Open Access Article
Open Access ArticleLive monitoring of cellular metabolism and mitochondrial respiration in 3D cell culture system using NMR spectroscopy†
Damian
Hertig
 abc,
Sally
Maddah
ab,
Roman
Memedovski
ab,
Sandra
Kurth
b,
Aitor
Moreno
d,
Matteo
Pennestri
e,
Andrea
Felser
b,
Jean-Marc
Nuoffer‡
*bf and
Peter
Vermathen‡
abc,
Sally
Maddah
ab,
Roman
Memedovski
ab,
Sandra
Kurth
b,
Aitor
Moreno
d,
Matteo
Pennestri
e,
Andrea
Felser
b,
Jean-Marc
Nuoffer‡
*bf and
Peter
Vermathen‡
 *a
*a
aDepartment of Biomedical Research and Radiology, University of Bern, Switzerland. E-mail: peter.vermathen@insel.ch
bInstitute of Clinical Chemistry, University Hospital Bern, Switzerland. E-mail: jean-marc.nuoffer@insel.ch
cGraduate School for Cellular and Biomedical Sciences, University of Bern, Switzerland
dBruker BioSpin AG, Application Science Department, CH-8117 Fällanden, Switzerland
eBruker UK Limited, Banner Lane, Coventry, UK
fDepartment of Pediatric Endocrinology, Diabetology and Metabolism, University Children's Hospital of Bern, Switzerland
First published on 9th June 2021
Abstract
Background: Because of the interplay between mitochondrial respiration and cellular metabolism, the simultaneous monitoring of both cellular processes provides important insights for the understanding of biological processes. NMR flow systems provide a unique window into the metabolome of cultured cells. Simplified bioreactor construction based on commercially available flow systems increase the practicability and reproducibility of bioreactor studies using standard NMR spectrometers. We therefore aim at establishing a reproducible NMR bioreactor system for metabolic 1H-NMR investigations of small molecules and concurrent oxygenation determination by 19F-NMR, with in depth description and validation by accompanying measures. Methods: We demonstrate a detailed and standardized workflow for the preparation and transfer of collagen based 3D cell culture of high cell density for perfused investigation in a 5 mm NMR tube. Self-constructed gas mixing station enables 5% CO2 atmosphere for physiological pH in carbon based medium and is perfused by HPLC pump. Results & Discussion: Implemented perfused bioreactor allows detection of perfusion rate dependent metabolite content. We show interleaved dynamic profiling of 26 metabolites and mitochondrial respiration. During constant perfusion, sequential injection of rotenone/oligomycin and 2-deoxy-glucose indicated immediate activation and deactivation of glycolytic rate and full inhibition of oxygen consumption. We show sensitivity to detect substrate degradation rates of major mitochondrial fuel pathways and were able to simultaneously measure cellular oxygen consumption.
Introduction
The mitochondrion is an organelle found in nearly all eukaryotic cells and is the main source of energy production. It serves as prime site of adaptation to nutrient availability and energy production with the stepwise catabolism of fuelled monosaccharides, fatty acids and amino acids from the cytosol.1 It is well known that the role of mitochondria in cell biology goes beyond ATP production through oxidative phosphorylation.2,3 Multiple metabolic signalling pathways have been described including release of mitochondrial metabolites, modulation of calcium fluxes or production of free radicals.4,5 Empirically most common outcomes of in vitro toxicity screening are mitochondrial perturbations.6 This highlights the need for methods enabling simultaneous observation of cellular metabolic profiles and mitochondrial oxidative function to better understand interplay and investigate patho-mechanisms. Numerous studies demonstrate strong interdependence of observed cellular metabolite content and energetic state.7–9 While most current in vitro monitoring methods detect individual metabolites or mitochondrial activity there are increasing efforts in detecting multiple metabolites in cell cultures.10Metabolic investigation of living cells using NMR spectroscopy
High resolution NMR has been established as a powerful tool for metabolic characterization of cells, extracts, biopsies or organisms.11–20 However, analysis of cell lysates or extracts renders longitudinal metabolic studies investigating response to challenges laborious, as repeated sample preparation is needed. Further the variability between the biological samples related to cell manipulation needs to be considered, when investigating cells longitudinally or upon intervention. The investigation of rapidly changing metabolic cell responses are challenging and require very efficient quenching of metabolism.These obstacles can be addressed by NMR measurements of intact cells in their “natural” environment, i.e. cell culture in presence of complete enzymatic and regulatory systems.21 However, this requires maintaining viable cell culture conditions with optimized parameters.22 Perfused cell and organ model systems for NMR have been developed already in the 1980s and 1990s for the investigation of biochemistry and physiology.23–27 These perfused cell and organ studies require a perfusion apparatus controlling nutrient delivery, temperature, pH, and gas delivery to keep cells close to natural conditions.
Only few systems employ standard 5 mm NMR tubes applicable for the most widespread standard 5 mm NMR probes.28–31 In order to increase the volume in the NMR sensitive region most use wide-bore probes (≥10 mm)32–36 which are however not routinely available at most centers. Furthermore, the disadvantage of the much lower filling volume is partly compensated by the higher filling factor of the smaller radio frequency coil. Other advantages of smaller volumes include easier nutrition and oxygen delivery to the cells, a more direct response of the smaller volume upon challenges due to faster medium exchange, and lower material costs of chemicals facilitating approaches of in vitro drug or toxicity screening.37,38 Other very interesting and elegant recent efforts involve microfluidic devices for 1H NMR metabolomic investigations with lab-on-a chip technology of cell cultures like single tumor spheroids.39,40 While very promising, these systems pose several technical and scientific challenges and are also not readily available. Furthermore, in cases where the number of available cells is of no concern, miniaturization may not be required. Very recently Bruker released a flow tube (InsightMR™, Bruker), which allows for online reaction monitoring41 and its further specialization InsightCell™. The InsightCell™ flow tube is compatible with standard 5 mm NMR probes, i.e. no special probe is required. The implementation is presented here.
The vast majority of NMR investigations on living cells in bioreactors have applied 31P NMR to investigate metabolism or 13C NMR of labelled (hyperpolarized) substrates, in order to trace metabolic pathways in real time and measure metabolic fluxes.42,43 Only few studies performed 1H NMR investigations for small metabolite detection25,29,30,39,44–48 and even less longitudinal 1H NMR metabolomical investigations of living cell cultures in bioreactors under controlled cell culture conditions. Besides assessment of metabolic changes, determination of oxygenation and oxygen consumption is highly valuable as a surrogate marker of aerobic energy production via mitochondrial oxidative phosphorylation.
Various non-NMR based approaches for oxygen consumption studies investigation have been established, such as oxygen measurement using amperometric oxygen sensors49–51 or optical sensors.52 Although these techniques provide detailed information on the oxygen consumption of cells, none of these methods provide data on the metabolome and do therefore not allow for studying interaction of mitochondrial function and metabolome.
