Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Current advances in the detection of COVID-19 and evaluation of the humoral response
Maria Ana Cristina
Huergo
 *a and
Nguyen Thi Kim
Thanh
*a and
Nguyen Thi Kim
Thanh
 *bc
*bc
aTheoretical and Applied Physical Chemical Research Institute (INIFTA), National Univesity of La Plata (UNLP), CONICET. Sucursal 4 Casilla de Correo 16, 1900 La Plata, Argentina. E-mail: mahuergo@inifta.unlp.edu.ar
bBiophysics Group, Department of Physics and Astronomy, University College London (UCL), London, WC1E 6BT, UK
cUCL Healthcare Biomagnetic and Nanomaterials Laboratories, 21 Albemarle Street, London W1S 4BS, UK. E-mail: ntk.thanh@ucl.ac.uk
First published on 9th December 2020
Abstract
The new outbreak caused by coronavirus SARS-CoV-2 started at the end of 2019 and was declared a pandemic in March 2020. Since then, several diagnostic approaches have been re-adapted, and also improved from the previous detections of SARS and MERS coronavirus. The best strategy to handle this situation seems to rely on a triad of detection methods: (i) highly sensitive and specific techniques as the gold standard method, (ii) easier and faster point of care tests accessible for large population screening, and (iii) serology assays to complement the direct detection and to use for surveillance. In this study, we assessed the techniques and tests described in the literature, their advantages and disadvantages, and the interpretation of the results. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) is undoubtedly the gold standard technique utilized not only for diagnostics, but also as a standard for comparison and validation of newer approaches. Other nucleic acid amplification methods have been shown to be adequate as point of care (POC) diagnostic tests with similar performance as RT-qPCR. The analysis of seroconversion with immunotests shows the complexity of the immune response to COVID-19. The detection of anti-SARS-CoV-2 antibodies can also help to detect previously infected asymptomatic individuals with negative RT-qPCR tests. Nevertheless, more controlled serology cohort studies should be performed as soon as possible to understand the immune response to SARS-CoV-2.
1. Introduction
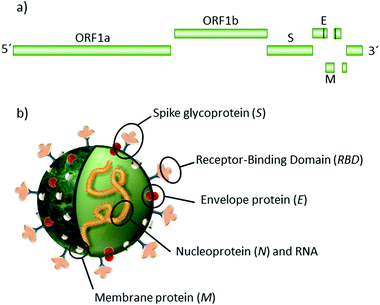
A new type of pneumonia, named coronavirus disease 2019 (COVID-19), appeared in Wuhan, Hubei Province, China at the end of 2019.1 The agent behind it is the new severe acute respiratory syndrome coronavirus (SARS-CoV-2) that emerged as the most major global healthcare challenge for a century. Since its appearance, over 86 million people have been infected, more than 1.86 million died so far, and the overstretched health systems caused additional excess deaths to those already caused by COVID-19.2–4SARS-CoV-2 is a single-stranded positive-sense RNA (+ssRNA) virus, surrounded by a lipidic envelope, with a diameter of 80–90 nm. The genome is organized into 3 main regions: two open reading frames (ORF), ORF1a and ORF1b, located at the 5′ end. These large genes encode 16 non-structural proteins, including RNA-dependent RNA polymerase (RdRp) and RNA helicase. The third region comprises genes encoding the structural and accessory proteins at the 3′ end.5Scheme 1 illustrates the virion and its main structural proteins: transmembrane proteins M and E, nucleoprotein N and trimeric glycoprotein spike S, composed of subunits S1 and S2. Unlike the common winter cold virus, SARS-CoV-2 does not primarily affect the upper respiratory tract. Instead, it invades and replicates in low respiratory tract tissues and lungs, where angiotensin-converting enzyme 2 (ACE2) receptors are abundant and permissive.5–9 The entrance into cells is mediated by the trimeric glycoprotein S, which recognises and binds to the ACE2 receptor via its receptor-binding domain (RBD).10 The RBD-ACE2 union induces the incorporation of the viral RNA into the cell, triggering the viral replication and virion assembly in double membrane vesicles.11
Until now, the key strategy that proved to be effective in limiting the virus spread is the use of masks, being in open air or permanently ventilated spaces, frequent hand washing and the restriction of individual contacts and mobility, guided by the early and accurate diagnosis of the infection.12
It has been estimated that about 86% of all infected patients have not been diagnosed, and half of them were contagious.12,13 False-negative rates occur in approximately 30% of patients with COVID-19.14,15 A false-negative diagnosis can have serious consequences as unknown contagious people will inevitably spread its infection, hampering efforts to contain the spread of the virus and exposing healthcare workers to the virus.13,16
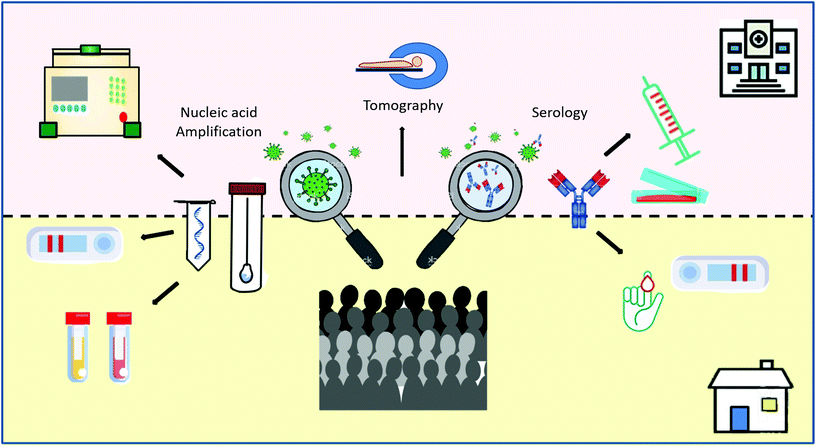
Current available diagnostic tests to handle the outbreak situation include a triad of complementary approaches, from rapid clinical decisions to the screening of large populations (Scheme 2).17 First, highly sensitive and specific molecular tests are needed to detect the presence of the viral nucleic acids. RT-qPCR is the gold standard technique because of its sensitivity and specificity.18–20 However, the pandemic has overwhelmed RT-qPCR testing capacity in most countries, an expected situation when contagious cases increase exponentially.21 Second, a triage of suspected cases is needed for large populations. For this purpose, POC tests and those analysed in basic laboratories can be ideal for the diagnosis of SARS-CoV-2 infection. These tests permit a fast and accurate diagnosis of a medical condition at the time and place of patient care. Ideal POC tests should be fast (in hours, rather than days), simple-to-use for non-specialized personnel, and able to be performed at the bedside or nearby basic facilities. Third, antibody detection is key to understanding the natural history of the infection and to determine the accurate rates of transmission, fatality and mortality, the correlation with possible unknown future sequelae, and the development of protective anti-SARS-CoV-2 antibodies.22 In particular, serology tests are especially useful to diagnose suspected patients with negative RT-qPCR results and to identify asymptomatic infections.23 Seroconversion implies current and past exposure to the virus. Therefore, it is an ideal tool for surveillance, as it can be tested in common laboratories or at the POC, depending on the type of sample required.24
Other diagnostic methods, such as changes in computed tomography images,25,26 voice changes and different predictive algorithms,27 associated or not with Artificial Intelligence methods, are being developed or are already in use.28,29 For surveillance of COVID-19, smartphones offer a fast and simple way to share epidemiological data, POC testing and patient monitoring in real time.30–35 Even though these technologies have been demonstrated to be very useful, we will focus on molecular and immunological techniques and their specific targets in this review. We will start with the direct confirmation of the virus by nucleic acid determination techniques and other approaches for viral antigen detection. We will assess the current serology tests and the information that has been available so far to comprehend the natural history of the antibody immune response.36,37
1. Direct virus detection
Since January 10, 2020, when the entire SARS-CoV-2 sequence was shared via the Global Initiative on Sharing All Influenza Data (GISAID) platform, hundreds of researchers and companies have developed different strategies to detect the virus. In this section, we will describe the different techniques that have been adapted to detect the virus. The strategies typically search for the presence of either viral nucleic acids or viral proteins, mostly in nasopharyngeal swab (NPS) samples.
1.1 Reverse transcription polymerase chain reaction (RT-PCR)
This method recognises tiny amounts of a specific RNA target by sequence-specific primers. The recognised RNA is then transcribed into cDNA by the reverse transcriptase enzyme. Once arriving to this point, the assay continues as a typical PCR assay, amplifying the transcribed sequence by several orders of magnitude. To this end, a thermal cycler generates repeated cycles of heating and cooling to permit nucleic acid melting, annealing and DNA replication by a DNA polymerase. For viral infections, quantitative RT-PCR (RT-qPCR) results are faster and more sensitive than conventional RT-PCR as it monitors the progress of the reaction in real time using fluorescent labels or electrical signals.38–41 This real time technique has shown the highest sensitivity, and currently is the gold standard assay for SARS-CoV-2 detection.18,42
Current RT-qPCR kits typically search for one or more of the following gene targets: RdRp and envelope protein E,18 RNA polymerase/Helicase (Hes),43 nucleocapsid protein N alone44 or in combination with ORF1b45 or glycoprotein S.46 It has been proposed that a multitarget gene amplification can play a key role in increasing sensitivity and preserving specificity, something of special importance for coronavirus diagnosis due to the high probability of mutations.47,48 However, other researchers did not find significant differences between the performance of the different multitarget and single-target strategies.49–51 Furthermore, the kit developed at Rutgers University, for ORF1ab, N and S genes, considers a sample as positive for SARS-CoV-2 if amplification is detected with at least two of the three target sequences. More specifically, the ORF1ab region shows 83% sensitivity compared to 100% for the S and N genes, and 100% specificity in all cases.52 The comparison between different kits did not show cross-reactivity with other respiratory viruses, except for the SARS-CoV E gene, as expected.53
Recently, the Centre of Disease Control of USA developed an Influenza – SARS-CoV-2 (Flu SC2) multiplex assay for the simultaneous RT-qPCR diagnosis of influenza and COVID-19, two frequent diseases with some similar symptoms.54 The approach utilizes protein N gene for the detection of SARS-CoV-2, matrix (M1) gene for the detection of influenza A, and nonstructural 2 (NS2) gene for the detection of influenza B. Of particular importance, the test includes the RNase P gene for the detection of human nucleic acid as an internal control.
An ultrafast molecular testing device developed by the start-up GNA Biosolutions from the Ludwig Maximilians University of Munich has recently received considerable attention.55 The fundamentals of this system rely on DNA detection driven by the localized heating of microcyclers immersed in the reaction solution. By keeping the excitation of the microcyclers to a microsecond time scale, the heat field generated in the denaturation step extends by only a few micrometres into the reaction solution. Consequently, only a minute fraction (≪1%) of the reaction solution is heated to the denaturation temperature, while the rest of the solution remains at the initial temperature, allowing for elongation and annealing. The functionalization of the microcyclers with DNA complementary to the amplicon permits the localization of the reaction onto the heating surface. This development is based on the foundational work by Stehr et al. on the optothermal melting of DNA attached to gold nanoparticles (AuNPs).56,57 The latest approach utilizes the Joule heating of conductive microheaters, such as micrometre-sized metal wires or ultrathin microstructured metal foils. The patented PCR technology operates with short pulses to control temperature cycles at the nano to micro scale, accelerating the PCR reaction times by a factor of ten.58 The reverse transcription, amplification and detection of the SARS-CoV-2 viral RNA can be performed simultaneously in up to 8 samples and within 15 min.
Table 1 shows a summary of several RT-qPCR kits and probes for the diagnoses of SARS-CoV-2 developed by different Institutions. The Food & Drug Administration, Test-Tracker and The Foundation for Innovative New Diagnostics (FIND) webpages provide updates on the available SARS-CoV-2 molecular tests.59–61
| Test and institution | Gene/region target | Limit of detection (LoD) | Ref. |
|---|---|---|---|
| Centre for disease control and prevention, China | ORF1ab and N | 1.5 copies per reaction | 62 |
| Charité virology, Berlin, Germany | RdRp, E and N | 5.2; 3.8; 8.3 copies per reaction respectively | 18 and 63 |
| School of Public Health, LKS Faculty of Medicine, University of Hongkong, China | ORF1b and N | <10 copies per reaction | 45 |
| Beijing Wantai Biological Pharmacy Enterprise Co. Lt | ORF1b and N | 1–10 copies per reaction | 61 |
| Seegene Inc. Allplex™ 2019-nCoV assay | RdRp, E and N | 1–10 copies per reaction | 61 |
| RT-qPCR, National Institute of Infectious Disease | N gene | 5 copies per reaction | 64 and 65 |
| CDC 2019 novel coronavirus (nCoV) real-time RT-PCR | N1 and N2 | ∼31 and 6 copies per reaction | 44 |
| Institut Pasteur | RdRp and E | 10 copies per reaction | 66 |
| Boditech Med. Inc ExAmplar COVID-19 real-time PCR kit | E gene | 10–50 copies per reaction | 54 and 61 |
| RdRp gene | 50–100 copies per reaction | ||
| COVID-19-RdRp/Hel RT-PCR assay | RdRP/Helicase | 11.2 copies per reaction | 43 |
| Simplexa | S and ORF1ab | 500 copies per mL | 67 |
| nCoV-QS (MiCo BioMed) | ORF3a and N | 1.8 and 4.24 copies per mL | 68 |
| Shanghai Kehua Bio-Engineering Co. Ltd KHB diagnostic kit | ORF1ab, N and E | 1–10 copies per reaction | 61 |
| BGI Health (HK) Co. Ltd, RT-PCR kit for detection 2019-nCOV (CE-IVD) | ORF1 gene | 1–10 copiesper reaction | 61 |
| The Rutgers Clinical Genomics Laboratory SARS-CoV-2 Assay | ORF1ab, S and N | 200 copies per mL | 52 |
| bioMérieux SA ARGENE® | RdRp and N | 1–50 copies per reaction | 61 |
| SARS-COV-2 R-GENE® | |||
| Tib Molbiol/Roche Diagnostic Multiplex RNA Virus Master | E gene | 1–10 copies per reaction | 61 |
| RealStar® SARS-CoV-2 RT-PCR kit (Altona Diagnostics) | E: betacoronavirus | 1–10 copies per reaction | 69 |
| E- S: SARS-CoV-2 | 1–10 copies per reaction | ||
| RT-PCR + restriction fragment length polymorphism | RdRp and E | 204 and 70 copies per reaction respectively | 70 |
| COVID-19-nsp2 | Non-structural protein 2 (nsp2) | 1.8 TCID50 per mL | 71 |
Most protocols are referred to NPS samples. However, RT-qPCR has also good sensitivity in saliva samples. The collection of saliva is currently being used as a simpler and safer strategy for self-sample collection and delivery.52,72–74
1.2 Isothermal amplification techniques
Based on the same idea of biological amplification of nucleic acids, newer molecular techniques have emerged, playing an important role in resource-limited regions. As previously mentioned, PCR assays require cycles of different temperatures to complete the different steps of melting, annealing and replication. Isothermal amplification substitutes the high temperature melting of nucleic acids by DNA polymerases with high strand displacement activity. These enzymes advance the separation of the DNA strands, while they synthesize the new complementary strands. When incubated at their optimal temperatures, these polymerases are extremely efficient, as multiple molecular reactions can proceed simultaneously, rather than being forced to operate sequentially.75
There are several different strategies: loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA) or transcription mediated amplification (TMA), strand displacement amplification (SDA), helicase-dependent amplification (HDA), isothermal recombinase polymerase amplification (RPA), rolling-circle amplification (RCA) and signal amplification by ternary initiation complexes (SATIC).76 Some of these approaches have already been adapted as POC testing for COVID-19.77,78 In these cases, as the target is an RNA molecule, the assays require a first step of enzymatic reverse transcription of the viral RNA into cDNA.
