), as suggested by previous reports.30,49,68 To build the multivariate model, Raman spectra were acquired via raster scanning, grouped using k-means clustering and labelled according to H&E staining obtained from the same section. The model was trained with 492 spectra, obtained by averaging the Raman spectra from a labelled k-means cluster (Fig. 2a).
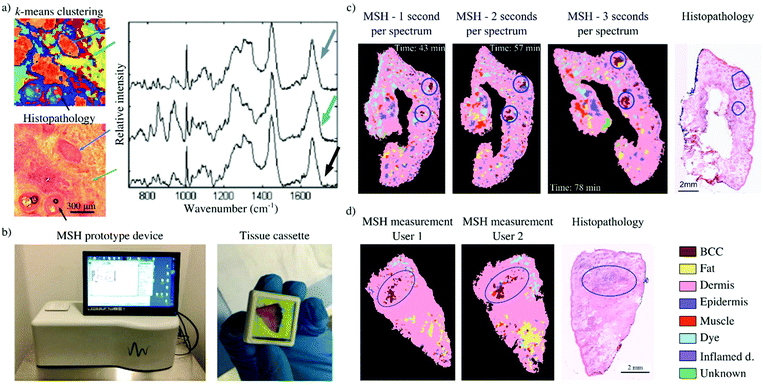 | ||
| Fig. 2 Multimodal spectral histopathology (MSH) for the detection of BCC in skin specimens: (a) k-means clustering of Raman raster scan of different tissue types (blue arrow – BCC, green arrow – dermis, black arrow – epidermis). The centroid of each cluster shows the spectral differences between these tissue types (reproduced from ref. 46 with permission from PNAS, copyright 2013); (b) the MSH prototype device and a tissue cassette loaded with a skin specimen; (c) evaluation of intra-user variability for the MSH prototype device: variation of tissue placement and Raman exposure time retains the location of true BCC-positive detections; (d) evaluation of inter-user variability for the MSH prototype device: measurements of tissue specimens by different users have produced comparable results (reproduced from ref. 58 with permission from © The Optical Society, copyright 2017). | ||
This proof-of-principle study also investigated 20 independent skin samples collected during Mohs micrographic surgery, washed, and stored frozen (10-BCC positive and 10-BCC negative). For each sample, the frozen section histopathology of the adjacent section to the investigated surface was used as the standard of reference. For the BCC-negative samples, an average of three false negative segments were observed, covering less than 1% of the surface area. These results correlated well with the observed 94% specificity. Out of the 10-BCC positive specimens, nine were correctly diagnosed. The one false negative diagnosis corresponded to a specimen that contained only a single BCC region, smaller than 200 μm. For a typical 1 cm x 1 cm sample, the diagnosis required 500–1500 Raman spectra, which corresponded to a measurement time of 20–60 minutes.
These promising results led to the development of a desktop prototype device.58 The MSH prototype was designed to capture both auto-fluorescence images (405 nm excitation, 450–500 nm detection) and Raman spectra (785 nm excitation) of the whole resection surface of tissue specimens loaded within custom cassettes (Fig. 2b). The operating software was developed to be used by clinical staff and to perform the tissue analysis without requiring user interaction or data interpretation. Results were displayed as false-colour maps, where residual BCC was highlighted as red segments within the detection map.
An important requirement for the analysis algorithm is inter-instrument transferability, as the implementation of such technology into clinical practice would require multiple devices that use the same software. For this, the segmentation algorithm described in Takamori et al. was adapted, to be minimally influenced by inter-instrument variations.67 The segmentation of auto-fluorescence images was performed by a watershed algorithm,69 to which the texture-enhanced gradient and the local intensity minima of morphologically flattened auto-fluorescence images were adopted.70 Sampling points for Raman spectroscopy were generated based on auto-fluorescence intensity variance and area of the segments. The transferability of the segmentation and sampling algorithms was investigated by Boitor et al.58 For a dataset of 97 frozen specimens resected via Mohs surgery from 70 patients, the median number of BCC tumours missed due to segmentation and sampling were the same for the two instruments. The Raman spectral classification model, developed with spectra from the laboratory instrument (Raman spectra were collected as raster maps and labelled as described above46), had a per spectrum sensitivity and specificity of 87.7% and 98.4% respectively, via 5-fold cross validation. The same model was then tested on spectra acquired on the prototype device, showing a sensitivity of 81.8%% and specificity of 92.5% in discriminating BCC from all other tissue types. Since the diagnosis of each segment requires multiple BCC spectral detections, the 6% decrease in classification accuracy did not influence the performance of the analysis algorithm.