Oxygen quantification in living cell culture using 19F NMR
In our approach, the provided oxygen content is quantified within perfused solid phase 3D cell cultures, using embedded fluorine atoms. We hypothesize that this in situ measurement enables detection of unwanted local hypoxia and facilitates setting a desired oxygen content inside the scaffold, rather than in systems placing the oxygen sensor outside solid phase 3D cell cultures.NMR based approaches for measuring oxygen content by detection of oxygen dependent T1 spin–lattice relaxation times of perfluorinated compounds, have been described frequently.53–55 These approaches mainly focus on long-term tissue or cell oxygenation or the investigation of oxygen consumption in cells or organisms cultured in suspension. One study performed real-time measurements of oxygen tension in two layers of perfluorotripropylamine-doped alginate beads placed before and after perfused cancer cells on microbeads.35
To our knowledge, no NMR-based protocol has been described which quantified oxygen concentration with high temporal resolution inside perfused solid phase based 3D cell cultures.
Human fibroblasts as 3D cell culture model
Fibroblasts are easy to cultivate, widely used in biochemical, genetic and toxicological studies, and can provide a model to study specific disease states or normal physiology.56,57 Fibroblast-collagen matrix cultures enable the analysis of cell physiology under conditions that more closely resemble an in vivo-like environment than conventional 2D cell culture.58 Therefore, collagen based 3D fibroblast cultures were used as cell culture model in this bioreactor study.Aim
To our knowledge, no study has been performed on living human 3D cell culture in a standard 5 mm NMR probe system that combined the measurement of metabolic profiles by 1H NMR with measurements of oxygen consumption using 19F NMR with a time resolution of a few minutes. The aims of this study are therefore (1.) to investigate the cell embedding strategies and analyze the viability of the cells, (2.) to establish the perfused NMR bioreactor using the InsightCell™ flow tube with a standard 5 mm NMR probe, (3.) to test the detectability, stability and reproducibility of longitudinal and fast dynamic metabolic processes (4.) to test the sensitivity of 19F T1 relaxation based oxygen quantification as surrogate marker of mitochondrial oxidative phosphorylation in order to perform combined metabolic measurements and (5.) to test sensitivity to pathway changes upon pathway-specific inhibitors.Materials & methods
Conventional 2D cell culture
Human fibroblasts (anonymous cell lines) were previously established from skin biopsy from healthy controls in routine diagnostics and kindly provided form the routine cell culture laboratory of the Centre of Laboratory Medicine of the University Hospital Bern. The control fibroblast cell line was cultured in minimal essential medium (MEM) supplemented with 10% fetal calf serum, 1× non-essential amino acids (100 μM glycine, L-alanine, L-asparagine, L-aspartic acid, L-glutamic acid, L-proline, L-serine) (GIBCO/Invitrogen, Carlsbad, CA), 2 mM L-glutamine, 200 μM uridine, 1 mM sodium pyruvate, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin and 10 μg ml−1 chlortetracycline at 37 °C and 5% CO2. The medium was exchanged between two and three times a week. Fibroblasts were cultured at 37 °C in a humidified 5% CO2 cell culture incubator and were passaged using 0.05% trypsin–EDTA. Cell numbers were determined in a Neubauer hemacytometer using trypan blue exclusion method.3D cell culture preparation
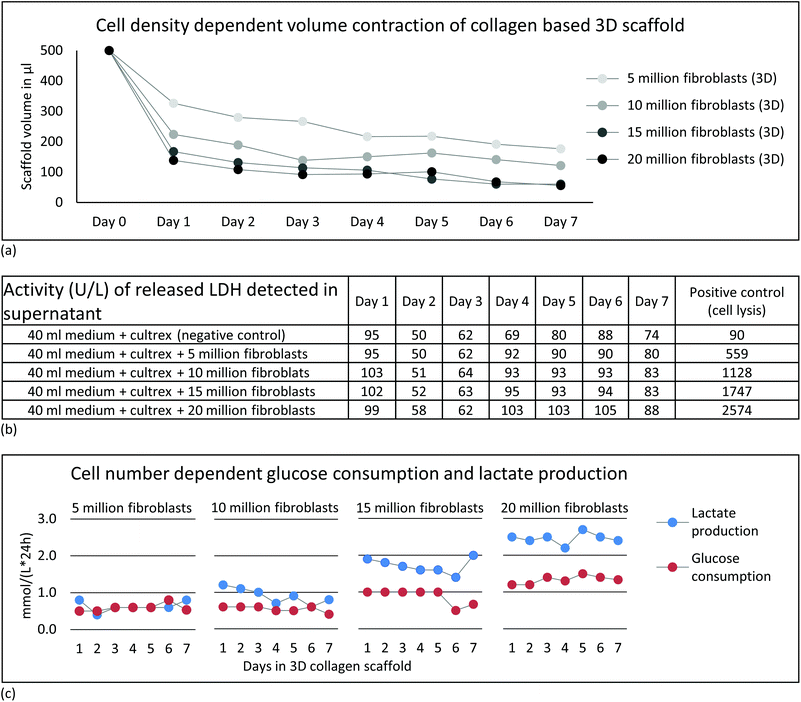
Based on previous NMR studies, a minimum of five million fibroblasts were estimated to provide sufficient signal to perform proton-based metabolic analyses within few minutes of acquisition.59 Therefore we evaluated a range of cell number from five million cells up to twenty million.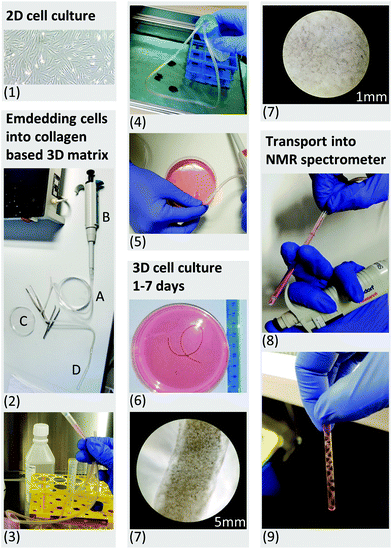 | ||