Loop-mediated isothermal amplification
LAMP assays utilize a DNA polymerase that requires a set of two or three pairs of primers to amplify the sequences at a constant temperature of 60–65 °C.79,80 In a typical assay, 4 primers are used to recognize different regions of the target sequence. Two additional primers and a recombinase enzyme are used to produce the amplification of the genetic material in a continuous loop or dumbbell structure (Scheme 3). The result can be determined by turbidimetry,81 caused by magnesium pyrophosphate precipitate (a byproduct of the amplification); by a change in colour with the addition of pH-sensitive dyes; by fluorescence based on fluorophores that recognize and bind to dsDNA in real time; or by immunochromatography.82,83 This technique is faster than PCR, taking <1 h and does not require complex technology, such as thermocyclers. However, the non-specificity of the detected signal, as it relies on the changes in the physicochemical properties of the system, can become a limitation. When the assay is left to proceed for longer periods, a false positive result can occur by nonspecific amplification.84
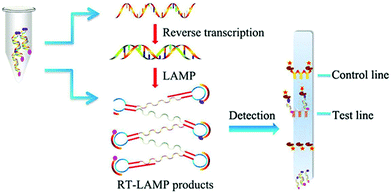 | ||
| Scheme 3 Schematic illustration of RT-LAMP assay. The amplification is performed in a constant temperature water bath. In this example, products are detected with a vertical flow visualization strip. Reproduced from ref. 82 with permission from Frontiers, copyright 2018. | ||
Several groups have already adapted this method to the detection of SARS-CoV-2 using different dyes (SYBR green,85 phenol red,86–89 WarmStart®90 and calcein91) for naked eye detection. Most of the protocols search for nucleoprotein N. However, some groups include different genes in the same test, i.e., ORF1ab and N or E and N, for simultaneous detection. This strategy permits the combination of a highly specific region, like ORF1ab or E, with the high sensitivity offered by gene N to guarantee both conditions.86,90,92 In general, the limit of detection (LoD) for the LAMP assay has shown lower but comparable performance to that of RT-qPCR, and it is highly recommended for the large-scale diagnostic testing of SARS-CoV-2.69,84
Recombinase polymerase amplification
RPA is the adaptation of the homologous recombination process of DNA. In a typical assay, a recombinase protein and recombinase factors form a nucleoprotein filament with the primer that searches for homologous sequences in DNA. This complex splits the dsDNA, the recombinase disassembles, and a DNA polymerase initiates the replication process following the primer. The incorporation of both forward and reverse primers enables the exponential accumulation of the amplified duplex DNA, consisting of the sequence between the forward and reverse primers.75 Several methods of detection have been applied in combination to RPA. However, for the detection of SARS-CoV-2, it has been mostly used in combination with the clustered regularly interspaced short palindromic repeats (CRISPR) assay (see subsection 1.3).
Transcription mediated amplification
TMA starts with the retroviral transcription of RNA into cDNA; after which, the RNA is degraded and the new cDNA replicated into double stranded DNA (dsDNA). dsDNA is then transcribed into new RNAs, which repeat the amplification cycle. Pham et al. developed an assay for the detection of two separate target sequences in the ORF1ab region by an automated system.93 A study to evaluate the capacity of the TMA assay to detect the virus in clinical samples has shown a concordance of almost 100% with the RT-qPCR method both in sensitivity and specificity.94 The assay takes 3.5 h to complete. However, the main limitation of this technique is the need of an automated set up, which increases the costs of the assay.
Signal amplification by ternary initiation complexes
SATIC is a one-step real-time assay for RNA detection. The reaction occurs at 37 °C, and it does not require reverse transcription steps. It involves the formation of a ternary initiation complex between a circular DNA template, a DNA primer and the target RNA. The ternary complex is amplified to generate multiple copies of G-quadruplex (G4) by the rolling-circle amplification (RCA) technology. RCA copies the natural process of the unidirectional nucleic acid replication of circular molecules of DNA or RNA, such as plasmids, bacteriophages and viroid genomes. After amplification, specific fluorophores, such as N3-hydroxyethyl thioflavin T (ThT-HE), show selective emission when interacting with G4s, allowing real-time detection.95 The method can be also adapted to direct visual detection by the formation of two different ternary initiation complexes. The SATIC assay has been recently approved for SARS-CoV-2 detection.96 The technique does not require specific set-ups, and virus determination can be made in about 25 min after sample collection. Authors claimed the reaction is highly specific, however there are no references about the sensitivity of the assay.
1.3 Clustered regularly interspaced short palindromic repeats
CRISPR-Cas enzymes and CRISPR sequences composed of a primitive immune system were developed to defend prokaryotes against bacteriophages and mobile genetic elements.97–100 This system has been adapted for genome editing by recent Nobel laureates, Emmanuelle Charpentier and Jennifer A. Doudna. The CRISPR-Cas editing method has revolutionized genetic engineering and in the last years, has been adapted for diagnostic strategies. The principle relies on the endonuclease activity of CRISPR-Cas enzymes that recognize the presence of specific sequences via programmable CRISPR-RNA (crRNA). When the crRNA binds the target sequence, the endonuclease activity is triggered, and the recognised nucleic acid is cut. For diagnostic purposes, Cas12 and Cas13a are the preferred enzymes, as they present a secondary activity of nonspecific cutting once the specific endonuclease activity has been triggered. Cas12 targets dsDNA and its secondary activity determines the multiple cuts of any ssDNA found.101 Cas 13a targets ssRNA and its secondary activity determines the cutting of cis and trans ssRNA.102,103 The use of linked quenched/fluorescent probes permits the detection of a fluorescent signal after the endonuclease activity cleaves the reporter molecule, indicating the presence of the nucleic sequence.104–106 The main difference with previously described methods relies on the ability of CRISPR assays to identify the amplified gene, making the strategy more specific.69
Broughton et al. have adapted this molecular test to detect SARS-CoV-2 in RNA-extracted samples in <40 min.16 The test is called SARS-CoV-2 DNA endonuclease-targeted CRISPR trans reporter (DETECTR), and is based on RT-LAMP amplification, followed by the detection of predefined coronavirus sequences by the enzyme Cas12. The primers target the E and N genes, and human RNase P for quality control. The test can be visualized by fluorescence or as a lateral flow assay, using 6-carboxyfluorescein-biotin conjugate as the reporter molecule that binds to the control line if uncleaved, and to the detection line when cleaved by Cas12. The estimated LoD for the DETECTR assay was 10 copies per μl reaction. The sensitivity and specificity relative to the RT-qPCR assay are 95% and 100%, respectively. A similar protocol was developed by Curti et al., in which primers are directed to recognize the RdRp, ORF1b and ORF1ab regions (Scheme 4).107 Based on the same enzyme, Ramahandran et al. developed an assay in which they achieve an electric field gradient using isotachophoresis (ITP) on a microfluidic chip.108 The strategy combines ITP for purification, LAMP for amplification, and ITP enhanced CRISPR for detection. The sensitivity and specificity are comparable to other CIRSPR assays, but the entire process, from raw sample to result, can be performed in 30 min. However, the need of a device with electrodes for ITF on microchips can become an economic limitation of the technique.
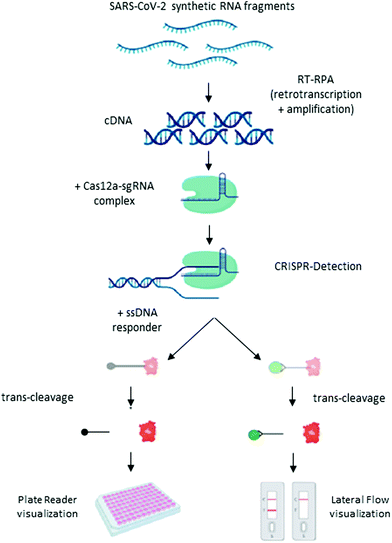 | ||
| Scheme 4 Schematic illustration of the CRISPR-Cas12 workflow. General scheme of the CRISPR detection procedure visualized by a fluorescent reader or lateral flow strip. Reproduced from ref. 107 with permission from bioRivx, copyright 2020. | ||
Several protocols have been developed based on Cas13a and isothermal amplification. The strategy named SHERLOCK,104–106 developed two years ago, has already been adapted by different groups for the diagnosis of COVID-19.109,110 The SHERLOCK COVID-19 test targets ORF1ab and glycoprotein S genes. It shows high sensitivity, reaching a LoD (of synthetic COVID-19 RNA sequence) in the range between 10–100 copies per μL in lateral flow assays.109 In the same line, the group of Hou et al. developed a similar assay to recognize the OFR1ab region, but used a fluorescence-based label for detection, achieving a LoD that was ten times lower.110 Rauch et al. developed another protocol based on Cas13a, named CREST, for the amplification and detection of protein N genes from SARS-CoV-2. This test can be utilized either for qualitative determinations associated with a lateral flow assay or for quantitative determinations based on fluorescence measurements, with similar LoD to the abovementioned CRISPR assays.111
1.4 Genomic hybridization
The ability of nucleic acids to recognize and hybridize with specific sequences can be utilized for direct genomic detection. Different strategies have been developed to generate a detectable signal once hybridization occurs. These tests are usually easier and can be performed faster, as they do not require previous amplification steps. However, the lack of an amplification strategy significantly decreases the sensitivity of the method.
Moitra et al. designed a AuNP colourimetric bioassay for the naked-eye detection of specific genetic sequences of SARS-CoV-2.112 In recent years, colourimetric biosensing based on AuNPs has garnered great attention due to their well-known optical properties: high extinction coefficient due to the presence of a localized surface plasmon resonance (LSPR) and photostability.113,114 In this work, authors functionalized AuNPs with thiol-terminated antisense-oligonucleotides (ASOs) complementary to different regions of the nucleoprotein gene. When ASO-capped AuNPs find their target sequence, stable aggregates form, changing their LSPR and colour (Scheme 5). The advantage of this test relies on the simplicity of naked-eye detection. To improve detection, a treatment with RNaseH at 65 °C for 5 min after incubation with the RNA extracted sample is utilized, generating a visual precipitate from the dispersion reaching a LoD of 0.18 ng μL−1.
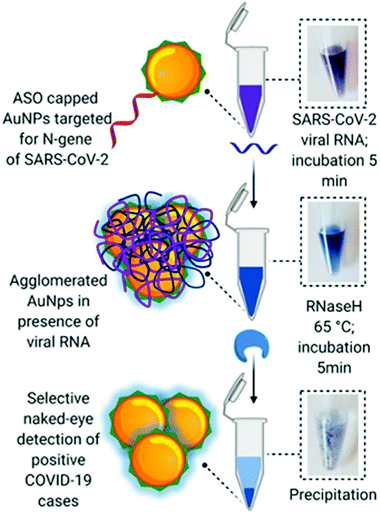 | ||
| Scheme 5 Schematic representation of a hybridization assay. Selective naked-eye detection of SARS-CoV-2 RNA mediated by ASO-capped AuNPs. Reproduced from ref. 112 with permission from ACS, copyright 2020. | ||
A similar approach, based on the LSPR of gold nanoislands (AuNIs), was developed by Qiu et al. In this case, the sample is located over a glass surface functionalized with ASO labelled AuNIs. The plasmonic chip is measured in an interferometric LSPR phase sensing system operated in attenuated total reflection mode at the interface between the glass substrate and liquid environment. When ASO-AuNIs detect the RdRp sequence of SARS-CoV-2, the local refractive index changes according to the binding events. To improve specificity, thermoplasmonic heat is used to increase the temperature of hybridization, facilitating the discrimination of similar sequences based on the principle proposed by Stehr et al.56 The LoD is 0.22 pM in a multigene mixture.115
Genomic microarrays have been developed in the past for the detection and differentiation of circulating coronaviruses.116–118 The principle relies on the reverse transcription of RNA and the posterior labelling of newly synthesized cDNA. Different surfaces in the array are functionalized with complementary sequences to bind and retain the labelled cDNA, thus indicating the presence of the virus nucleic acid. To the best of our knowledge, this strategy has not been developed yet for the recognition of SARS-CoV-2, probably due to its high cost. However, the detection of different emergent strains of the virus may become necessary if the pandemic continues spreading so fast, and microarrays are very efficient to this end.78
In section 2, we will assess protein microarrays developed for serologic determinations of COVID-19.
1.5 Antigen detection
Direct antigen detection refers to the specific immunorecognition and binding of a target molecule by antibody–antigen reaction. Assays are usually simple and take a few minutes. However, the main limitation of these techniques is the low sensitivity they present as a consequence of the absence of an amplification step.
Different methods are being developed to improve the direct antigen detection based on different technologies.