The optimised analysis algorithm was then used to analyse a set of 10 independent tissue specimens from 9 patients. In all cases, tumours were correctly detected, as observed by comparison with frozen section histopathology. A small number of false positive detections (∼6 per specimen) were observed in both BCC-positive and BCC-negative specimens. However, repeat measurements showed that true BCC detections were co-localised, while the false–positive BCC segments appeared at random locations (Fig. 2c). Repeat measurements by users of varying spectroscopy and clinical experiences were also performed for a small number of independent specimens. Overall, comparable results were obtained from measurements performed by a spectroscopist, a Mohs surgeon (∼1 hour of training) and a medical trainee (∼8 hours of training) (Fig. 2d). The instrument has been now integrated into the clinical workflow and has demonstrated the ability to measure fresh tissue specimens from the main anatomical sites relevant to Mohs surgery and detect the three main types of BCC (nodular, infiltrative and superficial).77
Selective-sampling Raman spectroscopy can also be implemented with multi-foci Raman microscopy. In multi-foci Raman microscopy, multiple laser beams are used to simultaneously measure Raman spectra from different locations of the sample. Sinjab et al. performed multi-foci Raman spectroscopy by splitting a high-powered laser beam (∼3 W at 785 nm) into multiple sampling points with the use of a liquid-crystal spatial light modulator (LC-SLM), creating a power-shared excitation pattern for Raman spectroscopy.71 The locations of these sampling points were controlled in real-time with the use of the LC-SLM, allowing simultaneous acquisition of spectra from multiple locations within a segment. A digital micromirror device (DMD) was programmed to match the laser excitation pattern and function as a slit, directing Raman-scattered beams towards a spectrometer and ensuring that high quality Raman spectra could be obtained from all beams (Fig. 3a). The LC-SLM is used to generate specific laser spot patterns for multi-foci Raman spectroscopy,72–75 while the DMD in the detection path allows for dynamic selection of the detection pattern depending on the segmented image.76 This instrument was used to perform measurements on frozen skin specimens, to determine whether multi-foci Raman spectroscopy could reduce measurements time. For this study, 15 skin specimens were investigated: 10 specimens were raster scanned to build a spectral classification model and five were measured using the selective sampling Raman spectroscopy approach based on six laser foci. Acquisition and segmentation of auto-fluorescence images were performed as previously described,46,67 while generation of sampling points was constrained to multiples of six within segments, as six spectra were acquired for each 2 second exposure (Fig. 3b). All spectra acquired within a single segment were averaged into a single spectrum, which was classified by a Raman spectral classification model. Each segment was therefore labelled according to the result of this classification, on a per segment basis.
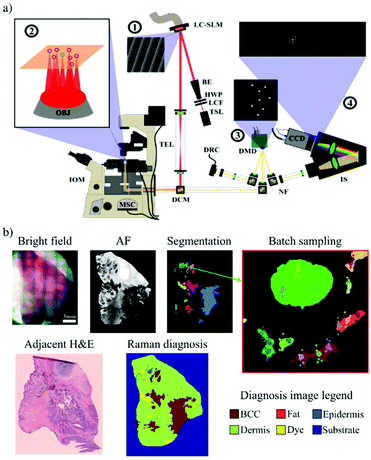 | ||
| Fig. 3 Multi-foci Raman spectroscopy to reduce multimodal spectral histopathology (MSH) measurement speeds: (a) the LC-SLM (1) generates multiple laser foci on the sample surface (2) and the DMD (3) generated conjugated reflective pinholes, with collected light being dispersed onto a CCD inside the spectrometer (4); (b) example of a multi-foci MSH measurement performed on a frozen BCC-positive skin specimen (Reproduced from ref. 71 with permission from © The Optical Society, copyright 2016). | ||
The multi-foci MSH measurement presented in Fig. 3b was captured in ∼11 minutes, with a total of 794 Raman spectra recorded with a 2 second exposure time. The BCC tumours seen in the adjacent frozen histology section were correctly detected in the Raman map (red segments), showing that multi-foci MSH could indeed reduce MSH measurements to timeframes that are ideal for clinical translation.
Assessment of surgical margins in breast cancer surgery
Breast cancer is one of the most common cancers among women, with more than 50,000 new cases reported in the UK in 2019.78 Randomized trials have reported that the survival rate of patients treated by breast conservative surgery (BCS) paired with adjuvant therapies is similar to patients who had radical or modified radical mastectomy.79 However, BCS has the advantage of preventing the complete loss of breast, improving the psychological impact on the patient.79 In case of BCS, the rate of re-operation caused by positive margins, detected by post-operative histology is 10–30%.80Considering the large surface areas of the excised BCS specimens (as large as several centimetres in diameter) and the high spatial resolution required to detect tumours as small as few hundred μm, faster alternatives to raster scanning Raman spectroscopy are required.
Kong et al. were among the first to apply selective sampling Raman spectroscopy to breast tissue samples.81 Firstly, by using frozen tissue sections (5 mm x 5 mm, 10 μm thin), the authors developed a classification model based on Raman spectra that provided diagnosis of mammary ductal carcinoma with 95.6% sensitivity and 96.2% specificity. The selective-sampling Raman spectroscopy was achieved by integrating auto-fluorescence (AF) imaging (365 nm excitation, 511 nm long-pass filter for detection) and Raman spectroscopy. This approach reduced the number of Raman spectra to a ∼20 spectra per mm2.