| Fig. 1 (1.) Up to 20 million primary human fibroblasts were cultured in 2D monolayers. (2.) A sterile infusion tube with an inner diameter of 4.2 mm (A) was attached to the tip of a conventional pipette (B), allowing transport of the cell suspension between the Petri dish (C) and the 5 mm NMR tube (D). (3.) Harvested cells were resuspended in liquid cell-collagen (cultrex™) suspension and injected into the sterile infusion tube. (4.) The open ends of the sterile infusion tube were sealed inside a sterile Falcon tube. The sterile infusion tube was subsequently placed in a water bath at 37 °C for 30 minutes. This allowed rapid polymerization of the cultrex™ under sterile conditions. (5.) After 30 minutes, the polymerized scaffold was released into a Petri dish using the air pressure of a connected pipette. (6.) Daily monitored cell morphology using light microscopy (7), glycolytic activity as well as LDH release (see Fig. 2) were performed to control for high viability of the 3D cell culture sample under normal culture condition in a CO2 incubator. (8.) For all NMR experiments the scaffold was taken up using an infusion tube and inserted into the standard 5 mm NMR tube after 2 days embedment in collagen. LDH measurement revealed no mechanically induced cell death after loading the 3D scaffold into the NMR tube. (9.) The filled standard NMR tube was connected to the InsightCell™ perfusion device and inserted into the NMR spectrometer within few minutes. | ||
Some of the preliminary test measurements were performed with emulsified PFTBA. 0.18% (vol/vol) of PFTBA emulsified with lecithin was added to Tyrode buffer containing 5% D2O pH 7.4.60
NMR bioreactor setup
NMR experiments were carried out on a 500 MHz Bruker Avance II spectrometer. HR-liquid state NMR experiments of cell culture samples were performed with a 5 mm ATM BBFO probe with z-gradient. The bioreactor was perfused using the same cell culture medium as described above for 2D cell culture but additionally supplemented with 5% D2O. For experiments with chemical inhibitors (glycolytic stress test), no fetal calf serum was added to the perfused medium to prevent possible binding of serum albumin with drugs used. Mitochondrial function was inhibited by combined addition of rotenone 5 μM (CI inhibitor) and oligomycin 5 μM (Inhibitor of mitochondrial phosphorylation). Competitive glycolysis inhibitor 2-deoxy-D-glucose (5 mM) was used for glycolysis inhibition.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz, a data size of 64 K points, 32 or 48 transients (≈4 or 5 min), an acquisition time of 3.28 s and a relaxation delay of 5 s. Spectral processing was performed using the Bruker Topspin software (version 3.2, patch level 5). The FIDs were exponentially multiplied using a line-broadening factor of 1 Hz and Fourier transformed. Phased and baseline corrected 1H NMR spectra were calibrated to the left peak of the lactate –CH3 doublet (1.324 ppm). Spectral assignments were based on literature references,62,63 our own additional 2D correlation spectroscopy (1H1H-TOCSY) measurements and data obtained from the Human Metabolome Database (HMDB).64 From the pre-processed fibroblast 1H spectra spectral regions (buckets) with a variable bucket size according to the peak width were selected and integrated. In control measurements of polymerizing collagen without cells a resonance at 3.7 ppm was measured, which almost completely disappeared after the first 30 minutes as polymerization progressed and was no longer detected after overnight incubation with cells.
000 Hz, a data size of 64 K points, 32 or 48 transients (≈4 or 5 min), an acquisition time of 3.28 s and a relaxation delay of 5 s. Spectral processing was performed using the Bruker Topspin software (version 3.2, patch level 5). The FIDs were exponentially multiplied using a line-broadening factor of 1 Hz and Fourier transformed. Phased and baseline corrected 1H NMR spectra were calibrated to the left peak of the lactate –CH3 doublet (1.324 ppm). Spectral assignments were based on literature references,62,63 our own additional 2D correlation spectroscopy (1H1H-TOCSY) measurements and data obtained from the Human Metabolome Database (HMDB).64 From the pre-processed fibroblast 1H spectra spectral regions (buckets) with a variable bucket size according to the peak width were selected and integrated. In control measurements of polymerizing collagen without cells a resonance at 3.7 ppm was measured, which almost completely disappeared after the first 30 minutes as polymerization progressed and was no longer detected after overnight incubation with cells.