Electrochemical biosensors
These devices provide specific analytical information using a bioreceptor in direct spatial contact with an electrochemical transduction element. Seo et al. developed a field-effect transistor-based biosensor (FET) that utilizes the high conductivity of graphene and the specificity of anti-SARS-CoV-2 spike antibodies to detect the presence of the virus, recognising and binding the spike protein in NPS samples from COVID-19 patients.119 It is a very simple POC test, as it does not require previous sample preparation. According to the authors, the sensor can detect the S protein without the need of any special treatment to separate it from the viral capsid. The assay has shown high sensitivity (242 copies per mL) compared to other antigen immunodetection tests (Scheme 6).120,121
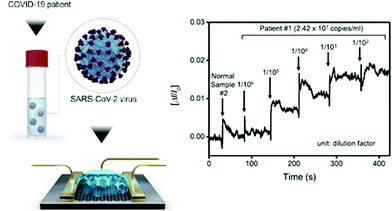 | ||
| Scheme 6 COVID-19 FET sensor operation procedure. Left: Schematic illustration of the device, in which SARS-CoV-2 spike antibodies are conjugated to graphene via a probe linker. Right: Real-time response of COVID-19 FET toward SARS-CoV-2 clinical sample. Reproduced from ref. 119 with permission from ACS, copyright 2020. | ||
Lateral flow immuno-assays (LFIA)
These are chromatographic paper POC tests designed for the direct determination of a target molecule by its recognition with labelled antibodies. The result can be recognized by the presence of one or more coloured lines in a paper strip (more details in section 2). These rapid POC tests have been adapted for the direct detection of nucleocapsid antigens present in respiratory samples using reporter antibodies labelled with fluorophores122 or AuNPs.123 The performance of these tests is around 56–68% relative to RT-qPCR determination in individuals with high viral loads, corresponding to symptomatic and hospitalized COVID-19 patients. However, when utilized in populations including asymptomatic individuals with low viral loads, the percentage of detection significantly decreases.121
As previously mentioned, genomic hybridization and antigenic detection assays have shown low performance compared to nucleic acid detection assays due to the lack of amplification. Other strategies, such as surface enhanced Raman spectroscopy (SERS) based systems and microfluidic chips that could increase sensitivity, are also considered for development and approvement.78,124,125 Yeh et al. developed a multivirus capture component with optical detection by SERS that has been validated for avian influenza A virus, and could be adapted to detect SARS-CoV-2.126 The implementation of electrical and optical biosensors, mainly oriented to protein recognition, to the detection of nucleic acids for sensitive POC tests is currently a hot topic of discussion.125,127–129
2. Serology determinations
Seroconversion is the appearance of antibodies against a pathogen, in our case SARS-CoV-2. Most serological tests search for antibodies against nucleoprotein N and glycoprotein S because of their high immunological reactivity as B-cell epitopes.130,131A variety of immunoassays have been developed for serology determinations, which show a technical advantage over molecular determinations: antigens and antibodies, present in samples and in detection kits, are much more stable than RNA (Scheme 7a).132 Another advantage is the longer period of time seroconversion offers to the indirect detection of the virus. In this section, we have mainly focused on enzyme-linked immuno-sorbent assay (ELISA) and the related techniques, LFIA and protein microarrays, as they are most adequate for first line screening due to their simplicity and high-throughput. Table 2 presents a list of different serological assays. The FIND webpage provides updates on the available SARS-CoV-2 tests.133
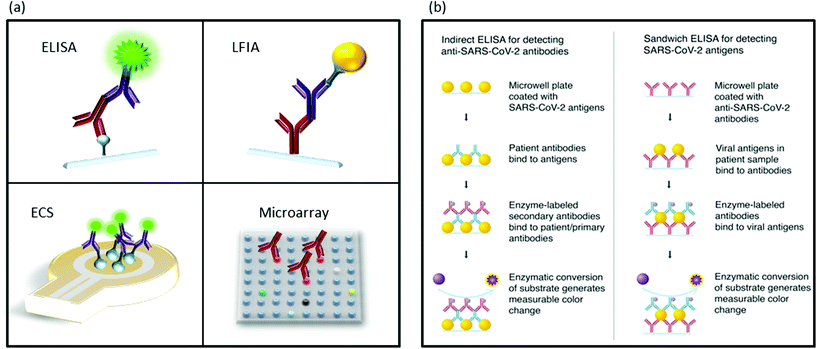 | ||
| Scheme 7 Schematic illustration of immuodetection assays. (a) Different methods for protein detection. ELISA, LFIA, ECS: electrochemical sensing and microarrays. (b) Indirect ELISA for antibody detection and sandwich ELISA for antigen detection assays. Scheme (b) is reproduced from ref. 78 with permission from ACS, copyright 2020. | ||
| Method | Antigen | Antibody | Signalling method | Ref. |
|---|---|---|---|---|
| Microarray | 67 antigens | Multitarget IgG | Fluorescence | 134 |
| Magnetic chemiluminescence enzyme immunoassay (MCLIA) | S and N proteins | IgM and IgG | Chemiluminescence | 135 and 136 |
| ELISA Euroimmun | S1 domain | IgA and IgG | Absorbance | 136–138 |
| ELISA, COVID-AR | Nucleoprotein N | IgM and IgG | Absorbance (HRP) | 139 |
| LFIA, Avioq | Not reported | IgM and IgG | Naked-eye with AuNPs | 136 |
| ELISA | Protein S | IgM and IgG | Absorbance (HRP) | 140 |
| LFIA | RBD | IgM and IgG | Naked-eye with AuNPs | 141 |
| CLIA Abbot | Nucleoprotein N | IgG | Chemiluminescence | 138 and 142 |
| Time-resolved fluorescence Immunochromatography assay | Not reported | IgM and IgG | Fluorescence | 143 |
| ELISA | RBD and trimeric protein S | IgM and IgG | Absorbance (HRP) | 144 and 145 |
| Double Sandwich ELISA | RDB | Total and IgM | Absorbance (HRP) | 146 |
| Indirect ELISA | Nucleoprotein N | IgG | ||
| Indirect ELISA | Nucleoprotein N | IgA, IgM and IgG | Absorbance | 13 |
| MCLIA | Nucleoprotein N and protein S | IgM and IgG | Chemiluminescence | 23 |
| MCLIA | ORF1ab, S, and N | IgM and IgG | Chemiluminescence | 147 |
| LFIA, NG-Test® | Not reported | IgM and IgG | Naked-eye with AuNPs | 138 |
| ELISA | RBD | Total antibody | Absorbance (HRP) | 137 |
| Indirect ELISA | S1, S, RBD and N | IgA and IgG | Absorbance (HRP) | 148 and 149 |
| Microarray | S1 | Betacoronavirus | Fluorescence | |
| ELISA | Trimeric protein S | IgM and IgG | Absorbance | 150 |
| MCLIA | RBD and N | IgA, IgM and IgG | Chemiluminescence | 151 |
| LFIA | Nucleoprotein N | IgG | Naked-eye with lanthanide-doped polystyrene nanoparticles | 70 |
| ELISA N | Nucleoprotein N | IgG | Absorbance (HRP) | 152 |
| ELISA tri-S | Trimeric protein S | IgA, IgM and IgG | Absorbance (HRP) | |
| LFIA, FarmaCoV | Nucleoprotein N | IgM and IgG | Naked-eye with AuNPs | 153 |
Viral genome sequencing, virus-culture and antibody-neutralization assays are paramount important studies to evaluate new SARS-CoV-2 mutations and to determine the capacity of antibody protection against the disease.148,154–157 Nevertheless, they are not strictly diagnostic tests, but more complex studies, and are out of the scope of this review.
2.1 Enzyme-linked immunosorbent assay
ELISA includes a family of assays, in which antibody–antigen recognition reactions take place. Since its development in the early 1970s, many different strategies involving primary and secondary antibodies labelled with fluorophores, enzymes that produce colourimetric reactions, and nanomaterials have been implemented with great success.158,159 Currently, most tests for anti-SARS-CoV-2 antibody detection follow the indirect strategy in which a surface is coated with viral proteins to be recognized by specific antibodies. After incubation with the sample, the plate is rinsed and filled with labelled anti-human antibodies that will bind to the anti-SARS-CoV-2 antibodies retained by viral antigens (Scheme 7b).78 Following this strategy, Amanat et al. utilized two different recombinant versions of the spike protein S: the full-length trimeric spike protein and the much smaller RBD.144,160 Banked serum samples (previous to COVID-19 pandemic) did not show cross-reactivity, while all patients positive for COVID-19 showed seroconversion with higher reactivity against the complete trimeric S protein compared to the RBD fragment. This was probably due to the major number of epitope regions.145,161 Bound antibodies were revealed using an anti-human-IgG horseradish peroxidase (HRP) conjugated secondary antibody. With similar strategies, other serologic assays developed recombinant N protein, antihuman IgM, and IgG conjugated to HRP or antibody-functionalized AuNPs.162–164Another strategy, named double-antibody sandwich magnetic chemiluminescence enzyme immunoassays (MCLIA), has been also adapted for the detection of COVID-19. This strategy utilizes magnetic beads functionalized with the selected antigen. When antibodies are exposed to the antigens, they will bind the beads via antibody–antigen reaction. These beads can be easily collected and washed for detection.
Long et al. used a recombinant protein containing the nucleocapsid N and a peptide from spike protein S.23 The recombinant proteins were conjugated with fluorescein isothiocyanate (FITC), and immobilized on anti-FITC antibody-conjugated magnetic particles to detect IgM and IgG. In a similar study, Cai et al. utilized synthetic antigens from ORF1ab, S, and N proteins conjugated to bovine serum albumin and a biotin–streptavidin system to measure IgM and IgG with MCLIA.147 In both cases, the authors showed cases of serology detection of COVID-19 in infected asymptomatic patients with negative RT-qPCR, highlighting the importance of serologic testing.
Most ELISAs utilize proteins N or S, alone or in combination with antigens from the ORF1ab region. In general, all assays have shown comparable sensitivities. However, differences regarding the specificity have been found. Fig. 1 shows the detection of cross-reactivity in samples of patients with COVID-19.23,134,156,165,166 Moreover, in this work, Okba et al. developed an assay using nucleoprotein N from SARS-CoV, which detected anti-SARS-CoV-2 antibodies.148 Tian et al. compared the performance of SARS-CoV-2 RBD, S1, or S trimer antigens in ELISA determinations.167 The authors found that the S trimer cross-reacted at low levels with antibodies elicited by circulating HCoV-OC43 and HCoV-HKU1 Betacoronavirus, probably due to the higher homology of the S2 portion of the trimer compared to the S1 portion alone. In the same study, the authors found higher sensitivity for S1 and the S trimer compared to RBD. This result can be expected since S1 and the S trimer contain other epitopes besides RBD.
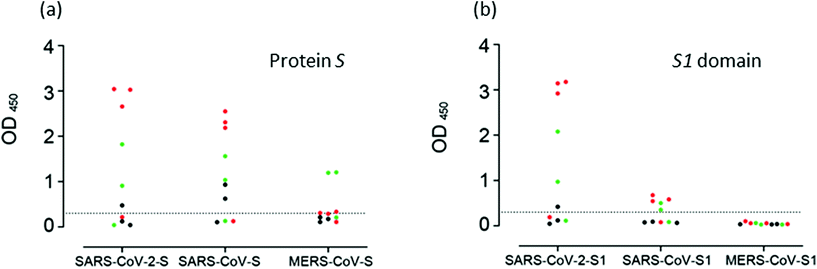 | ||
| Fig. 1 Cross-reactivity analysis of serum samples. Reaction of samples from patients with COVID-19 to (a) S proteins, and to (b) the S1 domain of protein S of SASR-CoV-2, SARS-CoV and MERS-CoV measured by ELISA. Reproduced from ref. 148 with permission from CDC, copyright 2020. | ||
2.2 Lateral flow immuno-assay
This technique follows the same principles as ELISA, adapted to a chromatographic paper strip for direct visualization.168 When the liquid sample is added at one end of the pad, it runs by capillary forces along a functionalized surface with reactive molecules. Typically, one line contains standard recognition molecules to control the quality of the test. In a second line, the specific reactive molecules recognize the target, indicating a positive result.169 New strategies for the simultaneous recognition of different targets are based on the development of multiplexed LFIA with several recognition lines.170 LFIAs are adequate for POC qualitative tests and inexpensive, as they require neither trained personnel nor any set up. Scheme 8 depicts a LFIA for the detection of anti-SARS-CoV-2 antibodies developed by Li Zhengtu et al. The test uses a recombinant protein from the RBD of protein spike to bind specific IgM and IgG. Trapped antibodies become visible with AuNPs-functionalized secondary antibody markers.141 The test can be utilized with blood samples from a finger prick and serum/plasma from venous blood with the same performance. The clinical detection sensitivity and specificity of this test was studied in more than 500 patients. The sensitivity was 88.66% and the specificity was 90.63% relative to RT-PCR determinations. However, in a recent study carried out by the National COVID Testing Scientific Advisory Panel from UK, the sensitivity and specificity of LFIA vs. ELISA were compared by means of RT-qPCR diagnosed samples as positive/negative for SARS-CoV-2.150 The authors used the trimeric spike protein for ELISA determinations. Antibodies binding to the S protein were detected with alkaline phosphatase-conjugated anti-human IgG or anti-human IgM. LFIA devices were designed to detect IgM, IgG or total antibodies to SARS-CoV-2 by nine manufacturers. The performances of LFIA and ELISA assays relative to RT-qPCR determinations were such that the sensitivity ranged from 65% to 85%, and the specificity ranged from 93% to 100%, respectively. Of the 50 designated negative samples tested by both ELISA and the nine different LFIA devices, 7 LFIA showed at least one false/positive result. Similar results have been presented in another comparative study between the ELISA and LFIA performances.171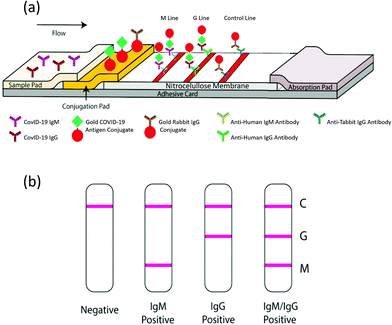 | ||
| Scheme 8 Schematic illustration of the rapid SARS-CoV-2 antibody test. (a) Detection device and (b) testing results. C: Control; G: IgG and M: IgM lines. Reproduced from ref. 141 with permission from Wiley, copyright 2020. | ||
Interestingly, several studies reported that the highest overall sensitivity from serological tests was obtained from total antibody determination protocols.141,143
2.3 Protein microarrays
This platform consists of a solid surface functionalized with selected proteins that provides support for the interaction between the immobilized and sample proteins. These chips enable the study of hundreds and even thousands biochemical properties of different proteins in parallel with high-throughput.172 Okba et al. developed a microarray for the study of antibodies against the betacoronavirus proteins N, S1 domain and S.149 Early this year, the authors adapted their assay to include SARS-CoV-2 in the analysis.148 Their study results are extremely important for avoiding false-positive results, as most people have been in contact with 4 endemic human coronaviruses. In a similar study, Khan et al. developed a protein microarray to analyse antibody response and cross-reactivity to SARS-CoV-2, SARS-CoV, MERS-CoV, the common human coronavirus, and other common respiratory viruses.134 Both groups identified the cross-reactivity of antibodies against nucleocapsid protein and glycoprotein spike (particularly at the S2 domain).134,1483. Discussion
3.1 Humoral response
Previous experiences with the SARS-CoV and MERS-CoV epidemics permitted a rapid response from the medical and scientific community to this outbreak.173,174 The SARS-CoV175 infection had shown that high antibody responses correlate with longer and more severe diseases. Fig. 2 depicts the levels of IgA, IgM and IgG in patients with mild, moderate and severe COVID-19. As can be seen, and most studies indicate, SARS-CoV-2 generates a similar humoral response compared to SARS-CoV, in which higher levels of antibodies are indicative of a more severe health condition.23,146,148,151 Two possible mechanisms have been proposed to explain the association of antibody levels and worse clinical outcome: a direct mechanism by antibody-dependent enhancement (ADE) of infection, and an indirect one via promotion of the cytokine storm.176 There are no clear indications of ADE disease at the moment, and passive immunization from convalescent patient plasma appears effective to treat COVID-19.177,178 Most studies point toward an uncontrolled inflammatory and impaired adaptive immune response as being responsible for local and systemic tissue damage.176,178 In order to prevent pneumonia and other clinical complications, the control of cytokine production and immune response is mandatory. However, this strategy is challenging as it can lead to an insufficient immune response, resulting in severe damage to patients.179 In this regard, interleukine-6 (IL-6) is a clear example of this complex regulation, as it stimulates both an inflammatory response and B-cell differentiation for antibody production.178 Furthermore, newborns from SARS-CoV-2-infected mothers have shown high levels of IL-6.178 Blanco-Melo et al. studied the host response to SARS-CoV-2 by analysing the transcriptional response and comparing it with other respiratory virus infections. It was observed that the transcriptional signature leading to COVID-19 is characterized by a fail in type I and type III interferon response, accompanied by high levels of chemokines for cell recruitment.180,181 Neufeldt et al. also studied the cytokine activation routes and arrived at the conclusion that an imbalance between immunoactivation and immunosuppression in host defenses seems to be the origin of the highly pathogenic coronavirus infection, and is one of the main targets to suppress severe disease symptoms.182 In a recent study, De Biasi et al. have proved that immune-regulation mediated by T cell exhaustion (a down regulation of immune response cause by the chronic stimulation generated by cancer or chronic viral infections) and IL-10 have a key role in breaking the excessive inflammation and helping to recover the homeostasis of the lungs.183,184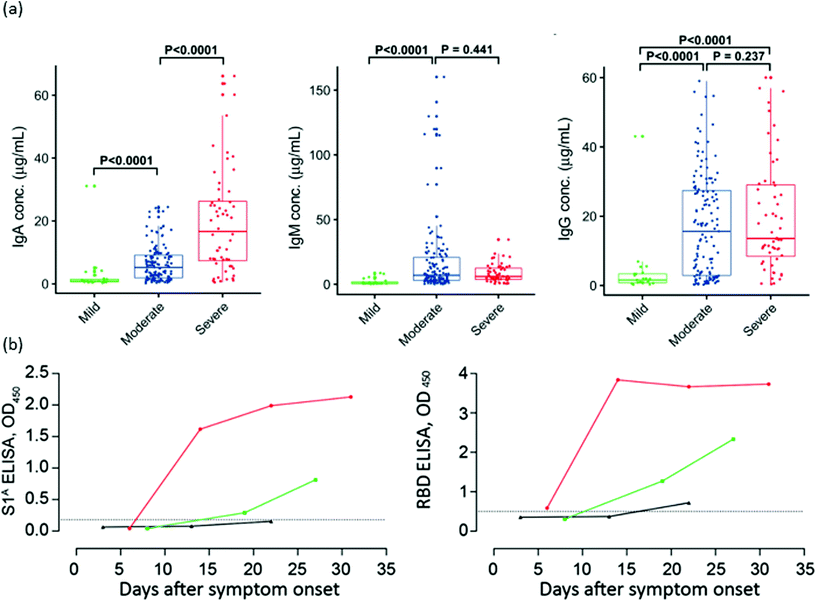 | ||
| Fig. 2 Antibody responses against SARS-CoV-2 after infection. (a) Serum levels of IgA, IgM and IgG anti-RBD in three distinct severity groups of COVID-19 patients. (b) Kinetics of IgG anti-S1A domain and anti-RBD in one severe (red) and two mild (green and black) COVID-19 patients. Reproduced from ref. 151 with permission from Wiley, copyright 2020 and ref. 148 with permission from CDC, copyright 2020. | ||
A higher proportion of IgG anti-nucleocapsid protein related to the anti-spike protein185 and elevated values of IL-6, IL-8, and TNF-α cytokines have been proposed as predictors of severity.186 However, other research studies disregard the possibility of the major role of cytokines in driving the severity of the disease.187 At present, we are still unable to differentiate severe viral infections from immune-enhanced diseases, either by clinical findings, immunological assays or biomarkers.188 Therefore, it is of utmost importance to identify the specific features and biomarkers that occur during the immune response to coronavirus infection.