Shipp et al. followed this approach and combined confocal auto-fluorescence microscopy (excitation at 405 nm, detection 450–520 nm) and Raman spectroscopy (785 nm laser) to allow assessment of larger fresh breast tissue specimens as arriving from surgery.24 The segmentation algorithm for the auto-fluorescence image was optimised to account for patient-to-patient variations and provided reliable, user-independent diagnosis across a broad range of tumour types and sizes.
This technique was tested on a total of 121 tissue specimens from 107 different patients.24 A classification model based on linear discriminant analysis (LDA) was developed using H&E staining as the standard of reference. The model included spectra from a broad range of tumours and normal tissues: invasive carcinoma (IC), DCIS, lobular carcinoma in situ (LCIS), malignant phyllodes, benign proliferative lesions (e.g. fibroadenoma, sclerosing adenosis, epithelial hyperplasia), inflammation, parenchyma, normal mammary stroma, and adipose tissue. For classification purposes, these tissue types were grouped into three classes based on spectral similarities: tumour (including invasive carcinoma, DCIS, LCIS, and malignant phyllodes (MP)), adipose tissue, and benign/normal non-adipose tissue. For measurements on large excision specimens, the number of Raman spectra was limited to 1000 to limit the acquisition time to 25 minutes (of which the AF imagining step was below 10 minutes). Fig. 4 shows typical examples of MSH measurements reporting two correctly identified specimen with positive margins (IC and small DCIS) and one example shows a specimen with true negative margins.
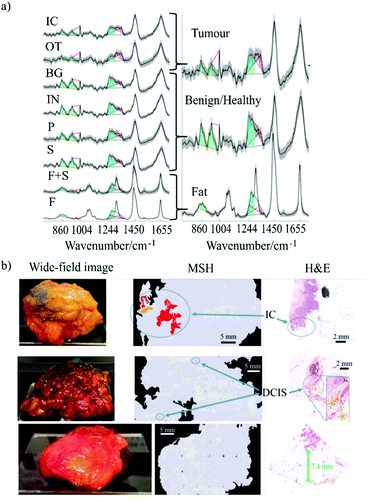 | ||
| Fig. 4 (a) Left: Mean of Raman spectra corresponding to normal breast tissue structures and cancer: fat (F), fat plus stroma (F+S), stroma (S), parenchyma (P), inflamed (IN), benign (BG), other tumour types (OT), invasive carcinoma (IC). Shadings represent the standard deviations. Right: simplified labelling of the tissue structures grouped as tumour, benign/healthy and fat. The spectral features used for classification are colour coded: blue for lipid-associated bands, green for protein-associated bands, and magenta for nucleic acid-associated bands. (b) Examples of MSH measurements of BCS specimens. The images include wide-field pictures of the specimens, the MSH measurements and the relevant H&E. The first two samples have positive margins, as confirmed by the histopathology analysis. The last example was correctly identified has having negative margins (Reproduced from ref. 24 with permission from Breast Cancer Research, copyright 2018). | ||
Overall, selective-sampling Raman spectroscopy detected breast carcinoma on the tissue resection surface with 95% sensitivity and 82% specificity in less than 25 minutes (total 121 samples, up to 4 cm × 6.5 cm in size). For the 51 whole specimens measured immediately after excision, selective-sampling Raman spectroscopy detected tumours in all scanned surfaces that had positive margins subsequently confirmed by histopathology, including those with DCIS.
Recently, Lizio et al.82 proposed the use of auto-fluorescence spectral imaging in a total internal reflection (TIR) imaging configuration in order to increase the efficiency of auto-fluorescence imaging and reduce the number of Raman spectra required for the diagnosis of large breast tissue specimens. The main aim was to improve the identification of adipose tissue by using TIR auto-fluorescence imaging, to reserve a larger number of Raman spectroscopy measurements to only non-adipose tissue areas. This is because, adipose tissue accounts for most of the tumour-clear area of excised breast tissue, therefore, fast identification of adipose tissue by auto-fluorescence could drastically reduce the number of Raman spectra required for investigation. The TIR configuration was proposed to enable faster wide-field imaging of the surface of the tissue specimen, while maintaining the required spatial resolution in the lateral plane.
The authors developed a prototype instrument based on four LEDs (365 nm wavelength, power 360 mW) aligned such that the excitation light was waveguided in the 1 mm thick quartz slide on which the tissue was placed. The internally reflected light generated an evanescent field that excited auto-fluorescence in the tissues. The emitted auto-fluorescence light was collected by a charge-coupled device (CCD) camera using two filters (442–488 nm and 480–562 nm), allowing for ratiometric imaging by dividing the image collected using the 442–488 nm filter by the image collected using the 480–562 nm (Fig. 5). The ratiometric analysis proved to be more reliable than intensity-based imaging because it was less sensitive to inter-patient variation and contact between tissue and quartz slide.