For chemometric analysis, the selected buckets were mean centred an analysed using PLS_Toolbox MatLab software (R2014b, The MathWorks Inc.). Principle Component Analysis (PCA) were performed to compare the metabolite content over time of fibroblast cell cultures under various perfusion rates.
All measurements were performed using a pseudo 2D inversion recovery 180°-τ-90° pulse sequence (t1pir) using 8 incremented delay times (0.005 s, 0.646 s, 1.317 s, 2.030 s, 2.804 s, 3.676 s, 4.732 s, 6.277 s) with 4–8 scans per time increment (total of 5–10 min). 1D spectra were manually phased and baseline corrected. Bruker Topspin software (version 3.2, patch level 5) was used for calculating T1 relaxation times based on I[t] = I[0] × (1 − 2A × exp(−t/T1)).
Results & discussion
We successfully built an NMR bioreactor system using a standard 5 mm NMR tube and commercially available flowtube apparatus. We evaluated the stability and reproducibility, optimized the 3D cell preparation and show the effect of cell number and perfusion rate on metabolism. Metabolic and mitochondrial respiration changes upon interventions were detected within minutes using 1H NMR for metabolite levels and 19F NMR for detection of oxygen level in 3D cultured human fibroblasts.3D cell culture sample preparation
The designed protocol for fibroblast embedding in a 3D collagen-based scaffold and subsequent rapid transfer into the NMR spectrometer within a few minutes is described step by step in Fig. 1. Five, 10, 15 and 20 million fibroblasts were incorporated into a 3D collagen-based matrix and cultured in standard cell culture incubator up to seven days (Fig. 1, steps 6 and 7). Documentation over a period of seven days showed a change in volume over time depending on cell density (Fig. 2a). Cellular contraction in collagen based matrix resulted in a decrease of scaffold volume from 35 to 70% after cell re-adhesion in the first 24 hours. Volume reductions declined to an average of 7% to 11% in the following six days. Observed contraction of collagen embedded fibroblasts as a function of cell density and its volume stabilization after days is consistent with the observations described in the literature.66,67 Standardization of cell density and days of incubation in collagen-based 3D cell culture before NMR experiment is therefore essential in order to get reproducible data in repeated experiments.Perfused NMR bioreactor experiments
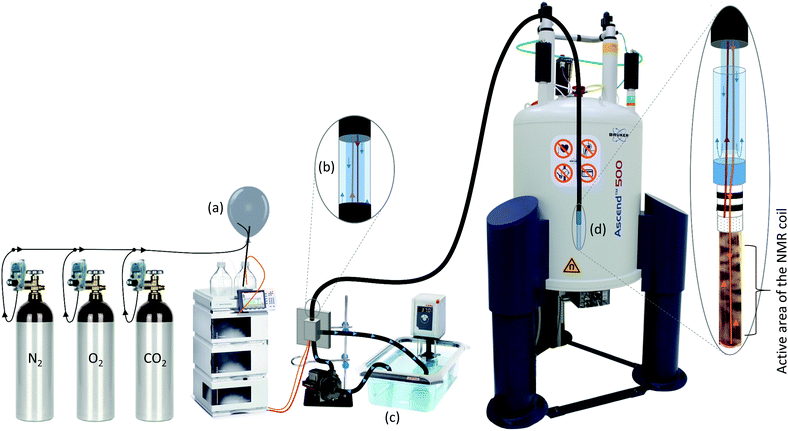
A detailed scheme of the perfused NMR bioreactor is described in Fig. 3. The carbon-based buffered glucose medium was controlled for a physiological pH of 7.4 using pH meter before perfusion.1H NMR for metabolic analysis
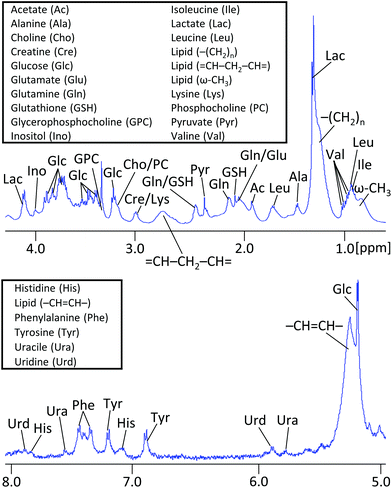
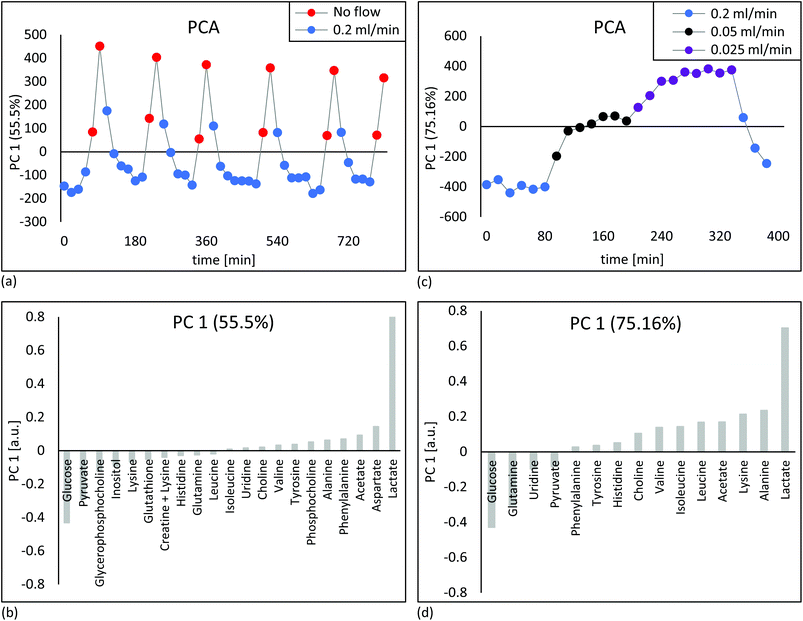
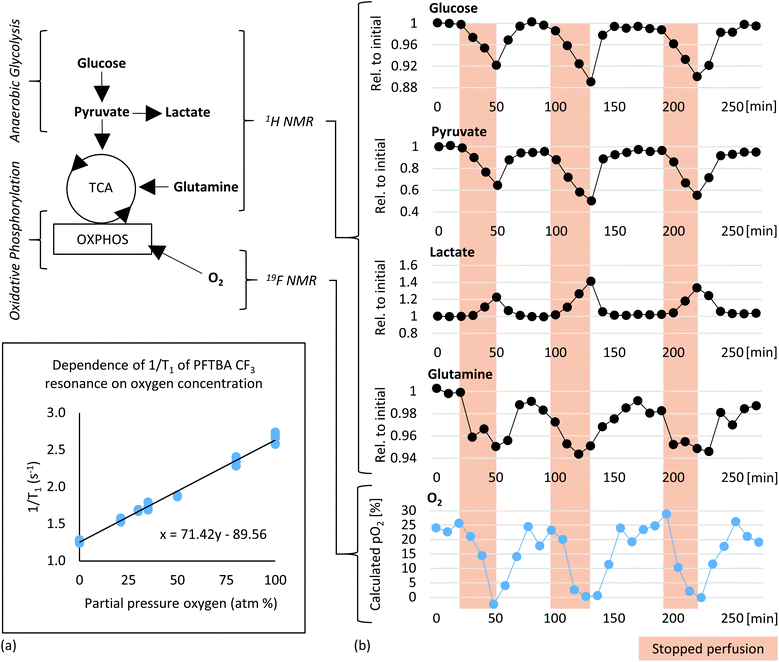
1H NMR spectra of embedded fibroblasts and cell medium were obtained in the bioreactor during perfusion. Resonances were assigned to overall 26 intra- and extracellular metabolites under applied standard cell culture condition (Fig. 4). In order to evaluate the sensitivity of the measurements detecting metabolic changes, experiments were performed with repeatedly changed perfusion rates for short- and long-term periods (Fig. 5). Using all resonances assigned to metabolites, PCA was performed for determination of overall spectral changes and demonstration of data stability.The feasibility to measure ongoing anaerobic glycolysis of living cells by proton NMR spectroscopy in the 5 mm flow tube has also been demonstrated by Cerofolini et al.29 for HEK293 T cells. We demonstrate in addition the feasibility for 1H NMR based metabolic investigation of fibroblasts cultured under controlled and stable conditions. Further, we show sensitivity to repeatedly measured flow rate dependent differences in a variety of metabolites, including glycolytic- and glutaminolytic- related metabolites with high temporal resolution. Importantly, metabolites returned repeatedly to the initial level over several hours of resumed flow.