A complementary strategy relies on the research of COVID-19 in rhesus macaques.189 These monkeys develop a similar disease as humans under exposure to SARS-CoV-2.190 Previously infected animals re-challenged with SARS-CoV-2 showed a protective neutralizing antibody response against re-exposure with 5 log reductions in median viral loads.191,192 In a recent study by Yu et al., a DNA vaccine encoding the full-length protein S has been tested. Following vaccination, animals were challenged with SARS-CoV-2, resulting in >3 log reduction in viral loads. Authors found titers of vaccine-triggered neutralizing antibodies correlated with protective efficacy. These data demonstrate antibody protection against SARS-CoV-2 in nonhuman primates.193
3.2 Interpretation of results
As previously mentioned, the real-time RT-PCR of swab samples collected from the upper respiratory tract is considered the gold standard laboratory diagnosis test of SARS-CoV-2 infection.20,194 Based on the reduced invasiveness and no need for swabs or trained personnel, prospective studies to evaluate the performance of saliva and sputum samples have been carried out. Results indicate that saliva samples have inferior sensitivity compared to NPS collection, and that the obtaining of sputum is rather low, as COVID-19 is characterized by dry cough.195–197 However, even in NPS samples, changes in the viral load during the course of COVID-19 are the main limitation of direct detection tests.17,198In an early analysis in China, Li Guo et al. demonstrated that the RT-qPCR positive rate was more than 90% on days 1–3 post-symptom onset (PSO), and then declined to less than 80% at day 6, and less than 50% after 14 d PSO.13 While positive results represent the confirmation of COVID-19, as cross-reactivity is negligible in the current available RT-qPCR tests, there are several possible interpretations for a negative result. Sample collection out of the time window for viral replication, when the viral load is insufficient, will also produce a false negative result.174 SARS-CoV-2 loads in upper respiratory tract have been shown to vary considerably, and negative determinations can be the result of a test that was taken too early, which would turn positive later, or a test taken too late in the course of the infection.14,199,200 In a recent review, authors analysed possible factors affecting nucleic acid test performance, suggesting the following strategies to prevent false-negative results: complementation with stool and blood samples when testing at late periods of illness; protecting RNA by adding guanidine salt to collected samples; setting proper positive, negative and inhibition controls to ensure high-quality results; simultaneous amplification of human RNase P gene. However, RT-qPCR negative results must be cautiously interpreted, especially in the case of close contact epidemiology or clinical feature suspicion for COVID-19.48
Seroconversion also depends on the moment of sample collection. It is widely accepted during the first 3 weeks. PSO that nearly 100% of patients develop anti-SARS-CoV-2 IgG, with a median of appearance at around day 14.13,24 The kinetics of IgM have shown more variability, appearing earlier,13,201 simultaneously, and later24 than IgG, or not being detectable at all.174 Unfortunately, IgA kinetics have not been widely studied (Fig. 3).23,151 Moreover, a recent study shows that between 2–8.5% of patients do not produce antibodies against the virus.202 It is also known that the performance of rapid tests can vary when utilized in routine testing laboratories. In UK, after an evaluation of serological tests, authorities concluded that many commercial antibody tests were not adequate for clinical use.150However, the existence of a detection window, in which an infected patient shows a negative result, has to be properly taken into account. This period corresponds to the time required for the immune system to produce the antibodies, which, is still under discussion in the case of COVID-19.36 In the same line of discussion, Deeks et al. reported the design of studies on the accuracy of COVID-19 tests, and the interpretation of the results still requires considerable improvement since most studies lack reliable sensitivity reports grouped by time PSO.37
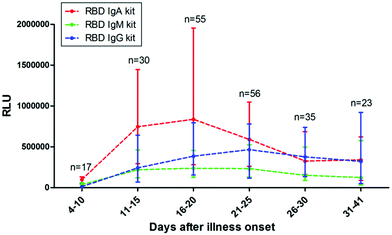 | ||
| Fig. 3 Antibody responses against SARS-CoV-2. The kinetics of anti-RBD IgA, IgM, and IgG levels in sera of COVID-19 patients at different time POS. RLU: relative light units. Reproduced from ref. 151 with permission from Wiley, copyright 2020. | ||
Most serology tests utilize proteins N and S because of their strong immunogenicity, with similar performances in terms of their sensitivity. Nevertheless, the comparison between the trimeric protein S, S1 subunit and RBD shows a decreasing reactivity, respectively.144,167 This observation is expected, as the number of epitopes decreases with the restriction of the antigen size. These results suggest that trimeric protein S should be the first option for serology determinations. However, different studies have shown cross-reactivity against protein S from other human betacoronaviruses.144,148,167 Antibodies against MERS-CoV and the winter cold viruses (HCoV-OC43 and HCoV-HKU1) react to protein S from SARS-CoV-2, while they do not recognise subunit 1 (S1) and RBD. This can be interpreted in terms of the high degree of conservation in the coronavirus of the S2 subunit relative to S1.148 The appearance of false-positive results can induce a misleading result of the predictive prevalence, and consequently to mistakes in the determination of public policies.22 This is especially important for the case of the endemic human coronaviruses, as most people present antibodies against them. On the other hand, cross-reactivity between antibodies against SARS-CoV and SARS-CoV-2 has been detected for proteins N, S and domains S1 and RBD. This finding is expected due to the similarity between these two virus.134,148 Nevertheless, as SARS-CoV has not been circulating among humans since 2003, the possibility of false-positive results seems negligible.
The delay in the antibody response and the uncertainty in the correlation between seropositivity and immune protection78 has called into question the usefulness of serology tests in acute SARS-CoV-2 infections. Some authors only recommend these tests for serological survey purposes.203 Wu et al. performed a study, in which 10% of asymptomatic individuals showed anti-SARSCoV-2 IgG in serum samples.204 The hospital staff from an eastern institution in France, diagnosed with COVID-19 by RT-qPCR, was tested for anti-SARS-CoV-2 antibodies using a commercial LFIA and a flow cytometry assay for comparison. Only 2 out of 162 participants had required hospitalization. The rapid test was positive for IgG/IgM determination in 153 (95.6%) samples, and the flow cytometry assay detected antibodies in 159 (99.4%). This finding supports the use of rapid serologic tests for the diagnosis of individuals recovered from SARS-CoV-2 infection.152 Moreover, different studies have shown that serology plays a key role in combination with molecular testing to improve sensitivity in cases presenting one more week PSO.23,24,146,150 Serology can be also applied to the prediction of severe cases, either by the quantification of antibodies or by the estimation of anti-N/anti-S IgG ratio.185 The Infectious Diseases Society of America (IDSA) Guidelines on the Diagnosis of COVID-19 identifies three potential indications for serologic testing, including: (1) evaluation of patients with a high clinical suspicion for COVID-19 when molecular diagnostic testing is negative, and at least 2 weeks have passed since symptom onset; (2) assessment of multisystem inflammatory syndrome in children; and (3) for conducting serosurveillance studies.205
Direct and indirect tests determine different aspects of the infection and also serve overlapping purposes, increasing the general sensitivity of detection.206 For these reasons, and especially for low and middle income countries (LMICs), aggressive testing is of paramount importance in pandemics such as COVID-19.17,207 Another strategy, especially useful for developing countries, is the pooling approach, which enables the simultaneous testing of dozens of samples.208–210 In order to facilitate access to testings in LMICs, the FIND developed a database of all COVID-19 diagnostic assays currently available or in development. It works by carrying out independent evaluations of some of these tests to assist in procurement decisions, ensuring that the assays meet WHO quality standards.211
The test positivity rate, i.e., number of tested individuals per positive result, varies enormously among countries, periods of time, and regions.212 Such differences disable any comparative study, hindering the development of prevention polices. Etchenique et al. proposed an alternative method for prevalence estimation that is especially useful for LMICs. The estimation relies on the number of deaths caused by COVID-19, which is much easier to collect, and the known proportion to infected cases is 1 death for every 200 infected people. This coefficient corresponds to the infection-fatality ratio (IFR), and has been estimated from large seroprevalence studies distributed by age and district.213,214 This strategy offers a reliable tool to corroborate the number of infected cases and to correct them if necessary, based on the IFR discriminated by age.215
The abovementioned study confirms the importance of developing active surveillance plans of complementary diagnostic approaches. We emphasise the advantages of organising diagnostic policies in terms of a triad of active search and tracing. First, a highly sensitive and specific molecular test is needed to detect the presence of the virus in clinically suspected cases. Secondly, rapid POC tests should be used to triage suspected cases within minutes. Several authors have confirmed the high sensitivity and simplicity advantages of their POC tests.216 Third, serological tests are needed to comprehend the natural history of infection, the secondary attack rates, and the correlation with possible unknown future sequelae. Determination of seroconversion allows for the identification of suspected cases with the negative results of direct detection tests, and are paramount to evaluate the quality and durability of immunogenicity produced with vaccination.24,36 The fact that many serologic assays showed no cross-reactivity demonstrates that humans are serologically naive to SARS-CoV-2, and explains the relatively high basic reproduction number (R0) of SARS-CoV-2 compared to that of other respiratory viruses, such as influenza virus.2,144 For this reason, antibody detection can be used as a sensitive marker for sentinel monitoring of imported cases in un-exposed communities.146,174 Many countries have started to emphasise the imperative to collect seroprevalence data.207,217
Finally, the unprecedented sample processing need has shown the value of innovative approaches and non-traditional developers, such as biofoundries, academic labs, start-ups, and small and medium enterprises to expand testing capacity and to offer new options.216,218
4. Conclusions and perspectives
Quantitative, real time RT-PCR is the gold standard and reference method for the direct determination of SARS-CoV-2 infection. Due to its requirements and the overwhelming situation produced by the COVID-19 pandemic, POC tests are very important. Isothermal amplification methods are a promising low-cost alternative to complement or even replace traditional PCR testing.After the 11th day PSO, serological assays may be more sensitive for the diagnosis of COVID-19 and should be performed, regardless of the RT-qPCR test results. Moreover, serology analysis results are also useful as a predictor of disease outcome. However, further studies, especially in asymptomatic patients, are necessary to better understand the humoral response to SARS-CoV-2 infection. The discussion around the protective or harmful role of the immune response remains open. A fundamental understanding of the role of acquired immunity to COVID-19 should be a priority to handle the 2nd wave and beyond strategies.
We have also learned from these times of the global pandemic that innovative approaches and non-traditional developers offered invaluable solutions to tackle the situation. Nevertheless, it is important to keep in mind that no laboratory tests, whether RT-qPCR or serology, can substitute clinical observation and practical experience. If there is a clinical suspicion for COVID-19, a negative response from tests cannot exclude the presence of the disease.
Research and development to enhance the sensitivity of the tests will have long-term benefits not just for COVID-19, but also for any emergent infectious diseases.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thankfully acknowledge Monica Florín-Christensen, Jochen Feldmann and Federico Bürsgens for helpful discussions. N. T. K. T. thanks EPSRC, UK (EP/M015157/1) and UCL Global Challenges Research Fund (GCRF) for financial support. M. A. H. thanks CONICET for travel assistance and ANPCyT (PICT 2016-0679).References
- P. Zhou, X. L. Yang, X. G. Wang, B. Hu, L. Zhang, W. Zhang, H. R. Si, Y. Zhu, B. Li, C. L. Huang, H. D. Chen, J. Chen, Y. Luo, H. Guo, R. Di Jiang, M. Q. Liu, Y. Chen, X. R. Shen, X. Wang, X. S. Zheng, K. Zhao, Q. J. Chen, F. Deng, L. L. Liu, B. Yan, F. X. Zhan, Y. Y. Wang, G. F. Xiao and Z. L. Shi, Nature, 2020, 579, 270–273 CrossRef CAS.
- Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K. S. M. Leung, E. H. Y. Lau, J. Y. Wong, X. Xing, N. Xiang, Y. Wu, C. Li, Q. Chen, D. Li, T. Liu, J. Zhao, M. Liu, W. Tu, C. Chen, L. Jin, R. Yang, Q. Wang, S. Zhou, R. Wang, H. Liu, Y. Luo, Y. Liu, G. Shao, H. Li, Z. Tao, Y. Yang, Z. Deng, B. Liu, Z. Ma, Y. Zhang, G. Shi, T. T. Y. Lam, J. T. Wu, G. F. Gao, B. J. Cowling, B. Yang, G. M. Leung and Z. Feng, N. Engl. J. Med., 2020, 382, 1199–1207 CrossRef CAS.
- N. H. L. Leung, D. K. W. Chu, E. Y. C. Shiu, K.-H. Chan, J. J. McDevitt, B. J. P. Hau, H.-L. Yen, Y. Li, D. K. M. Ip, J. S. M. Peiris, W.-H. Seto, G. M. Leung, D. K. Milton and B. J. Cowling, Nat. Med., 2020, 26, 676–680 CrossRef CAS.