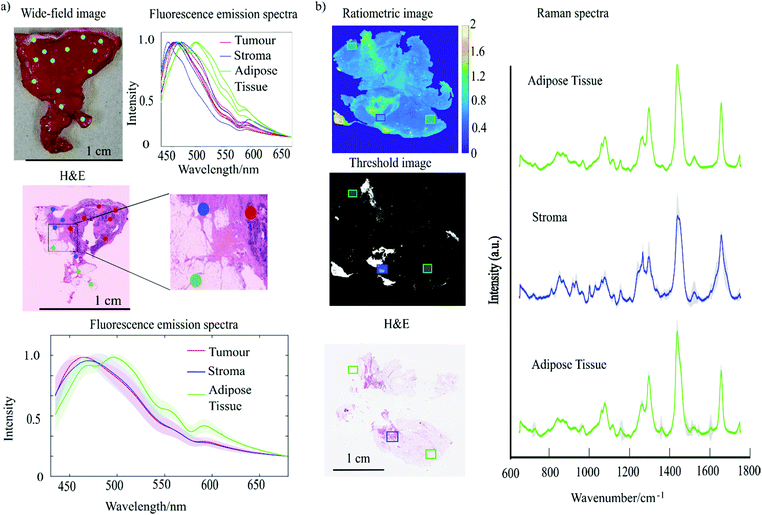 | ||
| Fig. 5 (a) Fluorescence emission spectra of breast tissue obtained using a 365 nm excitation wavelength. The figure reports: wide-field image of breast tissue sample with overlaid blue dots to indicate the locations from which spectra were collected. The normalised fluorescence emission spectra corresponding to each location measured. The H&E stained section of the sample presented with the colour coded dots corresponding to the location from where the spectra were collected (red = tumour, blue stroma, green = adipose tissue). (b) Experiment combining TIR and Raman spectroscopy. The figure reports: ratiometric AF image, threshold image (T < 0.8), H&E staining and the average of Raman spectra acquired from different location of the tissue indicated by the rectangles (Reproduced from ref. 82 with permission from © The Optical Society, copyright 2020). | ||
The TIR-Raman prototype was first tested on small tissue samples cut out from mastectomies and the images were compared with the H&E staining (typical examples showed in Fig. 5). Adipose tissue was consistently characterised by low ratio values in the ratiometric AF images compared to stroma and tumour. An algorithm was used to screen out the adipose tissue using a set threshold value of the ratio-images (0.8) and Raman spectroscopy measurements were distributed in the remaining regions of stroma or tumour. While still requiring further development, as the analysis is currently done in 45 minutes with the semi-automated set up, the authors estimated that analysis of whole breast resection specimens could be completed in approximately 15 minutes once the system will be fully automated.
While the initial studies in selective-sampling Raman spectroscopy employed a second optical modality for determining the spatial properties of the sample, Liao et al. investigated the combination of high wavenumber (HW) Raman imaging (spectral range 2500–3500 cm−1) and fingerprint Raman spectroscopy (spectral range 600–1800 cm−1). HW-Raman imaging was used to identify adipose tissue by targeting the C–H stretching bands of lipids at 2700–2950 cm−1, after which the areas identified as non-adipose were further investigated by fingerprint Raman spectroscopy in order to provide an accurate classification.54
The authors investigated several laser wavelengths to minimise the interference of auto-fluorescence while attempting to maximise the use of the spectral range where the CCD sensitivity was highest. Fig. 6 shows a comparison of Raman spectra of normal breast tissue and tumour obtained using lasers at different wavelengths: 532 nm, 660 nm and 785 nm. The 532 nm laser generated the larger fluorescence background that caused significant distortions of the Raman bands both in the fingerprint and HW regions. This was particularly apparent on the CH bands between 2850–3010 cm−1 in the HW region which was just detectable above the fluorescence background. The fluorescence interference was found to be stronger for non-adipose tissue for which only the water band ∼ 3400 cm−1 was clearly detectable. Similarly, in the fingerprint region, only the bands at 1157 cm−1 and 1525 cm−1, arising from carotenoids, were registered in the adipose tissue above the fluorescence background. The 660 nm laser excitation presented significant reduction of the fluorescence background, with Raman bands detectable in both high-wavenumber and fingerprint regions. The HW spectra of adipose tissue showed intense bands in the region between 2700–3000 cm−1 which are assigned to CH2 and CH3 stretching vibrations. Non-adipose tissue reported intense Raman bands in the 3000–3550 cm−1 region assigned to water stretching vibrations. Lastly, the authors tested the 785 nm laser for which the background noise was further reduced, however, the Raman bands were not strong enough in the HW region (corresponding to 1000–1100 nm) because of the low quantum efficiency of the CCD. Based on these preliminary results, selective-sampling Raman spectroscopy was performed using a 671 nm laser acquiring HW Raman imaging followed by fingerprint Raman point measurements at tissue locations that showed hight intensity in the HW Raman images. Fig. 6, presents typical examples of HW Raman images of breast tissue, showing that high intensity regions in the HW-Raman images correspond to adipose tissue, indicated by the green circle in the H&E staining.