19F NMR for oxygen quantification using perfluorotributylamine (PFTBA)
Obtaining cell culture oxygenation simultaneously to metabolic content would allow deeper insight into dynamic metabolic processes and the energetic state of cultured cells. Oxygenation determination via19F NMR based detection of T1 spin–lattice relaxation times was therefore implemented as surrogate marker of mitochondrial activity detected in collagen embedded cells.Combined 1H metabolic and 19F oxygen quantification
In order to test the sensitivity for detecting changes in oxygen tension in cell cultures upon challenges, “stop and go” measurements were performed analogous to flow-rate interruption experiments previously described. Collagen based cell culture scaffold with 20 million embedded fibroblasts and PFTBA carriers were perfused under constant flow at 0.2 ml min−1 (Fig. 6b). Alternately, 19F NMR based T1 relaxation times for oxygen measurement as a marker of mitochondrial oxidative phosphorylation and 1H NMR for measuring changes in metabolite content were measured with an acquisition time of 6 and 4 minutes respectively. Interruption of the perfusion rate for ∼30 minutes resulted repeatedly in a decrease of the remaining oxygen concentration due to continuous cellular respiration. Both, PFTBA coated thread and beads in 3D cell culture scaffold enabled detection of adapting T1 times in response to oxygen changes within minutes. A similar approach was presented by Pilatus et al. using 10 mm NMR flow tubes.35 Furthermore, the acquisition time (5–10 min) is in a comparable time frame as the oxygen quantification time required using conventional extracellular flux analyzer (7 min)72 and longer than bionas system (1–4 min)49 and O2k-FluoRespirometer or lab-on-a-chip system, which determine decrease of oxygen in real time during long phases of closed cell chambers.50,51Individual peak analysis of metabolites directly related to aerobic energy production via TCA fuel are displayed in Fig. 6b. As observed in the “stop and go” experiment shown in Fig. 5a, changes in glycolytic metabolites were observed (decrease in glucose and pyruvate and increase in lactate during stopped flow). The relative quantity of produced lactate is almost equivalent to observed proportion of decrease in pyruvate. A minor quantity of pyruvate might enter tricarboxylic acid (TCA) cycle for aerobic energy production. Pyruvate and glutamine are considered to be the main anaplerotic substrates among others.73 A decrease in glutamine was observed during the prolonged stop periods (Fig. 6) which is consistent with the observations during reduced flow (Fig. S1c†). Decreases in glutamine were not detected within shorter flow rate interruptions of 15 minutes as shown in ESI Fig. S1b.† We hypothesize the intervention period of 15 minutes are too short to measure significant degradation of glutamine. The observed decrease of glutamine goes along increasing evidence suggesting fibroblasts to degrade glutamine in significant amounts for energy production.74 The demonstrated sensitivity to study glutamine metabolism is of importance as glutamine is involved in multiple other pathways such as redox homeostasis, macromolecular synthesis, and signalling in cancer cells.75,76
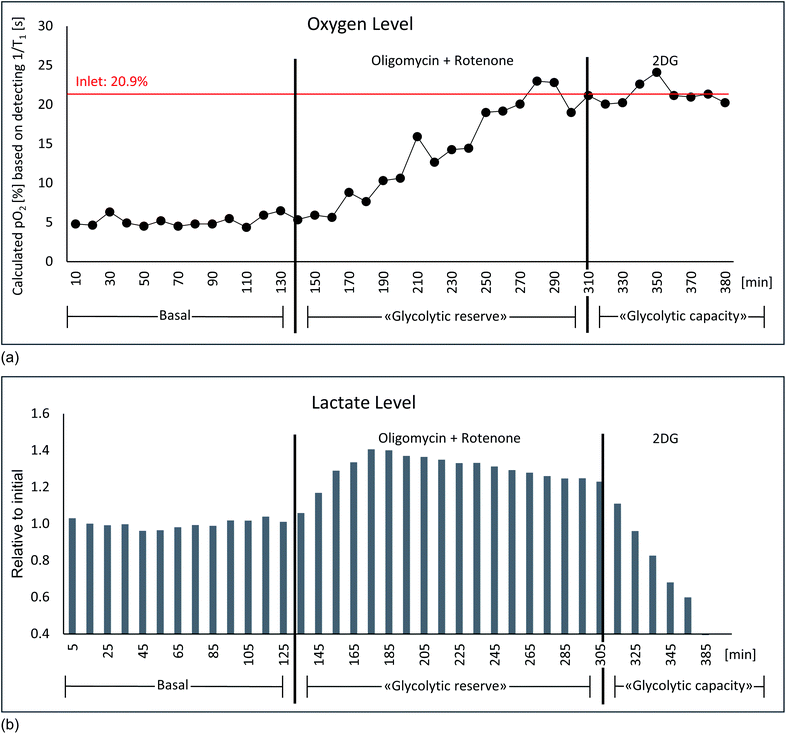 | ||
| Fig. 7 Glycolytic stress test: culture medium saturated with air contains a partial pressure of 21% oxygen. (a) With a flow rate of 0.2 ml min−1 a residual amount of approx. 7% oxygen was measured under basal conditions. After the addition of 5 μM oligomycin + 5 μM rotenone no more oxygen consumption takes place, consequently the oxygen content rises to the value of the admitted amount of oxygen. The oxygen values correspond to the T1 relaxation times of the δ(–CF3) resonance of PFTBA measured by 19F, which were converted into oxygen levels via generated fitting curve (Fig. 6a). (b) Simultaneous detection of lactate via1H NMR shows an increase in lactate production after oligomycin + rotenone addition, which was completely inhibited by the subsequent addition of 5 mM 2-deoxy-glucose. Described experiment revealing similar result was conducted in n = 2. | ||
While oxygen content is provided in absolute values, metabolite levels are currently reported only in relative quantity. However, absolute metabolite quantification is possible with the addition of an external reference and assumptions regarding exact number of cells or the ratio of cells to medium in the NMR sensitive volume (Fig. 3) of the bioreactor.
Presented experiments demonstrate the feasibility of detecting metabolic and mitochondrial respiration in living cells by combined in vitro metabolic profiling with 1H and 19F NMR based oxygen consumption studies. The presented bioreactor system using a standard NMR probe provides new possibilities for the simultaneous characterization of metabolic and mitochondrial functions of living cultured cells.
Conclusions
In this paper we show the feasibility to measure alternately and in real-time mitochondrial respiration and metabolic data combining a commercially available flow tube system with a standard 5 mm NMR probe. We describe in detail a reproducible collagen based 3D culture system for fibroblasts and a bioreactor setup. Using high cell densities of 5–20 million we demonstrate the viability of the cells and describe the effect of the flow rate and cell density on the metabolic activity. Importantly we show the needed sensitivity to detect substrate degradation rates of major mitochondrial fuel pathways and ability to measure rapid O2 and lactate changes as surrogate marker of oxidative phosphorylation and anaerobic glycolysis.Author contributions
Damian Hertig: conceptualization, data curation, formal analysis, investigation, methodology, project administration, writing – original draft; Sally Maddah: conceptualization, data curation, formal analysis, investigation, methodology, project administration; Roman Memedovski: data curation, formal analysis, investigation; Sandra Kurth: conceptualization, writing – review & editing; Aitor Moreno: rescources, writing – review & editing; Matteo Pennestri: resources, writing – review & editing; Andrea Felser: conceptualization, writing – review & editing; Jean-Marc Nuoffer: conceptualization, formal analysis, funding acquisition, resources, supervision, writing – review & editing; Peter Vermathen: conceptualization, formal analysis, funding acquisition, resources, supervision, writing – review & editing.Conflicts of interest
Within the framework of a research agreement, Bruker BioSpin AG allowed us to use the InsightCell™ Kit throughout the study period.Acknowledgements
Financial Supports from the Swiss National Science Foundation SNF (Nr. 310030_192691).Notes and references
- J. Nunnari and A. Suomalainen, Mitochondria: In sickness and in health, Cell, 2012, 148(6), 1145–1159 CrossRef CAS PubMed.
- N. S. Chandel, Mitochondria as signaling organelles, BMC Biol., 2014, 12, 34 CrossRef PubMed.