- COVID-19 CORONAVIRUS PANDEMIC, https://www.worldometers.info/coronavirus/, (accessed November 2020).
- L. Alanagreh, F. Alzoughool and M. Atoum, Pathogens, 2020, 9, 1–12 CrossRef.
- I. Hamming, W. Timens, M. L. C. Bulthuis, A. T. Lely, G. J. Navis and H. van Goor, J. Pathol., 2004, 203, 631–637 CrossRef CAS.
- M. Letko, A. Marzi and V. Munster, Nat. Microbiol., 2020, 5, 562–569 CrossRef CAS.
- T. Tang, M. Bidon, J. A. Jaimes, G. R. Whittaker and S. Daniel, Antiviral Res., 2020, 178, 104792 CrossRef CAS.
- J. Shang, Y. Wan, C. Luo, G. Ye, Q. Geng, A. Auerbach and F. Li, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 202003138 Search PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280 CrossRef CAS.
- E. J. Snijder, R. W. A. L. Limpens, A. H. de Wilde, A. W. M. de Jong, J. C. Zevenhoven-Dobbe, H. J. Maier, F. F. G. A. Faas, A. J. Koster and M. Bárcena, PLoS Biol., 2020, 18, 1–25 CrossRef.
- R. Li, S. Pei, B. Chen, Y. Song, T. Zhang, W. Yang and J. Shaman, Science, 2020, 493, 489–493 CrossRef.
- L. Guo, L. Ren, S. Yang, M. Xiao, D. Chang, F. Yang, C. S. Dela Cruz, Y. Wang, C. Wu, Y. Xiao, L. Zhang, L. Han, S. Dang, Y. Xu, Q. Yang, S. Xu, H. Zhu, Y. Xu, Q. Jin, L. Sharma, L. Wang and J. Wang, Clin. Infect. Dis., 2020, 71, 778–785 CrossRef CAS.
- T. Ai, Z. Yang and L. Xia, Radiology, 2020, 1–8 Search PubMed.
- Y. Yang, M. Yang, J. Yuan, F. Wang, Z. Wang, J. Li, M. Zhang, L. Xing, J. Wei, L. Peng, G. Wong, H. Zheng, W. Wu, C. Shen, M. Liao, K. Feng, J. Li, Q. Yang, J. Zhao, L. Liu and Y. Liu, The Innovation, 2020, 1(3), 100061 CrossRef.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS.
- R. Weissleder, H. Lee, J. Ko and M. J. Pittet, Sci. Transl. Med., 2020, 12, 1–6 Search PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. W. Chu, T. Bleicker, S. Brünink, J. Schneider, M. L. Schmidt, D. G. J. C. Mulders, B. L. Haagmans, B. Van Der Veer, S. Van Den Brink, L. Wijsman, G. Goderski, J. L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. G. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 1–8 Search PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS.
- World Health Organization, Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases, 2020, https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 Search PubMed.
- R. S. Srikrishna, D. Dhillon and D. Beier, We Need a Cheap Way to Diagnose Coronavirus, https://hbr.org/2020/02/we-need-a-cheap-way-to-diagnose-coronavirus, (February 2020).
- G. Gronvall, N. Connel, A. Kobokovich, R. West, K. Warmbrod, M. Shearer, L. Mullen and T. Inglesby, Developing a National Strategy for Serology (Antibody Testing) in the United States, The Johns Hopkins Center for Health Security, Baltimore, 2020 Search PubMed.
- Q. X. Long, B. Z. Liu, H. J. Deng, G. C. Wu, K. Deng, Y. K. Chen, P. Liao, J. F. Qiu, Y. Lin, X. F. Cai, D. Q. Wang, Y. Hu, J. H. Ren, N. Tang, Y. Y. Xu, L. H. Yu, Z. Mo, F. Gong, X. L. Zhang, W. G. Tian, L. Hu, X. X. Zhang, J. L. Xiang, H. X. Du, H. W. Liu, C. H. Lang, X. H. Luo, S. B. Wu, X. P. Cui, Z. Zhou, M. M. Zhu, J. Wang, C. J. Xue, X. F. Li, L. Wang, Z. J. Li, K. Wang, C. C. Niu, Q. J. Yang, X. J. Tang, Y. Zhang, X. M. Liu, J. J. Li, D. C. Zhang, F. Zhang, P. Liu, J. Yuan, Q. Li, J. L. Hu, J. Chen and A. L. Huang, Nat. Med., 2020, 26, 845–848 CrossRef CAS.
- M. Lipsitch, R. Kahn and M. J. Mina, Nat. Med., 2020, 26, 818–819 CrossRef CAS.
- A. A. Farid, G. I. Selim and H. A. A. Khater, Int. J. Sci. Eng. Res., 2020, 11, 1141–1149 Search PubMed.
- C. Shen, N. Yu, S. Cai, J. Zhou, J. Sheng, K. Liu, H. Zhou, Y. Guo and G. Niu, J. Pharm. Anal., 2020, 10, 123–129 CrossRef.
- F. A. Binti Hamzah, C. H. Lau, H. Nazri, D. C. Ligot, G. Lee and C. L. Tan, et al., CoronaTracker: World-wide Covid-19 outbreak data analysis and prediction, Bull World Health Organ, 2020, DOI:10.2471/BLT.20.255695.
- A. Kumar, P. K. Gupta and A. Srivastava, Diabetes, Metab. Syndr.: Clin. Res. Rev., 2020, 14, 569–573 CrossRef.
- L. Wynants, B. Van Calster, M. M. J. Bonten, G. S. Collins, T. P. A. Debray, M. De Vos, M. C. Haller, G. Heinze, K. G. M. Moons, R. D. Riley, E. Schuit, L. J. M. Smits, K. I. E. Snell, E. W. Steyerberg, C. Wallisch and M. Van Smeden, Br. Med. J., 2020, 369, m1328 CrossRef.
- R. D. Smith, Soc. Sci. Med., 2006, 63, 3113–3123 CrossRef.
- C. S. Wood, M. R. Thomas, J. Budd, T. P. Mashamba-Thompson, K. Herbst, D. Pillay, R. W. Peeling, A. M. Johnson, R. A. McKendry and M. M. Stevens, Nature, 2019, 566, 467–474 CrossRef CAS.
- A. Malekjahani, S. Sindhwani, A. M. Syed and W. C. W. Chan, Acc. Chem. Res., 2019, 52, 2406–2414 CrossRef CAS.
- B. Gates, N. Engl. J. Med., 2020, 382, 1677–1679 CrossRef CAS.
- Z. Allam and D. S. Jones, Healthcare, 2020, 8(1), 46 CrossRef.
- A. S. R. Srinivasa Rao and J. A. Vazquez, Infect. Control Hosp. Epidemiol., 2020, 41, 826–830 CrossRef.
- A. T. Huang, B. Garcia-Carreras, M. D. T. Hitchings, Y. Bingyi, L. C. Katzelnick, S. M. Rattigan, B. A. Borgert, C. A. Moreno, B. D. Solomon, L. Trimmer-Smith, V. Etienne, I. Rodriguez-Barraquer, J. Lessler, H. Salje, D. S. Burke, A. Wesolowski and D. A. T. Cummings, Nat. Commun., 2020, 11 Search PubMed , 4704.
- J. J. Deeks, J. Dinnes, Y. Takwoingi, C. Davenport, R. Spijker, S. Taylor-Phillips, A. Adriano, S. Beese, J. Dretzke, L. Ferrante di Ruffano, I. M. Harris, M. J. Price, S. Dittrich, D. Emperador, L. Hooft, M. M. Leeflang and A. Van den Bruel, Cochrane Database Syst. Rev., 2020, 6, CD013652 Search PubMed.
- M. L. Wong and J. F. Medrano, BioTechniques, 2005, 39, 75–85 CrossRef CAS.
- H. D. VanGuilder, K. E. Vrana and W. M. Freeman, BioTechniques, 2008, 44, 619–626 CrossRef CAS.
- Z. Wan, Y. Zhang, Z. He, J. Liu, K. Lan, Y. Hu and C. Zhang, Int. J. Mol. Sci., 2016, 17, 1–11 Search PubMed.
- J. Y. Noh, S. W. Yoon, D. J. Kim, M. S. Lee, J. H. Kim, W. Na, D. Song, D. G. Jeong and H. K. Kim, Arch. Virol., 2017, 162, 1617–1623 CrossRef CAS.
- M. Shen, Y. Zhou, J. Ye, A. A. Abdullah AL-maskri, Y. Kang, S. Zeng and S. Cai, J. Pharm. Anal., 2020, 10, 97–101 CrossRef.
- J. F. W. Chan, C. C. Y. Yip, K. K. W. To, T. H. C. Tang, S. C. Y. Wong, K. H. Leung, A. Y. F. Fung, A. C. K. Ng, Z. Zou, H. W. Tsoi, G. K. Y. Choi, A. R. Tam, V. C. C. Cheng, K. H. Chan, O. T. Y. Tsan and K. Y. Yuen, J. Clin. Microbiol., 2020, 58, e00310–e00320 CrossRef CAS.
- CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. https://www.cdc.gov/csels/dls/locs/2020/published-assay-information-for-the-2019-novel-coronavirus.html.
- D. K. W. Chu, Y. Pan, S. M. S. Cheng, K. P. Y. Hui, P. Krishnan, Y. Liu, D. Y. M. Ng, C. K. C. Wan, P. Yang, Q. Wang, M. Peiris and L. L. M. Poon, Clin. Chem., 2020, 66, 549–555 CrossRef.
- K. K.-W. To, O. T.-Y. Tsang, C. C.-Y. Yip, K.-H. Chan, T.-C. Wu, J. M.-C. Chan, W.-S. Leung, T. S.-H. Chik, C. Y.-C. Choi, D. H. Kandamby, D. C. Lung, A. R. Tam, R. W.-S. Poon, A. Y.-F. Fung, I. F.-N. Hung, V. C.-C. Cheng and J. F.-W. Chan, Clin. Infect. Dis., 2020, 71, 841–843 CrossRef CAS.
- Y. J. Jung, G.-S. Park, J. H. Moon, K. Ku, S.-H. Beak, S. Kim, E. C. Park, D. Park, J.-H. Lee, C. W. Byeon, J. J. Lee, J. Maeng, S. J. Kim, S. Il Kim, B.-T. Kim, M. J. Lee and H. G. Kim, bioRxiv, 2020 DOI:10.1101/2020.02.25.964775.
- A. Tahamtan and A. Ardebili, Expert Rev. Mol. Diagn., 2020, 20, 453–454 CrossRef CAS.
- C. B. F. Vogels, A. F. Brito, A. L. Wyllie, J. R. Fauver, I. M. Ott, C. C. Kalinich, M. E. Petrone, A. Casanovas-Massana, M. C. Muenker, A. J. Moore, J. Klein, P. Lu, A. Lu-Culligan, X. Jiang, D. J. Kim, E. Kudo, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takehashi, M. Taura, M. Tokuyama, A. Venkataraman, O.-E. Weizman, P. Wong, Y. Yang, N. R. Cheemarla, E. White, S. Lapidus, R. Earnest, B. Geng, P. Vijayakumar, C. Odio, J. Fournier, S. Bermejo, S. Farhadian, C. Dela Cruz, A. Iwasaki, A. I. Ko, M.-L. Landry, E. F. Foxman and N. D. Grubaugh, medRxiv, 2020 DOI:10.1101/2020.03.30.20048108.
- A. K. Nalla, A. M. Casto, M. L. W. Huang, G. A. Perchetti, R. Sampoleo, L. Shrestha, Y. Wei, H. Zhu, K. R. Jerome and A. L. Greninger, J. Clin. Microbiol., 2020, 58, e00557–e00520 CrossRef.
- M. J. Loeffelholz and Y. W. Tang, Emerging Microbes Infect., 2020, 9, 747–756 CrossRef CAS.
- Rutgers Clinical Genomics Laboratory TaqPath SARS-CoV-2 Assay EUA. Summary, 2020, https://www.fda.gov/media/136875/download Search PubMed.
- P. B. van Kasteren, B. van der Veer, S. van den Brink, L. Wijsman, J. de Jonge, A. van den Brandt, R. Molenkamp, C. B. E. M. Reusken and A. Meijer, J. Clin. Virol., 2020, 128, 104412 CrossRef CAS.
- Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay - Centers for Disease Control and Prevention, 2020, https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html Search PubMed.
- GNA Biosolutions, 2020, https://www.gna-bio.com/solutions/.
- J. Stehr, C. Hrelescu, R. A. Sperling, G. Raschke, M. Wunderlich, A. Nichtl, D. Heindl, K. Kürzinger, W. J. Parak, T. A. Klar and S. J. Feldmann, Nano Lett., 2008, 8, 619–623 CrossRef CAS.
- C. Hrelescu, J. Stehr, M. Ringler, R. A. Sperling, W. J. Parak, T. A. Klar and J. Feldmann, J. Phys. Chem. C, 2010, 114, 7401–7411 CrossRef CAS.
- L. Ullerich, S. Campbell, F. Krieg-Schneider, F. Bürsgens and J. Stehr, J. Lab. Med., 2017, 41, 239–244 Search PubMed.
- Emergency Use Authorization (EUA) information, and list of all current EUAs, 2020, https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#2019-ncov Search PubMed.
- Coronavirus Test Tracker: Commercially Available COVID-19 Diagnostic Tests, 2020, https://www.360dx.com/coronavirus-test-tracker-launched-covid-19-tests?page=8 Search PubMed.
- Find Evaluation Update: SARS-CoV-2 Molecular Diagnostics, 2020, http://www.finddx.org/covid-19/sarscov2-eval-molecular/ Search PubMed.
- Novel Coronavirus National Science and Technology Resource Service System, 2020, http://nmdc.cn/nCov/en Search PubMed.
- V. Corman, T. Bleicker, S. Brünink, C. Drosten, O. Landt, M. Koopmans and M. Zambon, Diagnostic detection of Wuhan coronavirus 2019 by real-time RT- PCR, Berlin, Germany, 2020 Search PubMed.
- K. Shirato, N. Nao, H. Katano, I. Takayama, S. Saito, F. Kato, H. Katoh, M. Sakata, Y. Nakatsu, Y. Mori, T. Kageyama, S. Matsuyama and M. Takeda, Jpn. J. Infect. Dis., 2020, 73(4), 304–307 CrossRef CAS.
- N. Nao, K. Shirato, H. Katano, S. Matsuyama and M. Takeda, Detection of second case of 2019-nCoV infection in Japan, National Institute of Infectious Diseases, Tokyo, p. 2020 Search PubMed.
- Protocol: Real-time RT-PCR assays for the detection of SARS-CoV-2, 2020, https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2 Search PubMed.
- L. Bordi, A. Piralla, E. Lalle, F. Giardina, F. Colavita, M. Tallarita, G. Sberna, F. Novazzi, S. Meschi, C. Castilletti, A. Brisci, G. Minnucci, V. Tettamanzi, F. Baldanti and M. R. Capobianchi, J. Clin. Virol., 2020, 128, 104416 CrossRef CAS.
- B. Freire-Paspuel, P. Vega-Mariño, A. Velez, P. Castillo, M. Cruz and M. A. Garcia-Bereguiain, J. Clin. Virol., 2020, 128, 104454 CrossRef CAS.