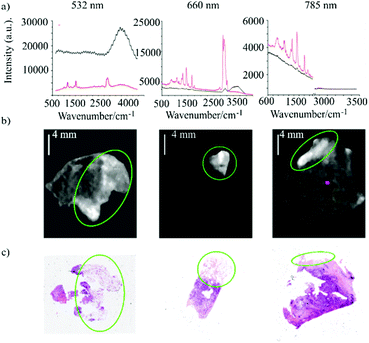 | ||
| Fig. 6 (a) Fingerprint and high–wavenumber Raman spectra of breast tissue samples using three different excitation wavelengths: 532 nm, 660 nm and 785 nm (red line represents adipose tissue, black line represents non adipose tissue). (b) HW–Raman images of breast tissues (intensity scale: 0–255) and the relevant H&E section. The green circles correspond to adipose tissue ((Reproduced from ref. 54 with permission from Journal of Raman Spectroscopy, copyright 2020). | ||
The authors demonstrated that HW-Raman imaging is effective as well as fast in screening adipose tissue, with typical imaging times of 2 minutes for 2 cm2 tissue samples. The authors also estimated that to analyse the whole surface of a WLE (5 cm2) will require ∼25 min, which will make this multimodal approach compatible with intraoperative analysis during BCS. This study demonstrates proof-of-principle, that faster methods for HW Raman imaging could be developed (using high transmission interference filters and single-element detectors rather than spectrometers with CCD) to enable diagnosis of larger breast tissue specimens within practical times (10–20 minutes).
Conclusions and future perspectives
While the ability of Raman spectroscopy to discriminate between normal tissue and cancerous tissue is well established, the assessment of surgical margins for whole tissue specimens has been limited by the long acquisition times required for imaging large specimens. While surface enhanced Raman spectroscopy and coherent Raman microscopy can provide an increase in signal intensity, selective-sampling Raman spectroscopy is a promising approach as it uses simple optical set-ups to measure high-resolution Raman spectra to detect endogenous molecular differences between tumour and normal tissue. By optimising the sampling density of the Raman spectra, using either adaptive sampling or multimodal imaging, the acquisition of diagnosis maps of large tissue samples can be completed within practical acquisition times (20–30 minutes) while retaining the required spatial resolution.As all steps in selective-sampling Raman spectroscopy can be fully automated (user-independent) and can produce objective and repeatable results, the technique requires no user interpretation and can be performed by clinical staff with minimal training, while also being consistent across patients and centres. Selective-sampling Raman spectroscopy results were shown to be concordant with frozen section histopathology for both skin and breast specimens across multiple devices. Depending on the tissue specimen size, typical imaging time was 30–60 minutes (for breast and skin cancers).
Further developments such as multi-foci Raman spectroscopy, TIR-Raman and HW-Raman imaging have the potential to further reduce the measurement time by providing a more tissue-specific selection of sampling points, thus enabling more efficient use of the Raman spectra (e.g. identification of adipose tissue in breast resections).
Nevertheless, the research so far has focused on technological developments, including development of prototype devices suitable for use in the clinic. The clinical integration of such prototypes is currently underway for Mohs micrographic surgery of BCC. Follow up studies with sufficiently large sample sizes are required for establishing the performance of these devices in the clinic.
Author contributions
Maria Giovanna Lizio, Writing – original draft preparation. Radu Boitor, Writing – original draft preparation. Ioan Notingher, SupervisionConflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the UK Engineering and Physical Sciences Research Council (grant number EP/L025620/1).Notes and references
- C. Global, Burden of Disease Cancer, Oncol., 2019, 5, 1749–1768 Search PubMed
.
- J. K. Chan, Int. J. Surg. Pathol., 2014, 22, 12–32 CrossRef PubMed
.
- H. Jaafar, Malays J. Med. Sci., 2006, 13, 4–12 Search PubMed
.
-
P. Dey, in Basic and Advanced Laboratory Techniques in Histopathology and Cytology, ed. P. Dey, Springer Singapore, Singapore, 2018, pp. 51–55, DOI:10.1007/978-981-10-8252-8_6
.
- D. L. Shriner, D. K. McCoy, D. J. Goldberg and R. F. Wagner Jr., J. Am. Acad. Dermatol., 1998, 39, 79–97 CrossRef CAS PubMed
.
- W. Menesi, E. W. Buchel and T. J. Hayakawa, Plast. Surg., 2014, 22, 179–182 CrossRef
.
- N. D. G. Ratnavelu, A. P. Brown, S. Mallett, R. J. Scholten, A. Patel, C. Founta, K. Galaal, P. Cross and R. Naik, Cochrane Database Syst. Rev., 2016, 3, CD010360–CD010360 Search PubMed
.
- K. H. Yoon, S. Park, J. Y. Kim, H. S. Park, S. I. Kim, Y. U. Cho and B.-W. Park, Ann. Surg. Treat. Res., 2019, 97, 49–57 CrossRef PubMed
.
- Y. T. Pan, T. Q. Xie, C. W. Du, S. Bastacky, S. Meyers and M. L. Zeidel, Opt. Lett., 2003, 28, 2485–2487 CrossRef CAS PubMed
.