- M. J. Goldenthal and J. Marín-García, Mitochondrial signaling pathways: A receiver/integrator organelle, Mol. Cell. Biochem., 2004, 262(1–2), 1–16 CrossRef CAS PubMed.
- D. C. Chan, Mitochondria: Dynamic Organelles in Disease, Aging, and Development, Cell, 2006, 125(7), 1241–1252 CrossRef CAS.
- C. Frezza, Mitochondrial metabolites: Undercover signalling molecules, Interface Focus, 2017, 7(2), 0–5 CrossRef.
- J. N. Meyer, J. H. Hartman and D. F. Mello, Mitochondrial Toxicity, Toxicol. Sci., 2018, 162(1, March), 15–23 CrossRef CAS.
- I. Martínez-Reyes and N. S. Chandel, Mitochondrial TCA cycle metabolites control physiology and disease, Nat. Commun., 2020, 11(1), 1–11 CrossRef PubMed.
- R. Shiratori, K. Furuichi, M. Yamaguchi, N. Miyazaki, H. Aoki and H. Chibana, et al., Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner, Sci. Rep., 2019, 9(1), 1–15 Search PubMed.
- J. Thompson Legault, L. Strittmatter, J. Tardif, R. Sharma, V. Tremblay-Vaillancourt and C. Aubut, et al., A Metabolic Signature of Mitochondrial Dysfunction Revealed through a Monogenic Form of Leigh Syndrome, Cell Rep., 2015, 13(5), 981–989 CrossRef CAS PubMed.
- J. Shi, L. Tong, W. Tong, H. Chen, M. Lan and X. Sun, et al., Current progress in long-term and continuous cell metabolite detection using microfluidics, TrAC, Trends Anal. Chem., 2019, 117, 263–279 CrossRef CAS.
- M. Čuperlović-Culf, D. A. Barnett, A. S. Culf and I. Chute, Cell culture metabolomics: Applications and future directions, Drug Discovery Today, 2010, 15(15–16), 610–621 CrossRef.
- I. F. Duarte, J. Marques, M. P. M. Marques, B. J. Blaise and A. M. Gil, Nuclear Magnetic Resonance (NMR) Study of the Effect of Cisplatin on the Metabolic Profile of MG-63 Osteosarcoma Cells research articles, J. Proteome Res., 2010, 9, 5877–5886 CrossRef CAS PubMed.
- J. L. Griffin, M. Bollard, J. K. Nicholson and K. Bhakoo, Spectral profiles of cultured neuronal and glial cells derived from HRMAS 1H NMR spectroscopy, NMR Biomed., 2002, 15(6), 375–384 CrossRef CAS PubMed.
- V. Righi, O. Andronesi, D. Mintzopoulos, P. Black and T. Aria, High-resolution magic angle spinning magnetic resonance spectroscopy detects glycine as a biomarker in brain tumors, Int. J. Oncol., 2010, 36, 301–306 CAS.
- E. Kaebisch, T. L. Fuss, L. A. Vandergrift, K. Toews, P. Habbel and L. L. Cheng, Applications of high-resolution magic angle spinning MRS in biomedical studies I - cell line and animal models, NMR Biomed., 2017, 30(6), 1–18 CrossRef.
- I. Lamego, I. F. Duarte, M. P. M. Marques and A. M. Gil, Metabolic markers of MG-63 osteosarcoma cell line response to doxorubicin and methotrexate treatment: Comparison to cisplatin, J. Proteome Res., 2014, 13(12), 6033–6045 CrossRef CAS.
- L. Mirbahai, M. Wilson, C. S. Shaw, C. McConville, R. D. G. Malcomson and J. L. Griffin, et al., 1H magnetic resonance spectroscopy metabolites as biomarkers for cell cycle arrest and cell death in rat glioma cells, Int. J. Biochem. Cell Biol., 2011, 43(7), 990–1001 CrossRef CAS PubMed.
- N. J. Serkova and K. Glunde, Metabolomics of cancer, Methods Mol. Biol., 2009, 520, 273–295 CrossRef CAS PubMed.
- B. Sitter, T. F. Bathen, M. Tessem and I. S. Gribbestad, Progress in Nuclear Magnetic Resonance Spectroscopy High-resolution magic angle spinning (HR MAS) MR spectroscopy in metabolic characterization of human cancer, Prog. Nucl. Magn. Reson. Spectrosc., 2009, 54(3–4), 239–254 CrossRef CAS.
- O. Beckonert, H. C. Keun, T. M. D. Ebbels, J. Bundy, E. Holmes and J. C. Lindon, et al., Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts, Nat. Protoc., 2007, 2(11), 2692–2703 CrossRef CAS PubMed.
- M. J. Smith, C. B. Marshall, F. X. Theillet, A. Binolfi, P. Selenko and M. Ikura, Real-time NMR monitoring of biological activities in complex physiological environments, Curr. Opin. Struct. Biol., 2015, 32, 39–47 CrossRef CAS PubMed.
- S. S. Santos, S. B. Leite, U. Sonnewald and P. M. Alves, Stirred Vessel Cultures of Rat Brain Cells Aggregates: Characterization of Major Metabolic Pathways and Cell Population Dynamics, J. Neurosci. Res., 2007, 85(15), 3386–3397 CrossRef CAS PubMed.
- A. Mancuso, E. J. Fernandez, H. W. Blanch and D. S. Clark, A Nuclear Magnetic Resonance Technique for Determining Hybridoma Cell Concentration in Hollow Fiber Bioreactors, Biotechnology, 1990, 8, 1282–1285 CAS.
- A. Mancuso, N. J. Beardsley, S. Wehrli, S. Pickup, F. M. Matschinsky and J. D. Glickson, Real-time detection of 13C NMR labeling kinetics in perfused EMT6 mouse mammary tumor cells and βHC9 mouse insulinomas, Biotechnol. Bioeng., 2004, 87(7), 835–848 CrossRef CAS PubMed.
- M. Milkevitch, E. A. Browning and E. J. Delikatny, Perfused cells, tissues and organs by magnetic resonance spectroscopy, eMagRes, 2016, 5(2), 1333–1346 CAS.
- D. L. Foxall and J. S. Cohen, NMR studies of perfused cells, J. Magn. Reson., 1983, 52(2), 346–349 CAS.
- J. J. Ackerman, P. J. Bore, D. G. Gadian, T. H. Grove and G. K. Radda, N.m.r. studies of metabolism in perfused organs, Philos. Trans. R. Soc., B, 1980, 289(1037), 425–436 CAS.
- M. Bastawrous, A. Jenne, M. Tabatabaei Anaraki and A. J. Simpson, In vivo NMR spectroscopy: A powerful and complimentary tool for understanding environmental toxicity, Metabolites, 2018, 8(2), 1–24 CrossRef PubMed.