- P. L. Bulterys, N. Garamani, B. Stevens, M. K. Sahoo, C. H. Huang, C. A. Hogan, J. Zehnder and B. A. Pinsky, J. Clin. Virol., 2020, 129, 104427 CrossRef CAS.
- Z. Chen, Z. Zhang, X. Zhai, Y. Li, L. Lin, H. Zhao, L. Bian, P. Li, L. Yu, Y. Wu and G. Lin, Anal. Chem., 2020, 92, 7226–7231 CrossRef CAS.
- C. C. Y. Yip, C. C. Ho, J. F. W. Chan, K. K. W. To, H. S. Y. Chan, S. C. Y. Wong, K. H. Leung, A. Y. F. Fung, A. C. K. Ng, Z. Zou, A. R. Tam, T. W. H. Chung, K. H. Chan, I. F. N. Hung, V. C. C. Cheng, O. T. Y. Tsang, S. K. W. Tsui and K. Y. Yuen, Int. J. Mol. Sci., 2020, 21, 2574 CrossRef CAS.
- M. Roberts, ‘Do it at home’ coronavirus saliva test trialled, 2020, https://www.bbc.com/news/health-53131237 Search PubMed.
- New Rutgers Saliva Test for Coronavirus Gets FDA Approval: Emergency Use Authorization Granted for New Biomaterial Collection Approach, 2020, https://www.rutgers.edu/news/new-rutgers-saliva-test-coronavirus-gets-fda-approval Search PubMed.
- S. Sharma, V. Kumar, A. Chawla and A. Logani, Int. Endod. J., 2020, 53, 1017–1019 CrossRef CAS.
- J. Li, J. Macdonald and F. Von Stetten, Analyst, 2019, 144, 31–67 RSC.
- J. Kim and C. J. Easley, Bioanalysis, 2011, 3, 227–239 CrossRef CAS.
- Y. Yan, L. Chang and L. Wang, Rev. Med. Virol., 2020, 30, e2106 CrossRef CAS.
- L. J. Carter, L. V. Garner, J. W. Smoot, Y. Li, Q. Zhou, C. J. Saveson, J. M. Sasso, A. C. Gregg, D. J. Soares, T. R. Beskid, S. R. Jervey and C. Liu, ACS Cent. Sci., 2020, 6, 591–605 CrossRef CAS.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS.
- P. Craw and W. Balachandran, Lab Chip, 2012, 12, 2469–2486 RSC.
- Y. Kitagawa, Y. Orihara, R. Kawamura, K. Imai, J. Sakai, N. Tarumoto, M. Matsuoka, S. Takeuchi, S. Maesaki and T. Maeda, J. Clin. Virol., 2020, 129, 104446 CrossRef CAS.
- P. Huang, H. Wang, Z. Cao, H. Jin, H. Chi, J. Zhao, B. Yu, F. Yan, X. Hu, F. Wu, C. Jiao, P. Hou, S. Xu, Y. Zhao, N. Feng, J. Wang, W. Sun, T. Wang, Y. Gao, S. Yang and X. Xia, Front. Microbiol., 2018, 9, 1101 CrossRef.
- H. Thi, C. Thai, M. Q. Le, C. D. Vuong, M. Parida, H. Minekawa, T. Notomi and F. Hasebe, J. Clin. Microbiol., 2004, 42, 1956–1961 CrossRef.
- V. L. Dao Thi, K. Herbst, K. Boerner, M. Meurer, L. P. M. Kremer, D. Kirrmaier, A. Freistaedter, D. Papagiannidis, C. Galmozzi, M. L. Stanifer, S. Boulant, S. Klein, P. Chlanda, D. Khalid, I. B. Miranda, P. Schnitzler, H. G. Kräusslich, M. Knop and S. Anders, Sci. Transl. Med., 2020, 12, eabc7075 CrossRef CAS.
- L. E. Lamb, S. N. Bartolone, E. Ward and M. B. Chancellor, medRxiv, 2020 DOI:10.2139/ssrn.3539654.
- D. Butler, C. Mozsary, C. Meydan, D. Danko, J. Foox, J. Rosiene, A. Shaiber, E. Afshinnekoo, M. MacKay, F. J. Sedlazeck, N. A. Ivanov, M. Sierra, D. Pohle, M. Zietz, U. Gisladottir, V. Ramlall, C. D. Westover, K. Ryon, B. Young, C. Bhattacharya, R. Phyllis, B. W. Langhorst, N. Tanner, J. Gawrys, D. Meleshko, D. Xu, P. A. D. Steel, A. J. Shemesh, J. Xiang, J. Thierry-Mieg, D. Thierry-Mieg, R. E. Schwartz, A. Iftner, D. Bezdan, J. Sipley, L. Cong, A. Craney, P. Velu, A. M. Melnick, I. Hajirasouliha, S. M. Horner, T. Iftner, M. Salvatore, M. Loda, L. F. Westblade, M. Cushing, S. Levy, S. Wu, N. Tatonetti, M. Imielinski, H. Rennert and C. Mason, bioRxiv, 2020 DOI:10.1101/2020.04.20.048066.
- N. Ben-Assa, R. Naddaf, T. Gefen, T. Capucha, H. Hajjo, N. Mandelbaum, L. Elbaum, P. Rogov, D. A. King, S. Kaplan, A. Rotem, M. Chowers, M. Szwarcwort-Cohen, M. Paul and N. Geva-Zatorsky, medRxiv, 2020 DOI:10.1101/2020.04.22.20072389.
- W. E. Huang, B. Lim, C. C. Hsu, D. Xiong, W. Wu, Y. Yu, H. Jia, Y. Wang, Y. Zeng, M. Ji, H. Chang, X. Zhang, H. Wang and Z. Cui, Microb. Biotechnol., 2020, 13, 950–961 CrossRef CAS.
- L. Yu, S. Wu, X. Hao, X. Li, X. Liu, S. Ye, H. Han, X. Dong, X. Li, J. Li, N. Liu, J. Liu, W. Zhang, V. Pelechano, W.-H. Chen and X. Yin, Clin. Chem., 2020, 66, 975–977 CrossRef.
- Y. Zhang, N. Odiwuor, J. Xiong, L. Sun, R. O. Nyaruaba, H. Wei and N. A. Tanner, medRxiv, 2020 DOI:10.1101/2020.02.26.20028373.
- W. Yang, X. Dang, Q. Wang, M. Xu, Q. Zhao, Y. Zhou, H. Zhao, L. Wang, Y. Xu, J. Wang, S. Han, M. Wang, F. Pei and Y. Wan, medRxiv, 2020 DOI:10.7143/jhep.47.248.
- X. Yin, W. Chen, V. Pelechano, X. Hao, S. Wu and L. Yu, Clin. Chem., 2020, 66, 975–977 CrossRef.
- J. Pham, S. Meyer, C. Nguyen, A. Williams, M. Hunsicker, I. Mchardy, I. Gendlina, D. Y. Goldstein, A. S. Fox, A. Hudson, P. Darby, P. Hovey, J. Morales, J. Mitchell, K. Harrington, M. Majlessi, J. Moberly, A. Shah, A. Worlock, M. Walcher, B. Eaton, D. Getman and C. Clark, J. Clin. Microbiol., 2020, 58, e01669–e01620 CrossRef CAS.
- P. Trémeaux, S. Lhomme, F. Abravanel, S. Raymond, C. Mengelle, J. M. Mansuy and J. Izopet, J. Clin. Virol., 2020, 129, 104541 CrossRef.
- H. Fujita, Y. Kataoka, S. Tobita, M. Kuwahara and N. Sugimoto, Anal. Chem., 2016, 88, 7137–7144 CrossRef CAS.
- Shionogi, 2020, https://www.shionogi.com/global/en/news/2020/06/200622.html.
- K. Murugan, K. Babu, R. Sundaresan, R. Rajan and D. G. Sashital, Mol. Cell, 2017, 68, 15–25 CrossRef CAS.
- R. P. Bhattacharyya, S. G. Thakku and D. T. Hung, ACS Infect. Dis., 2018, 4, 1278–1282 CrossRef CAS.
- Y. Li, S. Li, J. Wang and G. Liu, Trends Biotechnol., 2019, 37, 730–743 CrossRef CAS.
- A. C. Batista and L. G. C. Pacheco, J. Microbiol. Methods, 2018, 152, 98–104 CrossRef CAS.
- B. Zetsche, J. S. Gootenberg, O. O. Abudayyeh, I. M. Slaymaker, K. S. Makarova, P. Essletzbichler, S. E. Volz, J. Joung, J. Van Der Oost, A. Regev, E. V. Koonin and F. Zhang, Cell, 2015, 163, 759–771 CrossRef CAS.
- S. Shmakov, O. O. Abudayyeh, K. S. Makarova, Y. I. Wolf, J. S. Gootenberg, E. Semenova, L. Minakhin, J. Joung, S. Konermann, K. Severinov, F. Zhang and E. V. Koonin, Mol. Cell, 2015, 60, 385–397 CrossRef CAS.
- O. O. Abudayyeh, J. S. Gootenberg, S. Konermann, J. Joung, I. M. Slaymaker, D. B. T. Cox, S. Shmakov, K. S. Makarova, E. Semenova, L. Minakhin, K. Severinov, A. Regev, E. S. Lander, E. V. Koonin and F. Zhang, Science, 2016, 353, aaf5573 CrossRef.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang and P. C. Sabeti, Science, 2018, 360, 444–448 CrossRef CAS.
- M. J. Kellner, J. G. Koob, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, Nat. Protoc., 2019, 14, 2986–3012 CrossRef CAS.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS.
- L. Curti, F. Pereyra-Bonnet and C. A. Gimenez, bioRxiv, 2020 DOI:10.1101/2020.02.29.971127.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C.-H. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, PNAS, 2020, 117(47), 29518–29525 CrossRef CAS.
- F. Zhang, O. O. Abudayyeh, J. S. Gootenberg, C. Sciences and L. Mathers, Bioarchive, 2020, 1–8 Search PubMed.
- T. Hou, W. Zeng, M. Yang, W. Chen, L. Ren, J. Ai, J. Wu, Y. Liao, X. Gou, Y. Li, X. Wang, H. Su, J. Wang, B. Gu, T. Xu, J. Wang and T. Xu, medRxiv, 2020 DOI:10.1101/2020.02.22.20025460.
- J. N. Rauch, E. Valois, S. C. Solley, F. Braig, R. S. Lach, N. J. Baxter, K. S. Kosik, C. Arias, D. Acosta-Alvear and M. Z. Wilson, bioRxiv, 2020 DOI:10.1101/2020.04.20.052159.
- P. Moitra, M. Alafeef, K. Dighe, M. Frieman and D. Pan, ACS Nano, 2020, 14, 7617–7627 CrossRef CAS.
- N. T. K. Thanh and Z. Rosenzweig, Anal. Chem., 2002, 74, 1624–1628 CrossRef CAS.
- H. Aldewachi, T. Chalati, M. N. Woodroofe, N. Bricklebank, B. Sharrack and P. Gardiner, Nanoscale, 2018, 10, 18–33 RSC.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS.
- Q. Chen, J. Li, Z. Deng, W. Xiong, Q. Wang and Y. Q. Hu, Intervirology, 2010, 53, 95–104 CrossRef CAS.
- J. Hardick, D. Metzgar, L. Risen, C. Myers, M. Balansay, T. Malcom, R. Rothman and C. Gaydos, Diagn. Microbiol. Infect. Dis., 2018, 91, 245–247 CrossRef CAS.
- L. K. De Souza Luna, V. Heiser, N. Regamey, M. Panning, J. F. Drexler, S. Mulangu, L. Poon, S. Baumgarte, B. J. Haijema, L. Kaiser and C. Drosten, J. Clin. Microbiol., 2007, 45, 1049–1052 CrossRef CAS.
- G. Seo, G. Lee, M. J. Kim, S. H. Baek, M. Choi, K. B. Ku, C. S. Lee, S. Jun, D. Park, H. G. Kim, S. J. Kim, J. O. Lee, B. T. Kim, E. C. Park and S. Il Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS.
- Y. Kyosei, M. Namba, S. Yamura, R. Takeuchi, N. Aoki, K. Nakaishi, S. Watabe and E. Ito, Diagnostics, 2020, 10, 2–9 CrossRef.
- A. Scohy, A. Anantharajah, M. Bodéus, B. Kabamba-Mukadi, A. Verroken and H. Rodriguez-Villalobos, J. Clin. Virol., 2020, 129, 104455 CrossRef CAS.
- B. Diao, K. Wen, J. Chen, Y. Liu, Z. Yuan, C. Han, J. Chen, Y. Pan, L. Chen, Y. Dan, J. Wang, Y. Chen, G. Deng, H. Zhou and Y. Wu, medRxiv, 2020 DOI:10.1101/2020.03.07.20032524.
- P. Mertens, N. De Vos, D. Martiny, C. Jassoy, A. Mirazimi, L. Cuypers, S. Van den Wijngaert, V. Monteil, P. Melin, K. Stoffels, N. Yin, D. Mileto, S. Delaunoy, H. Magein, K. Lagrou, J. Bouzet, G. Serrano, M. Wautier, T. Leclipteux, M. Van Ranst, O. Vandenberg, B. Gulbis, F. Brancart, F. Bry, B. Cantinieaux, F. Corazza, F. Cotton, M. Dresselhuis, B. Mahadeb, O. Roels and J. Vanderlinden, Front. Med., 2020, 7, 225 CrossRef.
- C. A. Hogan, M. K. Sahoo, C. H. Huang, N. Garamani, B. Stevens, J. Zehnder and B. A. Pinsky, J. Clin. Virol., 2020, 128, 104410 CrossRef CAS.
- J. Zhuang, J. Yin, S. Lv, B. Wang and Y. Mu, Biosens. Bioelectron., 2020, 163, 112291 CrossRef CAS.
- Y. T. Yeh, K. Gulino, Y. H. Zhang, A. Sabestien, T. W. Chou, B. Zhou, Z. Lin, I. Albert, H. Lu, V. Swaminathan, E. Ghedin and M. Terrones, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895–901 CrossRef CAS.
- T. Suo, X. Liu, J. Feng, M. Guo, W. Hu, D. Guo, H. Ullah, Y. Yang, Q. Zhang, X. Wang, M. Sajid, Z. Huang, L. Deng, T. Chen, F. Liu, K. Xu, Y. Liu, Q. Zhang, Y. Liu, Y. Xiong, G. Chen, K. Lan and Y. Chen, Emerging Microbes Infect., 2020, 9, 1259–1268 CrossRef CAS.
- H. Zhu, H. Zhang, S. Ni, M. Korabečná, L. Yobas and P. Neuzil, TrAC, Trends Anal. Chem., 2020, 130, 115984 CrossRef CAS.
- A. Asghari, C. Wang, K. M. Yoo, H. Dalir and R. T. Chen, 2020, arXiv:2008.08572.
- M. Qiu, Y. Shi, Z. Guo, Z. Chen, R. He, R. Chen, D. Zhou, E. Dai, X. Wang, B. Si, Y. Song, J. Li, L. Yang, J. Wang, H. Wang, X. Pang, J. Zhai, Z. Du, Y. Liu, Y. Zhang, L. Li, J. Wang, B. Sun and R. Yang, Microbes Infect., 2005, 7, 882–889 CrossRef CAS.