- O. Assayag, M. Antoine, B. Sigal-Zafrani, M. Riben, F. Harms, A. Burcheri, K. Grieve, E. Dalimier, B. Le Conte de Poly and C. Boccara, Technol. Cancer Res. Treat., 2014, 13, 455–468 Search PubMed
.
- S. A. Alawi, M. Kuck, C. Wahrlich, S. Batz, G. McKenzie, J. W. Fluhr, J. Lademann and M. Ulrich, Exp. Dermatol., 2013, 22, 547–551 CrossRef PubMed
.
- M. Panjehpour, C. E. Julius, M. N. Phan, T. Vo-Dinh and S. Overholt, Lasers Surg. Med., 2002, 31, 367–373 CrossRef PubMed
.
- M. S. Kim, A. M. Lefcourt and Y.-R. Chen, Appl. Opt., 2003, 42, 3927–3934 CrossRef PubMed
.
- J. Ge, L. Fan, K. Zhang, T. Ou, Y. Li, C. Zhang, C. Dong, S. Shuang and M. S. Wong, Sens. Actuators, B, 2018, 262, 913–921 CrossRef CAS
.
- E. Bendau, J. Smith, L. Zhang, E. Ackerstaff, N. Kruchevsky, B. Wu, J. A. Koutcher, R. Alfano and L. Shi, J. Biophotonics, 2020, 13, e202000005 CrossRef CAS PubMed
.
-
A. Fast, A. Ganesan, K. Kelly, C. Zachary and M. Balu, High-speed, high-resolution mesoscopic multiphoton microscopy of human skin (Conference Presentation), SPIE, 2020
.
-
Biomed. Opt. Express, 2020, 11, 1851–1863 Search PubMed
.
- S. Weng, X. Xu, J. Li and S. T. Wong, J. Biomed. Opt., 2017, 22, 106017 Search PubMed
.
- C. L. Evans and X. S. Xie, Annu. Rev. Anal. Chem., 2008, 1, 883–909 CrossRef CAS PubMed
.
- K. Su and W. Lee, Cancers, 2020, 12, 115 CrossRef CAS PubMed
.
- J. Phipps, D. Gorpas, J. Unger, M. Darrow, R. Bold and L. Marcu, Phys. Med. Biol., 2017, 63, 015003 CrossRef PubMed
.
- K. Kong, C. Kendall, N. Stone and I. Notingher, Adv. Drug Delivery Rev., 2015, 89, 121–134 CrossRef CAS PubMed
.
- C. Kallaway, L. M. Almond, H. Barr, J. Wood, J. Hutchings, C. Kendall and N. Stone, Photodiagn. Photodyn. Ther., 2013, 10, 207–219 CrossRef CAS PubMed
.
- D. W. Shipp, E. A. Rakha, A. A. Koloydenko, R. D. Macmillan, I. O. Ellis and I. Notingher, Breast Cancer Res., 2018, 20, 69 CrossRef PubMed
.
-
Biomed. Opt. Express, 2017, 8, 5749–5766 Search PubMed
.
- B. Martin, E. Schäfer, E. Jakubowicz, P. Mayr, R. Ihringer, M. Anthuber, G. Schenkirsch, T. Schaller and B. Märkl, Virchows Arch., 2018, 473, 189–197 CrossRef PubMed
.
- T. A. Thomson, M. M. Hayes, J. J. Spinelli, E. Hilland, C. Sawrenko, D. Phillips, B. Dupuis and R. L. Parker, Mod. Pathol., 2001, 14, 1079–1086 CrossRef CAS PubMed
.
- C. Arguelles-Grande, C. A. Tennyson, S. K. Lewis, P. H. R. Green and G. Bhagat, J. Clin. Pathol., 2012, 65, 242 CrossRef PubMed
.
- A. Nijssen, K. Maquelin, L. Santos, P. Caspers, T. Bakker Schut, J. den Hollander, M. Neumann and G. Puppels, J. Biomed. Opt., 2007, 12, 034004 CrossRef PubMed
.
- M. Larraona-Puy, A. Ghita, A. Zoladek, W. Perkins, S. Varma, I. H. Leach, A. A. Koloydenko, H. Williams and I. Notingher, J. Biomed. Opt., 2009, 14, 054031 CrossRef PubMed
.
- A. S. Haka, K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari and M. S. Feld, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12371 CrossRef CAS PubMed
.
- M. D. Keller, E. Vargis, N. de Matos Granja, R. H. Wilson, M. A. Mycek, M. C. Kelley and A. Mahadevan-Jansen, J. Biomed. Opt., 2011, 16, 077006 CrossRef PubMed
.
- C. Kendall, N. Stone, N. Shepherd, K. Geboes, B. Warren, R. Bennett and H. Barr, J. Pathol., 2003, 200, 602–609 CrossRef PubMed
.
- Z. Huang, A. McWilliams, H. Lui, D. I. McLean, S. Lam and H. Zeng, Int. J. Cancer, 2003, 107, 1047–1052 CrossRef CAS PubMed
.
- N. Lakomkin and C. G. Hadjipanayis, Front. Surg., 2019, 6, 30 CrossRef PubMed
.