- L. Cerofolini, S. Giuntini, L. Barbieri, M. Pennestri, A. Codina and M. Fragai, et al., Real-Time Insights into Biological Events: In-Cell Processes and Protein-Ligand Interactions, Biophys. J., 2019, 116(2), 239–247 CrossRef CAS PubMed.
- R. D. Majumdar, M. Akhter, B. Fortier-McGill, R. Soong, Y. Liaghati-Mobarhan and A. J. Simpson, et al., In vivo solution-state NMR-based environmental metabolomics, eMagRes, 2017, 6(1), 133–148 Search PubMed.
- M. Tabatabaei Anaraki, R. Dutta Majumdar, N. Wagner, R. Soong, V. Kovacevic and E. J. Reiner, et al., Development and Application of a Low-Volume Flow System for Solution-State in Vivo NMR, Anal. Chem., 2018, 90(13), 7912–7921 CrossRef CAS PubMed.
- T. Harris, G. Eliyahu, L. Frydman and H. Degani, Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(43), 18131–18136 CrossRef CAS PubMed.
- S. Jeong, R. Eskandari, S. M. Park, J. Alvarez, S. S. Tee and R. Weissleder, et al., Real-time quantitative analysis of metabolic flux in live cells using a hyperpolarized micromagnetic resonance spectrometer, Sci. Adv., 2017, 3(6), 1–10 Search PubMed.
- K. R. Keshari, J. Kurhanewicz, R. E. Jeffries, D. M. Wilson, B. J. Dewar and M. Van Criekinge, et al., Hyperpolarized 13C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism, Magn. Reson. Med., 2010, 63(2), 322–329 CrossRef CAS PubMed.
- U. Pilatus, E. Aboagye, D. Artemov, N. Mori, E. Ackerstaff and Z. M. Bhujwalla, Real-Time Measurements of Cellular Oxygen Consumption, pH, and Energy Metabolism Using Nuclear Magnetic Resonance Spectroscopy, Magn. Reson. Med., 2001, 45, 749–755 CrossRef CAS.
- A. A. Shestov, A. Mancuso, S. C. Lee, L. Guo, D. S. Nelson and J. C. Roman, et al., Bonded cumomer analysis of human melanoma metabolism monitored by 13C NMR spectroscopy of perfused tumor cells, J. Biol. Chem., 2016, 291(10), 5157–5171 CrossRef CAS PubMed.
- J. Carvalho, S. Alves, M. M. C. A. Castro, C. F. G. C. Geraldes, J. A. Queiroz and C. P. Fonseca, et al., Development of a bioreactor system for cytotoxic evaluation of pharmacological compounds in living cells using NMR spectroscopy, J. Pharmacol. Toxicol. Methods, 2019, 95, 70–78 CrossRef CAS PubMed.
- G. Siegal and P. Selenko, Cells, drugs and NMR, J. Magn. Reson., 2019, 306(July), 202–212 CrossRef CAS PubMed.
- A. Kalfe, A. Telfah, J. Lambert and R. Hergenröder, Looking into Living Cell Systems: Planar Waveguide Microfluidic NMR Detector for in Vitro Metabolomics of Tumor Spheroids, Anal. Chem., 2015, 87(14), 7402–7410 CrossRef CAS PubMed.
- A. Yilmaz and M. Utz, Characterisation of oxygen permeation into a microfluidic device for cell culture by: In situ NMR spectroscopy, Lab Chip, 2016, 16(11), 2079–2085 RSC.
- D. A. Foley, A. L. Dunn and M. T. Zell, Reaction monitoring using online vs tube NMR spectroscopy: Seriously different results, Magn. Reson. Chem., 2016, 54(6), 451–456 CrossRef CAS PubMed.
- R. G. Shulman and D. L. Rothman, 13 C NMR of Intermediary Metabolism: Implications for Systemic Physiology, Annu. Rev. Physiol., 2001, 63(1), 15–48 CrossRef CAS PubMed.
- R. Sriram, M. Van Criekinge, A. Hansen, Z. J. Wang, D. B. Vigneron and D. M. Wilson, et al., Real-time measurement of hyperpolarized lactate production and efflux as a biomarker of tumor aggressiveness in an MR compatible 3D cell culture bioreactor, NMR Biomed., 2015, 28(9), 1141–1149 CrossRef CAS PubMed.
- K. Glunde, E. Ackerstaff, K. Natarajan, D. Artemov and Z. M. Bhujwalla, Real-time changes in 1H and 31P NMR spectra of malignant human mammary epithelial cells during treatment with the anti-inflammatory agent indomethacin, Magn. Reson. Med., 2002, 48(5), 819–825 CrossRef CAS PubMed.
- M. Milkevitch, N. J. Beardsley and E. J. Delikatny, Phenylbutyrate induces apoptosis and lipid accumulations via a peroxisome proliferator-activated receptor gamma-dependent pathway, NMR Biomed., 2010, 23(5), 473–479 CrossRef CAS PubMed.
- J. Xue, N. G. Isern, R. J. Ewing, A. V. Liyu, J. A. Sears and H. Knapp, et al., New generation NMR bioreactor coupled with high-resolution NMR spectroscopy leads to novel discoveries in Moorella thermoacetica metabolic profiles, Appl. Microbiol. Biotechnol., 2014, 98(19), 8367–8375 CrossRef CAS PubMed.
- Y. Mardor, O. Kaplan, M. Sterin, J. Ruiz-Cabello, E. Ash and Y. Roth, et al., Noninvasive Real-Time Monitoring of Intracellular Cancer Cell Metabolism and Response to Lonidamine Treatment Using Diffusion Weighted Proton Magnetic Resonance Spectroscopy, Cancer Res., 2000, 60(18), 5179–5186 CAS.
- M. Milkevitch, H. Shim, U. Pilatus, S. Pickup, J. P. Wehrle and D. Samid, et al., Increases in NMR-visible lipid and glycerophosphocholine during phenylbutyrate-induced apoptosis in human prostate cancer cells, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2005, 1734(1), 1–12 CrossRef CAS PubMed.
- P. Mestres and A. Morguet, The Bionas technology for anticancer drug screening, Expert Opin. Drug Discovery, 2009, 4(7), 785–797 CrossRef CAS PubMed.
- M. Makrecka-Kuka, G. Krumschnabel and E. Gnaiger, High-resolution respirometry for simultaneous measurement of oxygen and hydrogen peroxide fluxes in permeabilized cells, tissue homogenate and isolated mitochondria, Biomolecules, 2015, 5(3), 1319–1338 CrossRef CAS PubMed.
- F. Alexander, S. Eggert and J. Wiest, A novel lab-on-a-chip platform for spheroid metabolism monitoring, Cytotechnology, 2018, 70(1), 375–386 CrossRef CAS PubMed.
- D. A. Ferrick, A. Neilson and C. Beeson, Advances in measuring cellular bioenergetics using extracellular flux, Drug Discovery Today, 2008, 13(5–6), 268–274 CrossRef CAS PubMed.
- B. A. Berkowitz, C. A. Wilson, D. L. Hatchell and R. E. London, Quantitative determination of the partial oxygen pressure in the vitrectomized rabbit eye in Vivo using 19F NMR, Magn. Reson. Med., 1991, 21(2), 233–241 CrossRef CAS PubMed.
- K. A. McGovern, J. S. Schoeniger, J. P. Wehrle, C. E. Ng and J. D. Glickson, Gel–entrapment of perfluorocarbons: A fluorine–19 NMR spectroscopic method for monitoring oxygen concentration in cell perfusion systems, Magn. Reson. Med., 1993, 29(2), 196–204 CrossRef CAS PubMed.
- J. Taylor and C. Deutsch, 19F-nuclear magnetic resonance: measurements of [O2] and pH in biological systems, Biophys. J., 1988, 53(2), 227–233 CrossRef CAS.
- R. H. Haas, S. Parikh, M. J. Falk, R. P. Saneto, N. I. Wolf and N. Darin, et al., The in-depth evaluation of suspected mitochondrial disease, Mol. Genet. Metab., 2008, 94(1), 16–37 CrossRef CAS PubMed.
- J. Villegas and M. McPhaul, Establishment and Culture of Human Skin Fibroblasts, Curr. Protoc. Mol. Biol., 2005, 71(1), 1–9 Search PubMed.
- K. Duval, H. Grover, L. H. Han, Y. Mou, A. F. Pegoraro and J. Fredberg, et al., Modeling physiological events in 2D vs. 3D cell culture, Physiology, 2017, 32(4), 266–277 CrossRef CAS PubMed.
- G. Diserens, D. Hertig, M. Vermathen, B. Legeza, C. E. Flück and J. M. Nuoffer, et al., Metabolic stability of cells for extended metabolomical measurements using NMR. A comparison between lysed and additionally heat inactivated cells, Analyst, 2017, 142(3), 465–471 RSC.
- P. Joseph, J. Fishman, B. Mukherji and H. Sloviter, In Vivo 19F NMR Imaging of the Cardiovascular System, J. Comput. Assist. Tomogr., 1985, 9(6), 1012–1019 CrossRef CAS PubMed.
- J. A. Aguilar, M. Nilsson, G. Bodenhausen and G. A. Morris, Spin echo NMR spectra without J modulation, Chem. Commun., 2012, 48(6), 811–813 RSC.
- D. Hertig, A. Felser, G. Diserens, S. Kurth, P. Vermathen and J. M. Nuoffer, Selective galactose culture condition reveals distinct metabolic signatures in pyruvate dehydrogenase and complex I deficient human skin fibroblasts, Metabolomics, 2019, 15(3), 0 CrossRef PubMed.
- M. Vermathen, L. E. H. Paul, G. Diserens, P. Vermathen and J. Furrer, 1H HR-MAS NMR based metabolic profiling of cells in response to treatment with a hexacationic ruthenium metallaprism as potential anticancer drug, PLoS Biol., 2010, 8(19), e1000514 Search PubMed.
- D. S. Wishart, Y. D. Feunang, A. Marcu, A. C. Guo, K. Liang and R. Vázquez-Fresno, et al., HMDB 4.0: The human metabolome database for 2018, Nucleic Acids Res., 2018, 46(D1), D608–D617 CrossRef CAS PubMed.
- A. A. Ribeiro and K. Umayahara, 19F, 13C NMR analysis of an oxygen carrier, perfluorotributylamine, and perfluoropentanoic acid, Magn. Reson. Chem., 2003, 41(2), 107–114 CrossRef CAS.
- E. Bell, B. Ivarsson and C. Merrill, Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro, Proc. Natl. Acad. Sci. U. S. A., 1979, 76(3), 1274–1278 CrossRef CAS PubMed.
- K. Bott, Z. Upton, K. Schrobback, M. Ehrbar, J. A. Hubbell and M. P. Lutolf, et al., The effect of matrix characteristics on fibroblast proliferation in 3D gels, Biomaterials, 2010, 31(32), 8454–8464 CrossRef CAS PubMed.
- F. K. Chan, K. Moriwaki and M. J. De Rosa, Detection of Necrosis by Release of Lactate Dehydrogenase Activity, In: Immune Homeostasis: Methods and Protocols, 2013, p. 65–70 Search PubMed.
- J. M. S. Lemons, H. A. Coller, X. J. Feng, B. D. Bennett, A. Legesse-Miller and E. L. Johnson, et al., Quiescent fibroblasts exhibit high metabolic activity, PLoS Biol., 2010, 8(10), e1000514 CrossRef PubMed.
- G. Park, D. S. Oh, Y. U. Kim and M. K. Park, Acceleration of Collagen Breakdown by Extracellular Basic pH in Human Dermal Fibroblasts, Skin Pharmacol. Physiol., 2016, 29(4), 204–209 CrossRef CAS PubMed.
- J. Y. Park, S. J. Yoo, L. Patel, S. H. Lee and S. H. Lee, Cell morphological response to low shear stress in a two-dimensional culture microsystem with magnitudes comparable to interstitial shear stress, Biorheology, 2010, 47(3–4), 165–178 CAS.
- J. K. Salabei, A. A. Gibb and B. G. Hill, Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis, Nat. Protoc., 2014, 9(2), 421–438 CrossRef CAS.
- H. Brunengraber and C. R. Roe, Anaplerotic molecules: Current and future, J. Inherited Metab. Dis., 2006, 29(2–3), 327–331 CrossRef PubMed.
- C. Nieweld and R. Summer, Activated fibroblasts: Gluttonous for glutamine, Am. J. Respir. Cell Mol. Biol., 2019, 61(5), 554–555 CrossRef CAS PubMed.
- R. J. Deberardinis and T. Cheng, Q's next: The diverse functions of glutamine in metabolism, cell biology and cancer, Oncogene, 2010, 29(3), 313–324 CrossRef CAS PubMed.
- M. A. Medina, F. Sánchez-Jiménez, J. Márquez, A. Rodríguez Quesada and I. de Castro Núñez, Relevance of glutamine metabolism to tumor cell growth, Mol. Cell. Biochem., 1992, 113(1), 1–15 CrossRef CAS PubMed.
- B. K. Chacko, M. R. Smith, M. S. Johnson, G. Benavides, M. L. Culp and J. Pilli, et al., Mitochondria in precision medicine; linking bioenergetics and metabolomics in platelets, Redox Biol., 2019, 22, 101165 CrossRef CAS PubMed.
- S. A. Mookerjee and M. D. Brand, Measurement and Analysis of Extracellular Acid Production to Determine Glycolytic Rate, J. Visualized Exp., 2015, 2(106), 1–9 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00041a |
| ‡ Shared last author. |
| This journal is © The Royal Society of Chemistry 2021 |