- B. Meyer, C. Drosten and M. A. Müller, Virus Res., 2014, 194, 175–183 CrossRef CAS.
- R. Milo and R. Plilips, Cell biology by the numbers, Garland Science, Daft., 2016, vol. 53 Search PubMed.
- FIND Evaluation Update: SARS-CoV-2 Immunoassays, 2020, https://www.finddx.org/covid-19/pipeline/?section=immunoassays#diag_tab Search PubMed.
- S. Khan, R. Nakajima, A. Jain, R. R. de Assis, A. Jasinskas, J. M. Obiero, O. Adenaiye, S. Tai, F. Hong, D. K. Milton, H. Davies and P. L. Felgner, bioRxiv, 2020 DOI:10.1101/2020.03.24.006544.
- G. Lippi, G. L. Salvagno, M. Pegoraro, V. Militello, C. Caloi, A. Peretti, S. Gaino, A. Bassi, C. Bovo and G. Lo Cascio, Clin. Chem. Lab. Med., 2020, 58, 1156–1159 CAS.
- I. Montesinos, D. Gruson, B. Kabamba, H. Dahma, S. Van den Wijngaert, S. Reza, V. Carbone, O. Vandenberg, B. Gulbis, F. Wolff and H. Rodriguez-Villalobos, J. Clin. Virol., 2020, 128, 104413 CrossRef CAS.
- R. Lassaunière, A. Frische, Z. B. Harboe, A. C. Nielsen, A. Fomsgaard, K. A. Krogfelt and C. S. Jørgensen, medRxiv, 2020 DOI:10.1101/2020.04.09.20056325.
- T. Nicol, C. Lefeuvre, O. Serri, A. Pivert, F. Joubaud, A. Ducancelle, F. Lunel-Fabiani and H. Le Guillou-Guillemette, J. Clin. Virol., 2020, 129, 104511 CrossRef CAS.
- COVID-AR IgG and IgM, 2020, https://www.conicet.gov.ar/tag/covidar-igg/ Search PubMed.
- B. Freeman, S. Lester, L. Mills, M. A. U. Rasheed, S. Moye, O. Abiona, G. B. Hutchinson, M. Morales-Betoulle, I. Krapinunaya, A. Gibbons, C.-F. Chiang, D. Cannon, J. Klena, J. A. Johnson, S. M. Owen, K. S. C. Barney, S. Graham and N. J. Thornburg, bioRxiv, 2020 DOI:10.1101/2020.04.24.057323.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu, W. Chen, Y. Zhang, J. Wang, B. Huang, Y. Lin, J. Yang, W. Cai, X. Wang, J. Cheng, Z. Chen, K. Sun, W. Pan, Z. Zhan, L. Chen and F. Ye, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS.
- A. Bryan, G. Pepper, M. H. Wener, S. L. Fink, C. Morishima, A. Chaudhary, K. R. Jerome, P. C. Mathias and A. L. Greninger, J. Clin. Microbiol., 2020, 58, e00941–e00920 CrossRef CAS.
- X. Jia, P. Zhang, Y. Tian and J. Wang, medRxiv, 2020 DOI:10.1101/2020.02.28.20029025.
- F. Amanat, D. Stadlbauer, S. Strohmeier, T. H. O. Nguyen, V. Chromikova, M. McMahon, K. Jiang, G. A. Arunkumar, D. Jurczyszak, J. Polanco, M. Bermudez-Gonzalez, G. Kleiner, T. Aydillo, L. Miorin, D. S. Fierer, L. A. Lugo, E. M. Kojic, J. Stoever, S. T. H. Liu, C. Cunningham-Rundles, P. L. Felgner, T. Moran, A. García-Sastre, D. Caplivski, A. C. Cheng, K. Kedzierska, O. Vapalahti, J. M. Hepojoki, V. Simon and F. Krammer, Nat. Med., 2020, 26, 1033–1036 CrossRef CAS.
- D. Stadlbauer, F. Amanat, V. Chromikova, K. Jiang, S. Strohmeier, G. A. Arunkumar, J. Tan, D. Bhavsar, C. Capuano, E. Kirkpatrick, P. Meade, R. N. Brito, C. Teo, M. McMahon, V. Simon and F. Krammer, Curr. Protoc. Microbiol., 2020, 57, 1–15 Search PubMed.
- J. Zhao, Q. Yuan, H. Wang, W. Liu, X. Liao, Y. Su, X. Wang, J. Yuan, T. Li, J. Li, S. Qian, C. Hong, F. Wang, Y. Liu, Z. Wang, Q. He, Z. Li, B. He, T. Zhang, Y. Fu, S. Ge, L. Liu, J. Zhang, N. Xia and Z. Zhang, Clin. Infect. Dis., 2020, 71(16), 2027–2034 CrossRef CAS.
- X. Cai, J. Chen, J. Hu, Q. Long, H. Deng, K. Fan, P. Liao, B. Liu, G. Wu, Y. Chen, Z. Li, K. Wang, X. Zhang, W. Tian, J. Xiang, H. Du, J. Wang, Y. Hu, N. Tang, Y. Lin, J. Ren, L. Huang, J. Wei, C. Gan, Y. Chen, Q. Gao, A. Chen, C. He, D.-X. Wang, P. Hu, F.-C. Zhou, A. Huang, P. Liu and D. Wang, J. Infect. Dis., 2020, 222, 189–193 CrossRef CAS.
- N. M. A. Okba, M. A. Müller, W. Li, C. Wang, C. H. GeurtsvanKessel, V. M. Corman, M. M. Lamers, R. S. Sikkema, E. de Bruin, F. D. Chandler, Y. Yazdanpanah, Q. Le Hingrat, D. Descamps, N. Houhou-Fidouh, C. B. E. M. Reusken, B. J. Bosch, C. Drosten, M. P. G. Koopmans and B. L. Haagmans, Emerging Infect. Dis., 2020, 26, 1478–1488 CrossRef CAS.
- N. M. A. Okba, V. S. Raj, I. Widjaja, E. de Bruin, F. D. Chandler, W. B. Park, N.-J. Kim, E. A. B. A. Farag, M. Al-Hajri, B. J. Bosch, M. Oh, M. P. G. Koopmans, C. B. E. M. Reusken and B. L. Haagmans, Emerging Infect. Dis., 2019, 25, 1868–1877 CrossRef CAS.
- E. Adams, M. Ainsworth, R. Anand, M. Andersson, K. Auckland, K. Baillie, E. Barnes, S. Beer, J. Bell, T. Berry, S. Bibi, M. Carroll, S. Chinnakannan, E. Clutterbuck, R. Cornall, D. Crook, T. de Silva, W. Dejnirattisai, K. Dingle, C. Dold, A. Espinosa, D. Eyre, H. Farmer, M. F. Mendoza, D. Georgiou, S. Hoosdally, A. Hunter, K. Jeffrey, P. Klenerman, J. Knight, C. Knowles, A. Kwok, U. Leuschner, R. Levin, C. Liu, C. López-Camacho, J. Martinez, P. Matthews, H. McGivern, A. Mentzer, J. Milton, J. Mongkolsapaya, S. Moore, M. Oliveira, F. Pereira, E. Perez, T. Peto, R. Ploeg, A. Pollard, T. Prince, D. Roberts, J. Rudkin, V. Sanchez, G. Screaton, M. Semple, D. Skelly, E. N. Smith, A. Sobrinodiaz, J. Staves, D. Stuart, P. Supasa, T. Surik, H. Thraves, P. Tsang, L. Turtle, S. Walker, C. Washington, N. Watkins and J. Whitehouse, Wellcome Open Res., 2020, 5, 139 Search PubMed.
- H. Ma, W. Zeng, H. He, D. Zhao, Y. Yang, D. Jiang, P. Zhou, Y. Qi, W. He, C. Zhao, R. Yi, X. Wang, B. Wang, Y. Xu, Y. Yang, A. J. K. Kombe, C. Ding, J. Xie, Y. Ga, L. Cheng, Y. Li, X. Ma and T. Jin, Cell. Mol. Immunol., 2020, 17, 773–775 CrossRef CAS.
- S. Fafi-Kremer, T. Bruel, Y. Madec, R. Grant, L. Tondeur, L. Grzelak, I. Staropoli, F. Anna, P. Souque, S. Fernandes-Pellerin, N. Jolly, C. Renaudat, M. N. Ungeheuer, C. Schmidt-Mutter, N. Collongues, A. Bolle, A. Velay, N. Lefebvre, M. Mielcarek, N. Meyer, D. Rey, P. Charneau, B. Hoen, J. De Seze, O. Schwartz and A. Fontanet, EBioMedicine, 2020, 59 CrossRef , 102915.
- National Univeristy of La Plata, FarmaCoV Test Search PubMed n.d., https://farmacov.com/#top.
- J. F. W. Chan, S. Yuan, K. H. Kok, K. K. W. To, H. Chu, J. Yang, F. Xing, J. Liu, C. C. Y. Yip, R. W. S. Poon, H. W. Tsoi, S. K. F. Lo, K. H. Chan, V. K. M. Poon, W. M. Chan, J. D. Ip, J. P. Cai, V. C. C. Cheng, H. Chen, C. K. M. Hui and K. Y. Yuen, Lancet, 2020, 395, 514–523 CrossRef CAS.
- D. Chandler-Brown, A. M. Bueno, O. Atay and D. S. Tsao, bioRxiv, 2020 DOI:10.1101/2020.04.07.029199.
- H. Lv, N. C. Wu, O. Tak-Yin Tsang, M. Yuan, R. A. P. M. Perera, W. Shing Leung, R. T. Y. So, J. Man Chun Chan, G. K. Yip, T. Shiu Hong Chik, Y. Wang, C. Yau Chung Choi, Y. Lin, W. W. Ng, J. Zhao, L. L. M. Poon, J. S. M. Peiris, I. A. Wilson and C. K. P. Mok, Cell Rep., 2020, 31, 107725 CrossRef CAS.
- W. Chan, J. D. Ip, A. W. Chu, C. C. Yip, L. Lo, K. Chan, A. C. Ng, R. W. Poon, W. To, O. T. Tsang, W. Leung, M. Y. Kwan, G. T. Chua, T. W. Chung, I. F. Hung, K. Kok, V. C. Cheng, J. F. Chan, K. Yuen and K. K. To, J. Med. Virol., 2020, 92, 2725–2734 CrossRef CAS.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS.
- G. Chen, M. Jin, P. Du, C. Zhang, X. Cui, Y. Zhang, J. Wang, F. Jin, Y. She, H. Shao, S. Wang and L. Zheng, Food Agric. Immunol., 2017, 28, 315–327 CrossRef CAS.
- D. Wrapp, N. Wang, K. S. Corbett, J. A. Goldsmith, C. L. Hsieh, O. Abiona, B. S. Graham and J. S. McLellan, Science, 2020, 367, 1260–1263 CrossRef CAS.
- J. Pallesen, N. Wang, K. S. Corbett, D. Wrapp, R. N. Kirchdoerfer, H. L. Turner, C. A. Cottrell, M. M. Becker, L. Wang, W. Shi, W. P. Kong, E. L. Andres, A. N. Kettenbach, M. R. Denison, J. D. Chappell, B. S. Graham, A. B. Ward and J. S. McLellan, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E7348–E7357 CrossRef CAS.
- L. Liu, W. Liu, Y. Zheng, X. Jiang, G. Kou, J. Ding, Q. Wang, Q. Huang, Y. Ding, W. Ni, W. Wu, S. Tang, L. Tan, Z. Hu, W. Xu, Y. Zhang, B. Zhang, Z. Tang, X. Zhang, H. Li, Z. Rao, H. Jiang, X. Ren, S. Wang and S. Zheng, Microbes Infect., 2020, 22, 206–211 CrossRef CAS.
- J. Xiang, M. Yan, H. Li, T. Liu, C. Lin, S. Huang and C. Shen, medRxiv, 2020 DOI:10.1101/2020.02.27.20028787.
- W. Zhang, R. H. Du, B. Li, X. S. Zheng, X. L. Yang, B. Hu, Y. Y. Wang, G. F. Xiao, B. Yan, Z. L. Shi and P. Zhou, Emerging Microbes Infect., 2020, 9, 386–389 CrossRef CAS.
- Z. Zheng, V. M. Monteil, S. Maurer-Stroh, C. W. Yew, C. Leong, N. K. Mohd-Ismail, S. C. Arularasu, V. T. K. Chow, R. T. P. Lin, A. Mirazimi, W. Hong and Y.-J. Tan, Eurosurveillance, 2020, 25, 2000291 CrossRef.
- W. W. Yee, S. H. Lim and E. H. Seng, Ann. Acad. Med. Singap., 2020, 49(7), 523–526 Search PubMed.
- Y. Tian, C. Lian, Y. Chen, D. Wei, X. Zhang, Y. Ling, Y. Wang and L. S. Yeap, Cell Discovery, 2020, 6, 75 CrossRef CAS.
- K. M. Koczula and A. Gallotta, Essays Biochem., 2016, 60, 111–120 CrossRef.
- Y. Huang, T. Xu, W. Wang, Y. Wen, K. Li, L. Qian, X. Zhang and G. Liu, Microchim. Acta, 2020, 187, 70 CrossRef CAS.
- L. Anfossi, F. Di Nardo, S. Cavalera, C. Giovannoli and C. Baggiani, Biosensors, 2018, 9, 2 CrossRef.
- J. Whitman, J. Hiatt, C. Mowery, B. Shy, R. Yu, T. N. Yamamoto, U. Rathore, A. E. Gallman, T. E. Miller, A. G. Levine, R. Apathy, M. J. Lipke and J. A. Smith, medRxiv, 2020 DOI:10.1101/2020.04.25.20074856.
- G.-Da Syu, J. Dunn and H. Zhu, Mol. Cell. Proteomics, 2020, 19, 916–927 CrossRef.
- L. Lin, L. Lu, W. Cao and T. Li, Emerging Microbes Infect., 2020, 9, 727–732 CrossRef CAS.
- M. Infantino, A. Damiani, F. L. Gobbi, V. Grossi, B. Lari, D. Macchia, P. Casprini, F. Veneziani, D. Villalta, N. Bizzaro, P. Cappelletti, M. Fabris, L. Quartuccio, M. Benucci and M. Manfredi, Isr. Med. Assoc. J., 2020, 22, 203–210 Search PubMed.
- J. Gu, E. Gong, B. Zhang, J. Zheng, Z. Gao, Y. Zhong, W. Zou, J. Zhan, S. Wang, Z. Xie, H. Zhuang, B. Wu, H. Zhong, H. Shao, W. Fang, D. Gao, F. Pei, X. Li, Z. He, D. Xu, X. Shi, V. M. Anderson and A. S. Y. Leong, J. Exp. Med., 2005, 202, 415–424 CrossRef CAS.
- X. Cao, Nat. Rev. Immunol., 2020, 20, 269–270 CrossRef CAS.
- M. R. Salazar, S. E. González, L. Regairaz, N. S. Ferrando, V. González Martínez, P. M. Carrera Ramos, L. Muñoz, S. A. Pesci, J. M. Vidal, N. Kreplak and E. Estenssoro, medRxiv, 2020 DOI:10.1101/2020.10.08.20202606v1.
- G. Siracusano, C. Pastori and L. Lopalco, Front. Immunol., 2020, 11, 1–9 CrossRef.