- O. Stevens, I. E. Iping Petterson, J. C. C. Day and N. Stone, Chem. Soc. Rev., 2016, 45, 1919–1934 RSC
.
- S. K. Teh, W. Zheng, K. Y. Ho, M. Teh, K. G. Yeoh and Z. Huang, Br. J. Cancer, 2008, 98, 457–465 CrossRef CAS PubMed
.
- A. Mahadevan-Jansen, M. F. Mitchell, N. Ramanujam, A. Malpica, S. Thomsen, U. Utzinger and R. Richards-Kortum, Photochem. Photobiol., 1998, 68, 123–132 CrossRef CAS PubMed
.
- L. Cheng, N. K. Al-Kaisi, F. Gebrail, R. R. Shenk, N. H. Gordon and A. Y. Liu, J. Natl. Cancer Inst., 1997, 89, 1356–1360 CrossRef CAS PubMed
.
- A. Otto, I. Mrozek, H. Grabhorn and W. Akemann, J. Phys.: Condens. Matter, 1992, 4, 1143–1212 CrossRef CAS
.
- M. Schütz, D. Steinigeweg, M. Salehi, K. Kömpe and S. Schlücker, Chem. Commun., 2011, 47, 4216–4218 RSC
.
- S. Kang, Y. Wang, N. P. Reder and J. T. C. Liu, PLoS One, 2016, 11, e0163473 CrossRef PubMed
.
- S. Cui, S. Zhang and S. Yue, J. Healthc. Eng., 2018, 2018, 8619342 Search PubMed
.
-
X. Chen, X. Wang, X. Xu, J. Cheng, Z. Liu, S. Weng, M. Thrall, A. Goh, D. McCormick, K. Wong and S. T. Wong, Miniaturized CARS microendoscope probe design for label-free intraoperative imaging, SPIE, 2014 Search PubMed
.
- B. F. M. Romeike, T. Meyer, R. Reichart, R. Kalff, I. Petersen, B. Dietzek and J. Popp, Clinical Neurology and Neurosurgery, 2015, 131, 42–46 CrossRef PubMed
.
- K. Kong, C. J. Rowlands, S. Varma, W. Perkins, I. H. Leach, A. A. Koloydenko, H. C. Williams and I. Notingher, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15189–15194 CrossRef CAS PubMed
.
- K. Kong, F. Zaabar, E. Rakha, I. Ellis, A. Koloydenko and I. Notingher, Phys. Med. Biol., 2014, 59, 6141–6152 CrossRef PubMed
.
- M. K. Jolly, M. Boareto, B. G. Debeb, N. Aceto, M. C. Farach-Carson, W. A. Woodward and H. Levine, npj Breast Cancer, 2017, 3, 21–21 CrossRef PubMed
.
- C. J. Rowlands, S. Varma, W. Perkins, I. Leach, H. Williams and I. Notingher, J. Biophotonics, 2012, 5, 220–229 CrossRef CAS PubMed
.
- K. Kong, C. J. Rowlands, H. Elsheikha and I. Notingher, Analyst, 2012, 137, 4119–4122 RSC
.
- K. Kong, C. J. Rowlands, S. Varma, W. Perkins, I. H. Leach, A. A. Koloydenko, A. Pitiot, H. C. Williams and I. Notingher, J. Mol. Struct., 2014, 1073, 58–65 CrossRef CAS
.
- S. Zhang, Z. Song, G. M. D. P. Godaliyadda, D. H. Ye, A. U. Chowdhury, A. Sengupta, G. T. Buzzard, C. A. Bouman and G. J. Simpson, Anal. Chem., 2018, 90, 4461–4469 CrossRef CAS PubMed
.
- S. Schlücker, M. D. Schaeberle, S. W. Huffman and I. W. Levin, Anal. Chem., 2003, 75, 4312–4318 CrossRef PubMed
.
- Z. Liao, M. G. Lizio, C. Corden, H. Khout, E. Rakha and I. Notingher, J. Raman Spectrosc., 2020, 51(10), 1986–1995 CrossRef CAS
.
- T. W. Bocklitz, F. S. Salah, N. Vogler, S. Heuke, O. Chernavskaia, C. Schmidt, M. J. Waldner, F. R. Greten, R. Bräuer, M. Schmitt, A. Stallmach, I. Petersen and J. Popp, BMC Cancer, 2016, 16, 534 CrossRef PubMed
.
- A. U. Chowdhury, D. H. Ye, Z. Song, S. Zhang, H. G. Hedderich, B. Mallick, S. Thirunahari, S. Ramakrishnan, A. Sengupta, E. J. Gualtieri, C. A. Bouman and G. J. Simpson, Anal. Chem., 2017, 89, 5958–5965 CrossRef CAS PubMed
.
- J. W. Kang, N. Lue, C.-R. Kong, I. Barman, N. C. Dingari, S. J. Goldfless, J. C. Niles, R. R. Dasari and M. S. Feld, Biomed. Opt. Express, 2011, 2, 2484–2492 CrossRef PubMed
.