- H. F. Florindo, R. Kleiner, D. Vaskovich-Koubi, R. C. Acúrcio, B. Carreira, E. Yeini, G. Tiram, Y. Liubomirski and R. Satchi-Fainaro, Nat. Nanotechnol., 2020, 15, 630–645 CrossRef CAS.
- D. Blanco-Melo, B. E. Nilsson-Payant, W. C. Liu, S. Uhl, D. Hoagland, R. Møller, T. X. Jordan, K. Oishi, M. Panis, D. Sachs, T. T. Wang, R. E. Schwartz, J. K. Lim, R. A. Albrecht and B. R. tenOever, Cell, 2020, 181, 1036–1045 CrossRef CAS.
- G. Li, Y. Fan, Y. Lai, T. Han, Z. Li, P. Zhou, P. Pan, W. Wang, D. Hu, X. Liu, Q. Zhang and J. Wu, J. Med. Virol., 2020, 92, 424–432 CrossRef CAS.
- C. J. Neufeldt, B. Cerikan, M. Cortese, J. Frankish, J.-Y. Lee, A. Plociennikowska, F. Heigwer, S. Joecks, S. S. Burkart, D. Y. Zander, M. Gendarme, B. El Debs, N. Halama, U. Merle, M. Boutros, M. Binder and R. Bartenschlager, bioRxiv, 2020 DOI:10.1101/2020.07.21.212639.
- I. S. Okoye, M. Houghton, L. Tyrrell, K. Barakat and S. Elahi, Front. Immunol., 2017, 8, 1–21 Search PubMed.
- S. De Biasi, M. Meschiari, L. Gibellini, C. Bellinazzi, R. Borella, L. Fidanza, L. Gozzi, A. Iannone, D. Lo Tartaro, M. Mattioli, A. Paolini, M. Menozzi, J. Milić, G. Franceschi, R. Fantini, R. Tonelli, M. Sita, M. Sarti, T. Trenti, L. Brugioni, L. Cicchetti, F. Facchinetti, A. Pietrangelo, E. Clini, M. Girardis, G. Guaraldi, C. Mussini and A. Cossarizza, Nat. Commun., 2020, 11, 1–17 Search PubMed.
- C. Atyeo, S. Fischinger, T. Zohar, M. D. Slein, J. Burke, C. Loos, D. J. McCulloch, K. L. Newman, C. Wolf, J. Yu, K. Shuey, J. Feldman, B. M. Hauser, T. Caradonna, A. G. Schmidt, T. J. Suscovich, C. Linde, Y. Cai, D. Barouch, E. T. Ryan, R. C. Charles, D. Lauffenburger, H. Chu and G. Alter, Immunity, 2020, 53, 524–532 CrossRef.
- D. M. Del Valle, S. Kim-Schulze, H. H. Huang, N. D. Beckmann, S. Nirenberg, B. Wang, Y. Lavin, T. H. Swartz, D. Madduri, A. Stock, T. U. Marron, H. Xie, M. Patel, K. Tuballes, O. Van Oekelen, A. Rahman, P. Kovatch, J. A. Aberg, E. Schadt, S. Jagannath, M. Mazumdar, A. W. Charney, A. Firpo-Betancourt, D. R. Mendu, J. Jhang, D. Reich, K. Sigel, C. Cordon-Cardo, M. Feldmann, S. Parekh, M. Merad and S. Gnjatic, Nat. Med., 2020, 26, 1636–1643 CrossRef CAS.
- M. Kox, N. J. B. Waalders, E. J. Kooistra, J. Gerretsen and P. Pickkers, JAMA, J. Am. Med. Assoc., 2020, E1–E3 Search PubMed.
- A. M. Arvin, K. Fink, M. A. Schmid, A. Cathcart, R. Spreafico, C. Havenar-Daughton, A. Lanzavecchia, D. Corti and H. W. Virgin, Nature, 2020, 584, 353–363 CrossRef CAS.
- B. Rockx, T. Kuiken, S. Herfst, T. Bestebroer, M. M. Lamers, B. B. Oude Munnink, D. de Meulder, G. van Amerongen, J. van den Brand, N. M. A. Okba, D. Schipper, P. van Run, L. Leijten, R. Sikkema, E. Verschoor, B. Verstrepen, W. Bogers, J. Langermans, C. Drosten, M. Fentener van Vlissingen, R. Fouchier, R. de Swart, M. Koopmans and B. L. Haagmans, Science, 2020, 368, 1012–1015 CrossRef CAS.
- P. Yu, F. Qi, Y. Xu, F. Li, P. Liu, J. Liu, L. Bao, W. Deng, H. Gao, Z. Xiang, C. Xiao, Q. Lv, S. Gong, J. Liu, Z. Song, Y. Qu, J. Xue, Q. Wei, M. Liu, G. Wang, S. Wang, H. Yu, X. Liu, B. Huang, W. Wang, L. Zhao, H. Wang, F. Ye, W. Zhou, W. Zhen, J. Han, G. Wu, Q. Jin, J. Wang, W. Tan and C. Qin, Anim. Model Exp. Med., 2020, 3, 93–97 CrossRef.
- L. Bao, W. Deng, H. Gao, C. Xiao, J. Liu, J. Xue, Q. Lv, J. Liu, P. Yu, Y. Xu, F. Qi, Y. Qu, F. Li, Z. Xiang, H. Yu, S. Gong, M. Liu, G. Wang, S. Wang, Z. Song, Y. Liu, W. Zhao, Y. Han, L. Zhao, X. Liu, Q. Wei and C. Qin, bioRxiv, 2020 DOI:10.1101/2020.03.13.990226.
- A. Chandrashekar, J. Liu, A. J. Martinot, K. McMahan, N. B. Mercado, L. Peter, L. H. Tostanoski, J. Yu, Z. Maliga, M. Nekorchuk, K. Busman-Sahay, M. Terry, L. M. Wrijil, S. Ducat, D. R. Martinez, C. Atyeo, S. Fischinger, J. S. Burke, M. D. Slein, L. Pessaint, A. Van Ry, J. Greenhouse, T. Taylor, K. Blade, A. Cook, B. Finneyfrock, R. Brown, E. Teow, J. Velasco, R. Zahn, F. Wegmann, P. Abbink, E. A. Bondzie, G. Dagotto, M. S. Gebre, X. He, C. Jacob-Dolan, N. Kordana, Z. Li, M. A. Lifton, S. H. Mahrokhian, L. F. Maxfield, R. Nityanandam, J. P. Nkolola, A. G. Schmidt, A. D. Miller, R. S. Baric, G. Alter, P. K. Sorger, J. D. Estes, H. Andersen, M. G. Lewis and D. H. Barouch, Science, 2020, 4776, eabc4776 Search PubMed.
- J. Yu, L. H. Tostanoski, L. Peter, N. B. Mercado, K. McMahan, S. H. Mahrokhian, J. P. Nkolola, J. Liu, Z. Li, A. Chandrashekar, D. R. Martinez, C. Loos, C. Atyeo, S. Fischinger, J. S. Burke, M. D. Slein, Y. Chen, A. Zuiani, F. J. N. Lelis, M. Travers, S. Habibi, L. Pessaint, A. Van Ry, K. Blade, R. Brown, A. Cook, B. Finneyfrock, A. Dodson, E. Teow, J. Velasco, R. Zahn, F. Wegmann, E. A. Bondzie, G. Dagotto, M. S. Gebre, X. He, C. Jacob-Dolan, M. Kirilova, N. Kordana, Z. Lin, L. F. Maxfield, F. Nampanya, R. Nityanandam, J. D. Ventura, H. Wan, Y. Cai, B. Chen, A. G. Schmidt, D. R. Wesemann, R. S. Baric, G. Alter, H. Andersen, M. G. Lewis and D. H. Barouch, Science, 2020, 369, 806–811 CrossRef CAS.
- Guidance-and-Sop-Covid-19-Virus-Testing-in-NHS-Laboratories-V1, 2020, https://www.england.nhs.uk/coronavirus/wp-content/uploads/sites/52/2020/03/guidance-and-sop-covid-19-virus-testing-in-nhs-laboratories-v1.pdf.
- K. Skolimowska, M. Rayment, R. Jones, P. Madona, L. S. P. Moore and P. Randell, Clin. Microbiol. Infect., 2020, 26(12), 1711–1713 CrossRef CAS.
- E. Pasomsub, S. P. Watcharananan, K. Boonyawat, P. Janchompoo, G. Wongtabtim, W. Suksuwan, S. Sungkanuparph and A. Phuphuakrat, Clin. Microbiol. Infect., 2020 DOI:10.1016/j.cmi.2020.05.001.
- C. K. C. Lai, Z. Chen, G. Lui, L. Ling, T. Li, M. C. S. Wong, R. W. Y. Ng, E. Y. K. Tso, T. Ho, K. S. C. Fung, S. T. Ng, B. K. C. Wong, S. S. Boon, D. S. C. Hui and P. K. S. Chan, J. Infect. Dis., 2020, 222, 1612–1619 CrossRef CAS.
- Y. Liu, L. M. Yan, L. Wan, T. X. Xiang, A. Le, J. M. Liu, M. Peiris, L. L. M. Poon and W. Zhang, Lancet Infect. Dis., 2020, 20, 656–657 CrossRef CAS.
- P. Winichakoon, R. Chaiwarith, C. Liwsrisakun, P. Salee, A. Goonna, A. Limsukon and Q. Kaewpoowat, J. Clin. Microbiol., 2020, 58, e00297–e00220 CrossRef CAS.
- L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, M. Kang, Y. Song, J. Xia, Q. Guo, T. Song, J. He, H.-L. Yen, M. Peiris and J. Wu, N. Engl. J. Med., 2020, 382, 1175–1177 CrossRef.
- B. Lou, T.-D. Li, S.-F. Zheng, Y.-Y. Su, Z.-Y. Li, W. Liu, F. Yu, S.-X. Ge, Q.-D. Zou, Q. Yuan, S. Lin, C.-M. Hong, X.-Y. Yao, X.-J. Zhang, D.-H. Wu, G.-L. Zhou, W.-H. Hou, T.-T. Li, Y.-L. Zhang, S.-Y. Zhang, J. Fan, J. Zhang, N.-S. Xia and Y. Chen, Eur. Respir. J., 2020, 2000763 CrossRef CAS , in press.
- H. M. Staines, D. Kirwa, D. Clark, E. Adams, Y. Augustin, R. Byrne, M. Cocozza, A. Cubas-Atienzar, L. Cuevas, M. Cusinato, B. Davies, M. Davis, P. Davis, A. Duvoix, N. Eckersley, C. Williams, W. Kevin, J. Wilkins, J. Fitchett, S. Krishna and T. Planche, medRxiv, 2020 DOI:10.1101/2020.06.07.20124636.
- K. H. Hong, S. W. Lee, T. S. Kim, H. J. Huh, J. Lee, S. Y. Kim, J. S. Park, G. J. Kim, H. Sung, K. H. Roh, J. S. Kim, H. S. Kim, S. T. Lee, M. W. Seong, N. Ryoo, H. Lee, K. C. Kwon and C. K. Yoo, Ann. Lab. Med., 2020, 40, 351–360 CrossRef.
- X. Wu, B. Fu, L. Chen and Y. Feng, J. Med. Virol., 2020, 2–3 Search PubMed.
- K. E. Hanson, A. M. Caliendo, C. A. Arias, J. A. Englund, M. K. Hayden, M. J. Lee, M. Loeb, R. Patel, O. Altayar, A. El Alayli, S. Sultan, Y. Falck-Ytter, V. Lavergne, R. L. Morgan, M. H. Murad, A. Bhimraj and R. A. Mustafa, Clin Infect Dis, 2020 Search PubMed , ciaa1343.
- Z. Yongchen, H. Shen, X. Wang, X. Shi, Y. Li, J. Yan, Y. Chen and B. Gu, Emerging Microbes Infect., 2020, 9, 833–836 CrossRef.
- Coronavirus (COVID-19) Scaling up our testing programmes, 2020, https://www.gov.uk/government/publications/coronavirus-covid-19-scaling-up-testing-programmes Search PubMed.
- I. Santiago, ChemBioChem, 2020, 21(20), 2880–2889 CrossRef CAS.
- H. Shani-Narkiss, O. D. Gilday, N. Yayon and I. D. Landau, medRxiv, 2020 DOI:10.1101/2020.04.06.20052159.
- I Yelin, N. Aharony, E. S. Tamar, A. Argoetti, E. Messer, D. Berenbaum, E. Shafran, A. Kuzli, N. Gandali, O. Shkedi, T. Hashimshony, Y. Mandel-Gutfreund, M. Halberthal, Y. Geffen, M. Szwarcwort-Cohen and R. Kishony, Clin. Infect. Dis., 2020, 71(16), 2073–2078 CrossRef CAS.
- M. Peplow, Developing countries face diagnostic challenges as the COVID-19 pandemic surges, 2020, https://cendigitalmagazine.acs.org/2020/06/26/developing-countries-face-diagnostic-challenges-as-the-covid-19-pandemic-surges-2/content.html?utm_email+=94FED514342B026EF5C49448C9 Search PubMed.
- J. D. Sachs, S A. Karim, L. Aknin, J. Allen, K. Brosbøl, G. Cuevas Barron, P. Daszak, M. F. Espinosa, V. Gaspar, A. Gaviria, A. Haines, P. Hotez, P. Koundouri, F. Larraín Bascuñán, J.-K. Lee, M. Pate, P. Polman, G. Ramos, S. Reddy, I. Serageldin, R. Shah, J. Thwaites, V. Vike-Freiberga, M. K. Were, L. Xue, M. Zhu, C. Wang, C. Bahadur, M. E. Bottazzi, Y. Ben Amor, L. Barredo, O. Karadag Caman, G. Lafortune, E. Torres, J. Bagenal, I. Ethridge and J. G. E. Bartels, Lancet, 2020, 396(10257), 1102–1124 CrossRef CAS.
- A. T. Levin, W. P. Hanage, N. Owusu-Boaitey, K. B. Cochran, S. P. Walsh and G. Meyerowitz-Katz, Eur J Epidemiol, 2020 DOI:10.1007/s10654-020-00698-1.
- M. Pollán, B. Pérez-Gómez, R. Pastor-Barriuso, J. Oteo, M. A. Hernán, M. Pérez-Olmeda, J. L. Sanmartín, A. Fernández-García, I. Cruz, N. Fernández de Larrea, M. Molina, F. Rodríguez-Cabrera, M. Martín, P. Merino-Amador, J. L. León Paniagua, J. F. Muñoz-Montalvo, F. Blanco and R. Yotti, Lancet, 2020, 396(10250), 535–544 CrossRef.
- R. Etchenique and R. Quiroga, medRxiv, 2020 DOI:10.1101/2020.09.21.20198416.
- M. A. Crone, M. Priestman, M. Ciechonska, K. Jensen, D. J. Sharp, P. Randell, M. Storch and P. Freemont, medRxiv, 2020 DOI:10.1101/2020.05.02.20088344.
- D. M. Altmann, D. C. Douek and R. J. Boyton, Lancet, 2020, 395, 1527–1529 CrossRef CAS.
- C. Sheridan, Nat. Biotechnol., 2020, 38, 509–511 CrossRef.
| This journal is © The Royal Society of Chemistry 2021 |