- R. Boitor, K. Kong, D. Shipp, S. Varma, A. Koloydenko, K. Kulkarni, S. Elsheikh, T. B. Schut, P. Caspers, G. Puppels, M. van der Wolf, E. Sokolova, T. E. C. Nijsten, B. Salence, H. Williams and I. Notingher, Biomed. Opt. Express, 2017, 8, 5749–5766 CrossRef PubMed
.
- D. W. Shipp, F. Sinjab and I. Notingher, Adv. Opt. Photonics, 2017, 9, 315–428 CrossRef
.
- K. Mosterd, G. A. Krekels, F. H. Nieman, J. U. Ostertag, B. A. Essers, C. D. Dirksen, P. M. Steijlen, A. Vermeulen, H. Neumann and N. W. Kelleners-Smeets, Lancet Oncol., 2008, 9, 1149–1156 CrossRef PubMed
.
- S. C. Flohil, A. M. J. M. van Dorst, T. Nijsten, H. A. Martino Neumann and K. Munte, J. Eur. Acad. Dermatol. Venereol., 2013, 27, 1228–1235 CrossRef CAS PubMed
.
- K. Mosterd, G. A. M. Krekels, F. H. M. Nieman, J. U. Ostertag, B. A. B. Essers, C. D. Dirksen, P. M. Steijlen, A. Vermeulen, H. A. M. Neumann and N. W. J. Kelleners-Smeets, Lancet Oncol., 2008, 9, 1149–1156 CrossRef PubMed
.
- J. M. Baxter, A. N. Patel and S. Varma, BMJ, 2012, 345, e5342 CrossRef PubMed
.
- R. Na, I. M. Stender and H. C. Wulf, Acta Derm.-Venereol., 2001, 81, 246–249 CrossRef CAS PubMed
.
- R. Gillies, G. Zonios, R. R. Anderson and N. Kollias, J. Invest. Dermatol., 2000, 115, 704–707 CrossRef CAS PubMed
.
- A. D. Eleni, D. Clio, J. S. Alexander, S. Carmen and A. Christina, J. Biomed. Opt., 2014, 19, 1–9 Search PubMed
.
- S. Takamori, K. Kong, S. Varma, I. Leach, H. C. Williams and I. Notingher, Biomed. Opt. Express, 2015, 6, 98–111 CrossRef PubMed
.
- T. C. Bakker Schut, P. J. Caspers, G. J. Puppels, A. Nijssen, F. Heule, M. H. A. Neumann and D. P. Hayes, J. Invest. Dermatol., 2002, 119, 64–69 CrossRef PubMed
.
- R. C. W. Gonzalez, Richard, Digital Image Processing, Pearson, 2007 Search PubMed
.
- R. J. O. Callaghan and D. R. Bull, IEEE Trans. Image Process, 2005, 14, 49–62 Search PubMed
.
- F. Sinjab, K. Kong, G. Gibson, S. Varma, H. Williams, M. Padgett and I. Notingher, Biomed. Opt. Express, 2016, 7, 2993–3006 CrossRef CAS PubMed
.
- Z. Liao, F. Sinjab, H. M. Elsheikha and I. Notingher, J. Raman Spectrosc., 2018, 49, 1660–1667 CrossRef CAS
.
- J. Qi and W.-C. Shih, Opt. Lett., 2012, 37, 1289–1291 CrossRef CAS PubMed
.
- J. Qi, J. Li and W.-C. Shih, Biomed. Opt. Express, 2013, 4, 2376–2382 CrossRef PubMed
.
- L. Kong and J. Chan, Anal. Chem., 2014, 86, 6604–6609 CrossRef CAS PubMed
.
- F. Sinjab, Z. Liao and I. Notingher, Appl. Spectrosc., 2019, 73, 727–746 CrossRef CAS PubMed
.
- R. Boitor, R C. de Wolf, F. Weesie, D. Shipp, S. Varma, D. Veitch, A. Wernham, A. Koloydenko, G. Puppels, T. Nijsten, H. Williams, P. Caspers and I. Notingher, Biomed. Opt. Express, 2021, 12, 2015–26 CrossRef PubMed
.
- Z. Momenimovahed and H. Salehiniya, Breast Cancer, 2019, 11, 151–164 Search PubMed
.
- A. A. Onitilo, J. M. Engel, R. V. Stankowski and S. A. R. Doi, Clin. Med. Res., 2015, 13, 65–73 CrossRef PubMed
.
- J. Landercasper, E. Whitacre, A. C. Degnim and M. Al-Hamadani, Ann. Surg. Oncol., 2014, 21, 3185–3191 CrossRef PubMed
.
- K. Kong, F. Zaabar, E. Rakha, I. Ellis, A. Koloydenko and I. Notingher, Phys. Med. Biol., 2014, 59, 6141–6152 CrossRef PubMed
.
- M. G. Lizio, Z. Liao, D. W. Shipp, R. Boitor, R. Mihai, J. S. Sharp, M. Russell, H. Khout, E. A. Rakha and I. Notingher, Biomed. Opt. Express, 2021, 12, 940–954 CrossRef PubMed
.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |
