Label-free lipidome study of paraventricular thalamic nucleus (PVT) of rat brain with post-traumatic stress injury by Raman imaging†
Ardalan
Chaichi
 a,
Syed Mohammad Abid
Hasan
a,
Syed Mohammad Abid
Hasan
 a,
Nishir
Mehta
a,
Nishir
Mehta
 a,
Fabrizio
Donnarumma
b,
Philip
Ebenezer
c,
Kermit K.
Murray
a,
Fabrizio
Donnarumma
b,
Philip
Ebenezer
c,
Kermit K.
Murray
 b,
Joseph
Francis
b,
Joseph
Francis
 c and
Manas Ranjan
Gartia
c and
Manas Ranjan
Gartia
 *a
*a
aDepartment of Mechanical and Industrial Engineering, Louisiana State University, Baton Rouge, LA 70803, USA. E-mail: mgartia@lsu.edu
bDepartment of Chemistry, Louisiana State University, Baton Rouge, LA 70803, USA
cComparative Biomedical Sciences, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA 70803, USA
First published on 5th October 2020
Abstract
Post-traumatic stress disorder (PTSD) is a widespread psychiatric injury that develops serious life-threatening symptoms like substance abuse, severe depression, cognitive impairments, and persistent anxiety. However, the mechanisms of post-traumatic stress injury in brain are poorly understood due to the lack of practical methods to reveal biochemical alterations in various brain regions affected by this type of injury. Here, we introduce a novel method that provides quantitative results from Raman maps in the paraventricular nucleus of the thalamus (PVT) region. By means of this approach, we have shown a lipidome comparison in PVT regions of control and PTSD rat brains. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry was also employed for validation of the Raman results. Lipid alterations can reveal invaluable information regarding the PTSD mechanisms in affected regions of brain. We have showed that the concentration of cholesterol, cholesteryl palmitate, phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, sphingomyelin, ganglioside, glyceryl tripalmitate and sulfatide changes in the PVT region of PTSD compared to control rats. A higher concentration of cholesterol suggests a higher level of corticosterone in the brain. Moreover, concentration changes of phospholipids and sphingolipids suggest the alteration of phospholipase A2 (PLA2) which is associated with inflammatory processes in the brain. Our results have broadened the understanding of biomolecular mechanisms for PTSD in the PVT region of the brain. This is the first report regarding the application of Raman spectroscopy for PTSD studies. This method has a wide spectrum of applications and can be applied to various other brain related disorders or other regions of the brain.
1. Introduction
Lipids play an important role in the brain through their homeostasis to maintain the integrity of cell membranes and control the signaling through the membranes.1 Changes in brain lipids have been correlated with many neurological diseases2 such as Alzheimer's disease,3 Parkinson's disease,4 and schizophrenia.5 Structurally, brain tissues are composed of about 5–15% lipids that may account for 50% of the dry weight of the brain.6 Mass spectrometry has been the gold standard for lipidomics analysis and has been accurately applied to cells, tissues and whole organisms.7–9 Most classical lipidome analysis requires extraction and homogenization of the sample, which result in a loss of spatial localization. Immunohistochemistry based methods preserve the spatial information. However, this is a targeted method and the target lipids must be known in advance to select an appropriate antibody for staining. Also, the number of dyes available for staining the lipids is limited. Furthermore, multiplexing is difficult with the histochemical approach. For example, only one or two antibodies can be applied simultaneously on the same sample.Both matrix-assisted laser desorption/ionization (MALDI) based mass spectrometry and Raman spectroscopy can alleviate many of the limitations listed above. Raman spectroscopy has additional advantages over MALDI such as being non-destructive and no need for matrix deposition. In addition, Raman spectroscopy requires minimal sample preparation. Raman spectroscopy has become a ubiquitous method for molecular level analysis of various biological samples such as brain,10,11 heart,12,13 kidneys,14,15 lipids,16,17 and proteins18,19 due to its non-invasive and label-free nature. Among these, Raman spectroscopy of lipids has attracted particular attention in the field because of the strong Raman scattering of lipids provided by long nonpolar acyl chains in their structure.16 It is well-established that lipids play a significant role in different cellular functions such as transport in cell membranes, signaling and energy storage.20 Therefore, the Raman signal obtained from the lipid bands of cells could be utilized as pathological biomarkers. Although there are similarities in the Raman spectrum of different lipids, individual lipids possesses unique spectra depending on numerous factors including geometry, phase, solubility, saturation, and polymorphism.21 Lipids have Raman bands in both the fingerprint (400–1800 cm−1) and higher wavenumber group frequency regions (2800–3800 cm−1).22,23 The most typical characteristics of lipids which originate from hydrocarbon chains manifest themselves in 1200–1050 cm−1 (C–C stretch), 1250–1300 cm−1 (CH3 scissor and twist) and 1400–1500 cm−1 (CH2 scissor and twist) ranges.24 At higher wavenumbers, strong Raman bands appear in the 2800–3100 cm−1 region which is assigned to C–H stretching of lipids.16
Post-traumatic stress disorder (PTSD) is considered a prevalent psychiatric disorder caused by exposure to repeated or single life-threatening events such as individuals involved in traffic accidents, combat veterans and rape victims.25 PTSD patients relive traumatic event by sudden remembrance of traumatic memories or flashbacks originating from the extreme horror and feelings of helplessness caused by the traumatic event. People with PTSD develop psychiatric disorders and symptoms such as severe depression, substance abuse, cognitive impairments, and persistent anxiety.26 Controlled biological investigations of PTSD in human subjects are mostly restricted due to ethical and logistical issues. Thus, indirect neurobiology studies on post-traumatic stress injured brains have become conventional by means of translational animal model approaches.27,28 As a result, many studies have been carried out by exposing animals to acute or chronic stress conditions in order to study their physiology and behavior changes which provide valuable knowledge under conditions similar to that experienced by traumatized human subjects. Although traumatic memories can develop into PTSD, it is not the only influential factor involved in constructing an effective animal model due to the multidimensional nature of this disease.29 Accordingly, various animal models such as predator stress, inescapable shocks, single prolonged stress, and unpredictable variable stress have been developed to ensure the occurrence of severe fear stress and production of human-like biological and behavioral symptoms in animals.30
It is well-known that severe stress exposure negatively affects various parts of the brain which are responsible for emotional responses, memory, and decision-making functions.31,32 Numerous morphological and functional deteriorations in various regions of the brain including the hippocampus, prefrontal cortex, amygdala and thalamus have been observed in animal models exposed to prolonged stress conditions.33,34 Physiological and behavioral symptoms of PTSD in the brain and their connection is crucial to thoroughly comprehend this type of disorder. However, most of the studies in this field have been directed toward the hippocampus and prefrontal cortex regions and the other parts are frequently neglected due to complexity.35,36 The paraventricular nucleus of the thalamus (PVT) is one of the stress sensors in mammalian brains that has been recently examined for its correlation to post-traumatic stress disorder.37 According to recent studies,38,39 both psychological and physical stressors can affect and activate this region. Meanwhile, the correlation between adaptive behavioral responses due to severe stress and the PVT region of the brain is still tangled. PVT is a part of the thalamic nuclei located at the midline and intralaminar region and it is commonly assumed to participate in the arousal system.37 According to neuroanatomical investigations,37–39 PVT collects autonomic and arousal projections of the brainstem and nervous system. These studies have experimentally demonstrated PVT activation by arousal and stress stimulators. Furthermore, it was recently shown that drug addiction behaviors are also associated with this region of brain.40 In addition, substance addiction projections to the prefrontal cortex, amygdala, and nucleus accumbens are likely to originate from the PVT region.41
Despite the large population of patients affected by PTSD worldwide, there are no available reports regarding the application of Raman spectroscopy or imaging for this type of brain disorder. In order to understand the biochemical effects of PTSD on different regions of the brain which are controlling vital functions, it is substantially important to monitor the lipid alterations in those regions.20 Accordingly, novel therapeutic approaches can be developed to suppress these stress-induced changes. Recent studies illustrate the importance of stress-induced lipid metabolism analysis and their huge impact in unravelling the functions of brain regions.42,43 As a tangible example, it has been reported that prolonged stress stimulates an important lipid enzyme (phospholipase A2) which directly influences inflammatory responses by altering cellular lipid signaling.44 Furthermore, stress-induced lipid modulations affect the PVT region by altering arachidonoylglycerol and diacylglycerol levels.34 Additionally, ceramide level changes have also been observed in the brain due to prolonged stress conditions.44
There is growing interest in discovering novel therapeutics for treatment and molecular imaging methods for PTSD. We present new findings regarding the use of Raman spectroscopy imaging for measuring lipid changes in the brain. Specifically, we describe an approach for analyzing changes in the lipid levels in the PVT region of the brain tissue by obtaining 2D images of the formalin fixed brain sections from PTSD induced and control rats. We have used both unbiased PCA analysis and targeted approaches using lipid standards to identify lipid classes. We have validated the Raman imaging results with MALDI MS data. We anticipate that similar studies can be accomplished in other regions of brain to better understand the impact of stress-induced modulations on behavioral and physiological responses in PTSD. However, we need to also consider that coexistence of other conditions like traumatic brain injury could potentially cause lipidomic alterations in the brain. Although we tried to minimize the chance of other concomitant conditions by choosing a well-proven model for PTSD induction, we need to address this, as a general potential limitation of lipidomic studies in the brain.
2. Materials and methods
2.1. Ethical statement
All the animal experiments in this study were carried out in strict accordance with the guidelines of Care and Use of Laboratory Animals of the National Institutes of Health (NIH), US and approved by the Institutional Animal Care and Use Committee (IACUC) for animal subjects research at Louisiana State University.2.2. Biochemicals and reagents
To build a Raman reference library, purified lipid standards such as phosphatidylinositol (P6636-1G), phosphatidylserine (P7769-25MG), phosphatidylethanolamine (P1348-25MG), cholesteryl palmitate (C6072-1G), cholesterol (C8667-1G), galactocerebroside (C4905-10MG), glyceryl tripalmitate (T5888-1G), phosphatidic acid (P4013-100MG) and sphingomyelin (S0756-100MG) were purchased from Sigma-Aldrich (St Louis, Missouri). Other lipids such as sulfatide (Avanti # 131305) and ganglioside (Avanti # 860053) were purchased from Avanti Polar Lipids (Alabaster, Alabama).2.3. Brain tissue sample preparation
Naive adult male Sprague-Dawley rats (n = 12) (Harlan Laboratories, Indianapolis, IN) weighing 325–350 g with the age of 10 weeks were used for this study. The animals were fed ad libitum and kept under standard laboratory conditions (temperature: 20 °C, humidity: 23–42%). Alternating dark and light cycles (lasting 12 h each) were maintained. All animal handling procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Louisiana State University School of Veterinary Medicine.We used an acute predator exposure model27,45–48 to induce traumatic stress in the animal. In this model, rats were exposed periodically to a cat (adult, 7 years old, Harlan Laboratories, Indianapolis, IN) followed by rotating the rats into different cages to eliminate any social support and induce chronic psychological stress. The experiments continued for 31 days and the rats were exposed to the cat on day 1 and day 11 for 1 h. The first exposure was performed during the daylight cycle (07:00–19:00), while the second exposure was performed during the night cycle (19:00–07:00). Between day 1 to day 31, the rats were subjected to random cage rotation to make sure that no rat was housed with the same set of rats on consecutive days or more than four times within the experimental period. Also, it is important to note that during the exposure period, the cat was not allowed to touch the rats by putting the rats in Plexiglas containers. Furthermore, during the cage rotation periods, care was taken not to allow the cat or cat material near the cages. The control group (n = 6) were not subjected to cat exposure or cage rotation and were kept in the same cages from day 1 to day 31.
The behavioral test for anxiety was performed on all the rats using an elevated plus maze (EPM) experiment (EB-Instruments, Bioseb, Tampa Bay, FL).27,47,48 In these experiments the rats were allowed to roam freely for 5 min and their movement was captured using an overhead camera (BioEPM3C, EB-Instruments, Tampa Bay, FL). From the captured video, the number of entries into each arm as well as the total time spent in the open vs. closed arms were extracted.27
The rats from the control and PTSD groups were humanely euthanized via inhalation of carbon dioxide. Immediately after, transcardial perfusion was performed using 10 mM phosphate-buffered saline (PBS) solution. To fix the tissue, this step was followed by transcardial perfusion with 4% phosphate-buffered paraformaldehyde (PB-PFA) solution. We immediately harvested the whole brain using cranial dissection. The brains were cryosectioned into 40 μm thick slices that were kept in 1× PBS at 4 °C. The tissue sections containing the paraventricular nucleus of the thalamus (PVT) region were found by using the rat brain stereotaxic coordinates in the Bregma number regions of −1.20 mm to −3.6 mm (see ESI Fig. S1 and S2†). In general, the brain size is different in every rat. Due to the differences in the overall size of the brains, the paraventricular thalamic nucleus (PVT) region of brain is also slightly smaller or bigger in each animal. However, this size difference does not affect the results of study since we are calculating the weighted mean of pixel values. By considering the weighted mean value as the basis of comparison, size dependence is not an issue anymore. Samples were stored in PBS in a well-plate. For all the Raman experiments, samples were mounted on a mirror-like surface stainless steel slide. For MALDI experiments, the samples were mounted on indium tin oxide (ITO) coated glass slides.
2.4. Histology
The hematoxylin and eosin (H&E) staining protocol was followed for the histology observations. First, the nuclei were stained with alum hematoxylin. After rinsing, the samples were dipped in acidified alcohol (1 ml concentrated HCl with 400 ml 70% ethanol). Afterwards, samples were rinsed again and stained with eosin for 2 min. Finally, they were dehydrated and mounted on standard 75 mm × 25 mm microscope slides.2.5. Raman spectroscopy and microscopy
The Raman spectra of standard lipids in powder form were obtained with a Renishaw inVia Reflex Raman spectrometer with a 785 nm laser with an exposure time of 20 s and 100% power (laser power on the sample is ∼90 mW). All the Raman spectra from brain samples were obtained under PBS (1×) immersion conditions (lens was not immersed in PBS, we put the sample in PBS to prevent drying) with an exposure time of 10 s, objective lens of Leica LWD 50× (long-working distance) with high confocality (objective specifications: N PLAN L 50×/0.50; material number: 11566036; objective type: N PLAN; magnification: 50; numerical aperture: 0.5; coverglass: 0; immersion: dry (without); free working distance: 8.2; objective thread: M25; corr. of cover glass thickness: no; Iris diaphragm: no; spring loaded: no; methods: brightfield, fluorescence, DIC (Nomarski), polarisation, transmitted darkfield). The laser spot size is 1.1 μm with a penetration depth of a few microns. In the spectral acquisition mode, from each sample at least ten spectra were obtained for statistical analysis. For lipids, we used the extended mode in the full range of 100 to 3200 cm−1. In the imaging mode, for PTSD and control samples 798 and 702 spectra were collected, respectively, to generate the Raman images with a step size of 20 μm. For brain sample imaging the spectra were acquired in static mode with the center of 1100 cm−1 (spectral range is from 497.92 to 1638.44 cm−1). The size of the acquired image for control is 1765 μm × 546 μm and for PTSD is 812 μm × 364 μm. The mapping was done in point-by-point mode. The machine was calibrated using the silicon peak at 520 cm−1.2.6. Data processing and statistical analysis
All the Raman spectra were baseline corrected using Renishaw's WiRE 4.4 (Windows-based Raman Environment) software. Intelligent fitting was used in WiRE 4.4 for baseline subtraction. By default, the intelligent fitting baseline is applied with a polynomial value of 11. Preprocessing of the data and the subsequent principal component analysis were performed using Origin 2018 (OriginLab, Northampton, MA). The Min–Max normalization approach was applied to the datasets in the [0,1] range and smoothing was performed by means of the Savitzky–Golay method (window size is 5 and polynomial order is 2). Direct classical least squares analysis (DCLS) method was used to generate the mapping data. ImageJ 1.8 software was used for the quantification of Raman based imaging data. Fig. S3† shows the steps for the quantification of the Raman maps by means of pixel intensity and distribution. Accordingly, all the images were first converted into 8-bit format. The weighted mean value of brightness (pixel value) and the corresponding histogram data were then calculated for all the pixels in an ROI by using the “measure” function under the “analyze” tab of ImageJ software. This value is chosen as a representative for the relative concentration of lipids (Fig. S3†). A color thresholding method was used to determine the distribution of lipids. Based on the inspection of all the images, a brightness value of 111 (in order to have a good contrast) was selected as a threshold in all the images. By keeping the threshold constant, we analyzed the distribution of lipids in each Raman map (Fig. S3†).The Raman spectral data were analyzed using principal component analysis (PCA).49,50 Multivariate analysis using the “principal component analysis for spectroscopy” toolbox of OriginLab 2018 was used to perform the PCA. The spectral differences among the data sets were described by the principal components (PC). For parameter settings, we selected the covariance matrix to analyze and the number of components to extract was set to 8. We calculated PCA from the covariance matrix. Since computing the covariance matrix implicitly executes centering, we do not need to do any further mean centering. Each of the Raman spectra is described as a point on a score plot when selecting two or more PCs. Finally, the clustering of the data on the score plot and their vibrational fingerprint assignment is obtained by the loading plot.
We performed one-way analysis of variance (ANOVA) using Fisher's least significant difference (LSD) test using Statistical Analysis System (SAS) v.9.4 software package. We performed Levene's test for checking the homoscedasticity assumption. We formed eleven groups of lipids PCA values and checked whether the means of PCA values were significantly different among each group. The null hypothesis was “there is no difference among the PCA means in different lipid groups” and the alternative hypothesis was “at least one of the lipid groups has a different PCA mean from the others”. We used the common cut-off value of 0.05 or equivalently the 95% confidence level.
2.7. MALDI analysis
A matrix assisted laser desorption/ionization MALDI-TOF/TOF mass spectrometer (UltrafleXtreme, Bruker Daltonics, Billerica, MA, USA) was utilized for mass spectrometry imaging (MSI) in positive ion mode. Brain slices were mounted on indium tin oxide (ITO) coated slides (University Wafer, Inc.). Super-DHB (Millipore-Sigma), 2,5-dihydroxybenzoic acid (DHB, Sigma-Aldrich), and α-cyano-4-hydroxycinnamic acid (Millipore-Sigma) were used as the matrix.51 The matrix was prepared by dissolving super-DHB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) in a methanol and water solution containing 0.1 vol% of trifluoroacetic acid, to a final concentration of 10 mg mL−1. Tissue sections were dried in a vacuum chamber for about 7 minutes. Afterwards, the matrix was uniformly deposited on the tissue by means of a pneumatic nebulizer (nitrogen gas pressure = 70 kPa; liquid flow rate = 100 μL min−1). Lamp heating was used to dry the matrix on the surface of the tissue. All the MS data were smoothed using B-spline fit. A custom code, ‘Decimator.vi’ written in LabVIEW VI (National Instruments) was utilized for baseline subtraction, averaging, decimation, and de-noising of the obtained data. Bruker FlexAnalysis 3.0 software and MSiReader,52 an open-source Matlab package, were employed for data analysis after baseline subtraction. Peak assignments were done by using the available databases on lipidmaps.org. To generate MALDI images, 1944 and 539 spectra were recorded from PTSD and Control samples, respectively. MALDI images were analyzed by using the same method used for Raman images, as described above.
1 ratio) in a methanol and water solution containing 0.1 vol% of trifluoroacetic acid, to a final concentration of 10 mg mL−1. Tissue sections were dried in a vacuum chamber for about 7 minutes. Afterwards, the matrix was uniformly deposited on the tissue by means of a pneumatic nebulizer (nitrogen gas pressure = 70 kPa; liquid flow rate = 100 μL min−1). Lamp heating was used to dry the matrix on the surface of the tissue. All the MS data were smoothed using B-spline fit. A custom code, ‘Decimator.vi’ written in LabVIEW VI (National Instruments) was utilized for baseline subtraction, averaging, decimation, and de-noising of the obtained data. Bruker FlexAnalysis 3.0 software and MSiReader,52 an open-source Matlab package, were employed for data analysis after baseline subtraction. Peak assignments were done by using the available databases on lipidmaps.org. To generate MALDI images, 1944 and 539 spectra were recorded from PTSD and Control samples, respectively. MALDI images were analyzed by using the same method used for Raman images, as described above.
3. Results and discussion
3.1. Raman spectra for the PTSD and control tissue
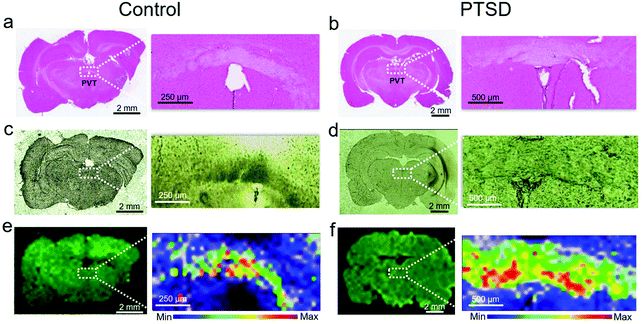
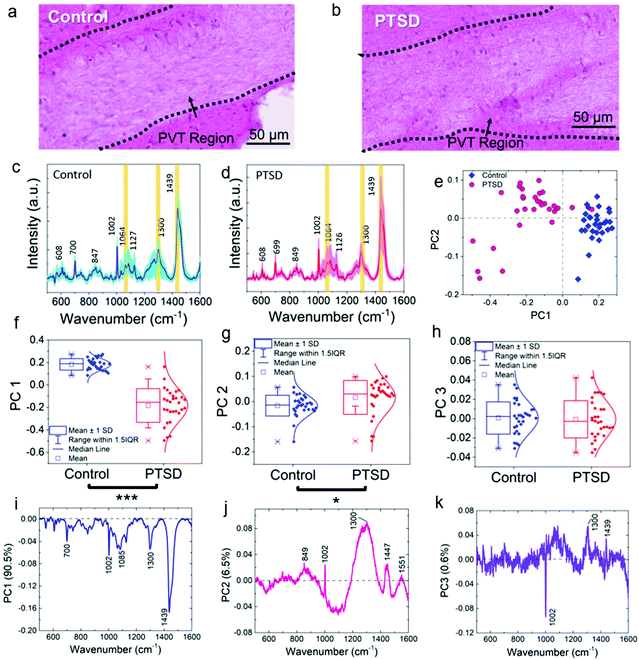
Fig. S1† shows an overview of this study from the animal model to output data. A predator exposure/psychosocial stress regimen was utilized to develop PTSD in the rat. For verification of PTSD induction, behavioral analysis, neurotransmitter changes, and oxidative stress analysis were performed as described in the previous study.27 In order to demonstrate the location of the PVT region in rat brain, Fig. 1 shows H & E (Fig. 1a and b), bright field (Fig. 1c and d) and Raman (Fig. 1e and f) images of the control and PTSD brain tissues. The magnified PVT region is shown next to the image of the full brain slice. The PVT region is visible with a white color in H & E images and a darker color in bright field images. Raman images (Fig. 1e and f) were plotted at the peak position of 1002 cm−1 for control and PTSD samples (color scales are the same). This peak is the most abundant peak in most of the biological samples. It is assigned to the aromatic ring C–C breathing mode and usually demonstrates the existence of phenylalanine amino acid.24 we can see about a 52% increase of phenylalanine in the PTSD sample compared to the control. Changes in concentration of phenylalanine can lead to arousal and alertness.53 Accordingly, the PVT region is distinguishable from the surrounding area due to the different contrast in all the three imaging methods.Fig. 2a and b show the magnified view (12×) of H & E images showing the PVT region and the different neuron cell structures inside and outside of the PVT. Accordingly, no morphological changes were observed in the aforementioned region. To probe the biochemical changes, Raman experiments were performed on the same region. Raman spectra of control (Fig. 2c) and PTSD (Fig. 2d) samples illustrate the biochemical changes in the brain in the PVT region due to the applied stress. The diagrams are achieved by plotting 30 different spectra inside the dotted area for each sample. The average value is highlighted by darker colors for both datasets. Raman bands of the brain spectra are assigned to cholesterol (608 cm−1), methionine (700 cm−1), DNA/RNA (847 cm−1), phenylalanine (1002 cm−1), acyl chains (C–C stretch; 1064 cm−1), proteins (C–N stretch; 1127 cm−1), lipids (CH2 twist; 1300 cm−1), and lipids (CH2 bend; 1439 cm−1).16 The Raman peak positions did not change significantly in the PTSD brain samples compared to control samples (the assignment of all the Raman peaks is listed in Table S1†). However, we found a consistent modulation of Raman intensities between control and PTSD samples. Therefore, we utilized the principal component analysis (PCA) method to reveal any possible biochemical changes in the PTSD brain samples. Significant changes were identified by the PCA and scatter plot that were able to discriminate both sample groups (PTSD and control). The most distinctive characteristics were observed in the lipid bands (acyl C–C stretch (1064 cm−1), CH2 twist (1300 cm−1), and lipid CH2 bend (1439 cm−1)) (also see Fig. S4 and S5†).
3.2. Raman fingerprint spectra discriminate PTSD and control
Fig. 2e shows the score plot from the first two principle components based on 30 spectra chosen from PTSD (pink) and control (blue) tissue samples. The score plot demonstrates a clear segregation and distinct clustering of data obtained from different sample groups. Fig. 2f–h show the box plot of the distribution of spectral data for the two groups. The spectral data of the PTSD samples were significantly shifted (P < 0.001) to the negative PC 1 range (median = −0.15) compared to the control (median = 0.18; Fig. 2f). Likewise, substantial changes (P < 0.05) were observed in the PC 2 (Fig. 2g) data distribution by shifting the median value from −0.14 in the control to 0.03 in the PTSD. However, the changes in PC 3 (Fig. 2h) were not significant (P > 0.05).To find the biochemical components in each spectral variation, loading plots (Fig. 2i–k) of the spectra obtained from the PVT region from PTSD and control are presented. The loading plots demonstrate the major spectral differences of the PCs. The overall contribution from the first three PCs is ∼97.6%. Characteristic peaks of each dataset are provided for PC 1, PC 2 and PC 3 (Fig. 2i–k). These characteristic peaks indicate the differences in the various Raman datasets. The loading plot for PC 1 (Fig. 2i) was obtained from PTSD and control samples and represents 90.5% of the spectral variation including the Raman peaks from all biomolecules (lipids, proteins, RNA and DNA). For example, 700 (methionine C–S trans), 1002 (phenylalanine C–C aromatic ring stretch), 1085 (phosphodiester groups in nucleic acids), 1300 (lipid CH2 twist) and 1438 cm−1 (lipid CH2 bend) are the prominent peaks for PC 1; 849 (amino acid stretch), 1002, 1300 and 1447 cm−1 (proteins and lipids CH2 bending) are the most distinctive bands for PC 2 (Fig. 2j). Moreover, the third component of PC (Fig. 2k), which contains only 0.6% of the spectral variation, can differentiate between proteins (1002 cm−1, negative value) and lipids (1300 cm−1, positive value). As can be seen, some of the most significant distinctive characteristics were observed in the lipid bands. As a result, a considerable change in the concentration of lipids can be derived from the Raman spectra of PTSD brain compared to the control sample. The in-depth discussion of lipid alterations is provided below.
3.3. Raman spectroscopy comparison of lipids relevant to brain tissue
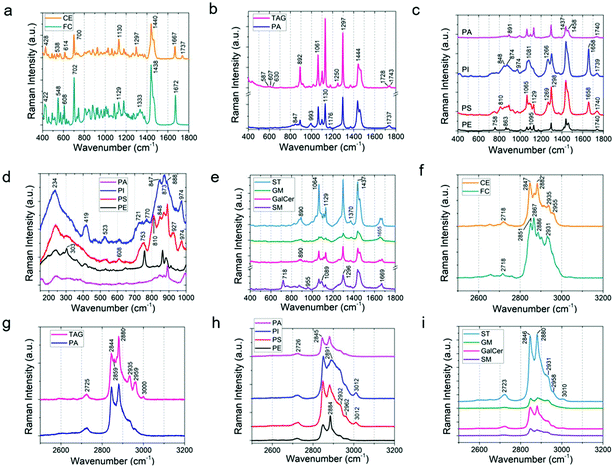
In order to track the lipid concentration changes in the rat brain due to acute stress exposure, Raman spectra of the eleven most common lipids in brain20,54 were acquired by collecting the Raman signal from standard reference lipid samples by means of a 785 nm excitation laser (Fig. S6†). As can be seen in Fig. 3a–e, different lipids have different Raman spectra which distinguish them from the other lipids. The spectral region from 400–1800 cm−1 shows the fingerprint region for each lipid. The peaks in the wavenumber region 2700–3500 cm−1 are due to the CH and OH stretch (Fig. 3f–i). For instance, peaks from 2845–2868 cm−1 are due to the![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 symmetric stretch, 2870–2904 cm−1 are from the
CH2 symmetric stretch, 2870–2904 cm−1 are from the ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2 asymmetric stretch, peaks from 2905–2940 cm−1 are due to the –CH3 symmetric stretch, 2941–2970 cm−1 are from the –CH3 asymmetric stretch, and the Raman peaks from 3000–3015 cm−1 are due to the unsaturated
CH2 asymmetric stretch, peaks from 2905–2940 cm−1 are due to the –CH3 symmetric stretch, 2941–2970 cm−1 are from the –CH3 asymmetric stretch, and the Raman peaks from 3000–3015 cm−1 are due to the unsaturated ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH bond stretch.16Fig. 3a shows the comparison of cholesterol (FC) and cholesteryl palmitate (or cholesterol ester, CE) (the molecular structure is shown in Fig. S7†). The Raman band due to the ester group at 1737 cm−1 is absent in cholesterol. Spectral bands at 1065, 1130, and 1297 cm−1 are due to the palmitic acid group in the cholesterol ester.54 The intense band at 1440 cm−1 is due to the acyl group (CH2 or CH3 scissor). Furthermore, the following band shifts were observed from cholesterol to cholesterol ester: 422 to 428, 548 to 538 (CH2 bend), 608 to 614 (ester group), and 1672 to 1667 cm−1 (C
CH bond stretch.16Fig. 3a shows the comparison of cholesterol (FC) and cholesteryl palmitate (or cholesterol ester, CE) (the molecular structure is shown in Fig. S7†). The Raman band due to the ester group at 1737 cm−1 is absent in cholesterol. Spectral bands at 1065, 1130, and 1297 cm−1 are due to the palmitic acid group in the cholesterol ester.54 The intense band at 1440 cm−1 is due to the acyl group (CH2 or CH3 scissor). Furthermore, the following band shifts were observed from cholesterol to cholesterol ester: 422 to 428, 548 to 538 (CH2 bend), 608 to 614 (ester group), and 1672 to 1667 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch).
C stretch).
Next, Fig. 3b shows a comparison of glyceryl palmitate (or triacylglyceride, TAG) and phosphatidic acid (PA) (Fig. S8†). They share quite similar structures with one palmitic acid chain in TAG replaced by a phosphate group for PA. The band due to the ester group appears at 1737 cm−1 for PA. The ester band for TAG splits into two at 1728 and 1743 cm−1, indicating that they are in a different surrounding environment compared to PA. The three small bands at 587, 607, and 630 cm−1 are due to glycerol.55 The band due to stretching of P–O vibration of the PO4 group is at 993 cm−1.56 The intense bands at 1061 (C–C stretch), 1130 (C–C stretch), 1297 (CH2 twist), and 1444 cm−1 (CH2/CH3 scissoring) are due to fatty acid chains. Fig. 3c and d show the comparison of different phospholipids. As shown in Fig. S9,† the parent group of phospholipids (PE, PI, PC, PS) is phosphatidic acid (PA). Therefore, the Raman bands of phospholipids share features similar to that of phosphatidic acid. The band for phosphatidylethanolamine (PE) at 758 cm−1 is assigned to ethanolamine (Fig. 3c). Similarly, the band at 1095 cm−1 of the PE Raman spectrum is due to phosphodioxy groups PO2− (P–O stretch). At low wavenumbers between 200–1000 cm−1, distinct differences among phospholipids were observed (Fig. 3d). Phosphatidylinositol (PI) shows peaks at 234, 419, 523, 721, 770, 847, 873, 888, and 974 cm−1; phosphatidylserine (PS) shows peaks at 234, 524, 608, 753, 810, 848, 873, 888, 927, and 974 cm−1. PI and PS have bands at 1658 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch) which are absent in PA and PE. All the phospholipids have bands for ester (1739 cm−1) and acyl chains (1065, 1130, 1298, and 1437 cm−1).
C stretch) which are absent in PA and PE. All the phospholipids have bands for ester (1739 cm−1) and acyl chains (1065, 1130, 1298, and 1437 cm−1).
Fig. 3e shows a comparison of the Raman spectra obtained from sphingolipids (see Fig. S10† for the molecular structures). Sphingolipids are composed of lipids, a ceramide backbone, and glucose rings with or without linker groups; the linker group for galactocerebroside (GalCer) is galactose, sulfate containing monosaccharide for sulfatide (ST), and oligosaccharides for ganglioside (GM). As can be seen in Fig. S10 and S11† (also Fig. 3e), the Raman signature of lipids (at 1064, 1129, 1298, and 1437 cm−1), ceramide backbone (1657, 1671 cm−1; C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch), and sugar chain (1370 cm−1; Fig. S11†) are clearly present. Fig. S11† also shows that the area under the peak at 1370 cm−1 is the highest for ganglioside (GM) and lowest for sphingomyelin (SM). This agrees well with the molecular structure of GM, which has the maximum saccharide content (Fig. S10†), and that of SM, which has no glucose group. Furthermore, we observed that the peak ratio of 1370 (sugar chain) to 1297 cm−1 (fatty acid) is the highest for GM. The Raman peak at 890 cm−1 (C–O–O skeletal mode) is present in all the sphingolipids except SM as SM does not have any C–O–O group (Fig. S11†). The sulfate band in ST is characterized by peaks at 614 and 995 cm−1 (Fig. S11†).57 Finally, sphingomyelin is composed of a phosphatidylcholine (PC) residue, which is characterized by Raman peaks at 718 and 875 cm−1 (Fig. 3e). Another key difference between the backbone of phospholipids and ceramide structure is the amide bond (1669 cm−1) in the ceramide backbone instead of the ester band (1739 cm−1) of phospholipids.
C stretch), and sugar chain (1370 cm−1; Fig. S11†) are clearly present. Fig. S11† also shows that the area under the peak at 1370 cm−1 is the highest for ganglioside (GM) and lowest for sphingomyelin (SM). This agrees well with the molecular structure of GM, which has the maximum saccharide content (Fig. S10†), and that of SM, which has no glucose group. Furthermore, we observed that the peak ratio of 1370 (sugar chain) to 1297 cm−1 (fatty acid) is the highest for GM. The Raman peak at 890 cm−1 (C–O–O skeletal mode) is present in all the sphingolipids except SM as SM does not have any C–O–O group (Fig. S11†). The sulfate band in ST is characterized by peaks at 614 and 995 cm−1 (Fig. S11†).57 Finally, sphingomyelin is composed of a phosphatidylcholine (PC) residue, which is characterized by Raman peaks at 718 and 875 cm−1 (Fig. 3e). Another key difference between the backbone of phospholipids and ceramide structure is the amide bond (1669 cm−1) in the ceramide backbone instead of the ester band (1739 cm−1) of phospholipids.
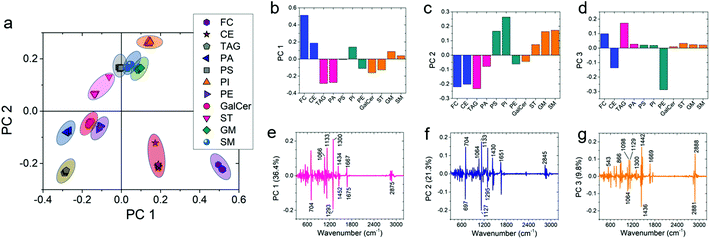
Furthermore, we utilized one-way multivariate statistical analysis coupled with the PCA approach to separate different populations of lipids. PCA analysis was performed on the 11 most common lipids found in brain tissue. Fig. 4a shows the PCA scatterplot results obtained from the first-derivative of the Raman data in various data sets of lipid spectra. The 95% confidence ellipse of the clustered group PCA distribution is also shown in the plots. The PCA analysis of the raw data did not cluster different classes of lipids separately and overlap among classes can be seen (Fig. S12a†). However, after normalization, smoothing, and taking the 1st derivative of the data, considerable separation among different lipid clusters was observed in the PCA results (Fig. S12b–d†). Meanwhile, the second-derivative did not further improve separation of lipid clusters. Therefore, it can be concluded that the first-derivative provides the best PCA results for our data set (Fig. 4a).
To include more variance among the data sets, PC 3 of the first-derivative was also calculated to plot the 3D PCA result (Fig. S12e†). Fig. 4b–d show the PC values of first-derivative for different lipids. Each bar shows the mean value of the data. By performing ANOVA on the PC 1 data set, it can be seen that the PC 1 value for every lipid is significantly different (P < 0.001) from the other lipids (see Table S2†). Similarly, the same significant difference (P < 0.001) was also observed for the PC 2 data set except for phosphatidylserine vs. sphingomyelin, phosphatidylserine vs. ganglioside, and ganglioside vs. sphingomyelin (see Table S2†). Furthermore, the PC 3 data also showed significant differences (P < 0.001) for most of the lipids (see Table S2†). The corresponding loading plots of PC 1, PC 2, and PC 3 show the major peaks responsible for the clustering of the data set (Fig. 4e–g). The percentage variance described by PC 1, PC 2, and PC 3 was 36.4%, 21.3%, and 9.8%, respectively. Analysis of the loading plots showed that the prominent Raman peaks of the lipids show the most variance at 704 (characteristic band for cholesterol), 1133 (acyl chain, palmitic and fatty acids), 1300 (lipid CH2 twist mode) and 1293 (methylene) cm−1 peak positions. As a result, it can be concluded that Raman spectra of the most abundant lipids in the brain are statistically different from each other. Therefore, we can use statistical methods to compare the lipid concentration in control and PTSD samples.
3.4. Raman identifies differences in lipid signature in PTSD and control brain tissues
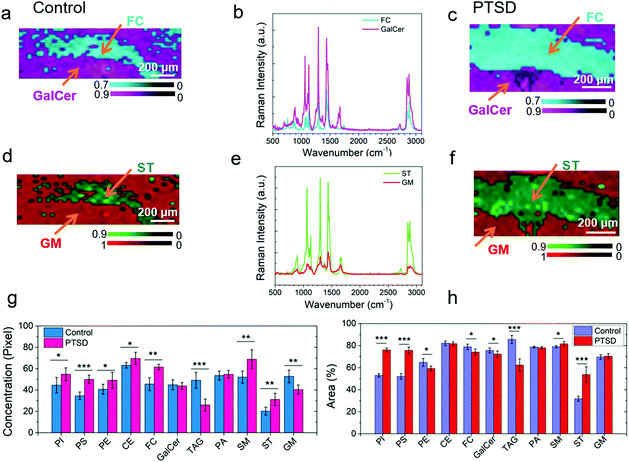
In order to compare the amount of lipid distribution in control and PTSD samples, we used direct classical least squares analysis (DCLS) method.58,59 According to this model, the mixture spectrum can be modeled as a linear mixture of various reference spectra. The DCLS model is useful when reference spectra are available from standard samples. Accordingly, we first acquired the Raman signal from the PVT region of both control and PTSD specimens. Afterwards, the Raman spectra of standard lipids (Fig. 3a–e) were used to quantify each lipid in terms of relative concentration and distribution. Fig. S13, S14 and S15† illustrate the Raman maps of eleven commonly found lipids in the PVT region of the rat brain (the bars beside each image indicate the value of the correlation coefficient between the corresponding lipid and brain spectra). After plotting the respective maps for each lipid, ImageJ software was used to analyze the relative concentration and distribution of lipids. To this end, the intensity value of brightness in each pixel and also distribution of pixels are needed. The intensity values give the relative concentration change of each lipid. The distribution of pixel intensity describes the distribution of lipids in the PVT region. For a tangible example, comparative Raman maps of cholesterol, galactocerebroside, sulfatide and ganglioside are shown in Fig. 5a–f. As can be seen, the relative concentration of cholesterol and sulfatide is greater in the PTSD sample compared to the control. In order to quantify the relative concentration values, we used the pixel value which is a number between 0–255. The value 0 indicates the minimum brightness and 255 indicates the maximum brightness of a pixel. These values were obtained from 8-bit images by means of ImageJ software. Fig. S16† shows the pixel value distributions of different lipids in control and PTSD samples. The relative shifts of the peaks to the right (higher pixel values), indicate an increase in the relative concentration of lipids. Therefore, we can acquire the weighted mean value for each lipid from this diagram that directly indicates the concentration changes of different lipids.Fig. 5g shows the acquired weighted mean values for different lipids in PTSD and control samples. Accordingly, the relative concentration of phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, cholesteryl palmitate, cholesterol, sphingomyelin and sulfatide experienced an increase by 23%, 44.6%, 20.6%, 10.2%, 35%, 32.2%, and 54% in the PTSD sample, respectively. On the other hand, the relative concentration of glyceryl tripalmitate and ganglioside decreased by 47.1% and 23.3%, respectively, relative to control tissue. Meanwhile, galactocerebroside and phosphatidic acid did not change noticeably. The stress response is followed by the release of corticosterone.44 This hormone, which is derived from cholesterol, mediates the pathologic responses of severe stress by binding to intercellular receptors. It is believed that the concentration of neutral lipids (cholesteryl palmitate and cholesterol) in the brain is correlated with the level of corticosterone.60 Therefore, a higher concentration of cholesterol indicates a higher level of corticosterone in brain. Accordingly, the changes in lipid concentration have numerous impacts on the physiological mechanisms of stress-related disorder. Changes in the concentration of phospholipids (phosphatidylinositol, phosphatidylserine and phosphatidylethanolamine) and sphingolipids (sphingomyelin, ganglioside, glyceryl tripalmitate and sulfatide) also reveal the alteration of phospholipase A2 (PLA2) which is associated with inflammatory processes in the brain.44,61
Apart from the relative concentration of lipids, we have also analyzed the areal distribution of lipids in the PVT region. The areal distribution is given as the percentage of occupied area by each lipid in the PVT region (Fig. 5h). By comparing the results shown in Fig. 5g and h, it can be observed that for some lipids (e.g. cholesterol and ganglioside), the distribution percentage of lipid does not follow the trend of relative concentration. For example, for cholesterol, the lipid areal distribution decreases slightly despite the increase in relative concentration of the lipid. Similarly, for ganglioside the distribution percentage is almost constant while the relative concentration is lower. The reason for the differences is due to the fact that we are not taking into account the pixel intensity values while calculating the areal distribution percentage. In other words, we are using a binary logic for distribution percentage to choose whether a pixel is ON (particular lipid is present) or OFF (lipid is absent). Therefore, the relative concentration and areal distribution should be utilized side by side in order to achieve a fair judgment of lipidome alterations in the brain tissues.
3.5. MALDI-MS imaging of brain tissue
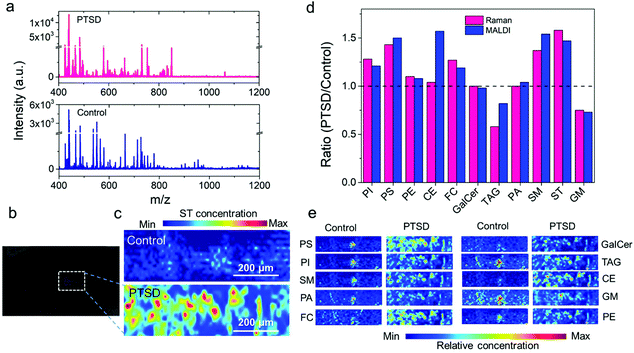
To validate the Raman imaging results, we utilized MALDI mass spectrometry imaging (MALDI-MSI) as a conventional lipidomic study method to elucidate the role of lipids in PTSD. The obtained lipidome profile is affected by the choice of ionization mode.62,63 Since a wide range of lipids can be ionized in positive mode, its widely used for lipidomics (Fig. S17†).62 However, phospholipids such as PI, PS, and PA yield better results in negative ionization mode.64 Although we used both modes, only the MALDI images obtained in positive mode are shown here. The MALDI-MS spectra obtained in the positive ion mode from the PVT region of the control and PTSD brain tissue sample are shown in Fig. 6a. The comparison of positive and negative ion mode spectra is shown in the ESI Fig. S18.† The MS spectra display both low molecular weight (400–650 Da) and high molecular weight (700–1200 Da) lipid species. Positive ion mode also allowed observation of low molecular weight lipids in the control brain tissue such as lysophosphatidylcholines (LPC O-15:1, m/z 466.3), which are generally masked by the signal from the matrix.65 The intense signal at m/z 663.4 for the control brain sample corresponds to PA 30:3;O3. Other observed species were phosphatidylglycerols (PG 28:2, m/z 663.4). For the PTSD sample, the intense mass peaks correspond to fatty acyls such as N-acyl taurines (NAT 20:4;O, m/z 466.2), glycerophospholipids (PA 39:8, PG 33:3, PA 37:5, PG 31:0, PA O-38:5, PG O-31:1, PG 30:1, m/z 731.5; PC O-42:5, PC 41:5, PE 44:5, PC O-40:2, m/z 850.7), glycerolipids such as di(acyl|alkyl)glycerols (DG 42:8, m/z 731.5; DG 44:3, m/z 753.6), tri(acyl|alkyl)glycerols (TAG 43:3, m/z 753.6), and sphingolipids such as hexosyl ceramides (HexCer 42:1;O3, m/z 850.7).Fig. 6b shows the bright field optical image of the MALDI-MS sample. The distribution of each lipid is shown in Fig. 6c and e (also see Fig. S19†). We can observe the pathological lipid changes in the PVT region of the brain due to PTSD. For example, the positive ion image of sulfatides (ST 38:1, m/z 890.5; Fig. 6c) showed an accumulation of sulfatides near the PVT of the PTSD rat. A similar elevated level of ST after traumatic brain injury was observed in an earlier study.66Fig. 6d shows a comparison of the Raman and MALDI image analysis results. Fig. 6d shows the ratio of PTSD to control calculated from the Raman images using the mean value of pixel intensity (pink bars) for different lipids. The corresponding PTSD to control ratio calculated from each MALDI-MS image (blue bars) is also compared with the Raman ratio. The ratio, R = 1 indicates no change, R > 1 indicates increase, and R < 1 indicates decrease of specific lipids in PTSD samples compared to the control samples. Accordingly, an increase in the relative concentration value of phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylethanolamine (PE), cholesterol (FC), sphingomyelin (SM) and sulfatide (ST) is observed by both Raman and MALDI imaging methods in the PTSD sample compared to the control. Moreover, the decrease in the ganglioside (GM) and glyceryl tripalmitate (TAG) in the PTSD sample was also confirmed by the MALDI images. No significant changes were observed for galactocerebroside (GalCer) and phosphatidic acid (PA) by any of the methods. Only cholesteryl palmitate and glyceryl tripalmitate displayed a difference in its relative concentration between MALDI detection and Raman detection. Therefore, the majority of the detected signals in MALDI are in agreement with the Raman imaging analysis.
Fig. 6e shows representative MALDI-MS images of selected lipid species in the control and PTSD brain samples. The images were constructed using the following mass peaks: phosphatidylserine (PS 34:2, m/z 760.5; PS 39:0, m/z 834.6; PS 39:7, m/z 842.5; PS 40:1, m/z 868.6; PS 43:4, m/z 882.6), phosphatidylinositol (PI 31:3, m/z 829.4; PI O-35:2, m/z 857.6; PI O-37:3, m/z 883.6; PI O-37:2, m/z 885.6), sphingomyelin (SM 34:2-O2, m/z 723.5; SM 33:0-O2, m/z 729.5; SM 34:1-O2, m/z 741.5; SM 36:2-O2, m/z 767.5), phosphatidic acid (PA O-31:0, m/z 643.5; PA O-33:1 & PA 32:1, m/z 647.5; PA O-35:3, m/z 671.5; PA 35:5, m/z 719.4), cholesterol (FC 24:5, m/z 401.2; FC 27:1-O, m/z 409.3; FC 26:2-O3, m/z 425.3; ST 27:1-O, m/z 425.3), galactocerebroside (GalCer 34:1-O2, m/z 722.5; GalCer 40:2-O2, m/z 804.6; GalCer 40:1-O2, m/z 806.6; GalCer 41:2-O2, m/z 834.6), triacylglyceride (TAG 50:9, m/z 817.6; TAG 52:9, m/z 845.7; TAG 50:2, m/z 869.7; TAG 52:3, m/z 895.7; TAG 58:13, m/z 921.7), cholesterol ester (CE 18:3, m/z 669.6; CE 20:5 & CE 18:2, m/z 671.6; CE 22:0, m/z 731.7; CE 24:1, m/z 757.7), ganglioside (GM or Hex(3)-HexNAc-KDN-Cer 36:1-O2, m/z 1543.8; GM 36:1-O2, m/z 1562.9; GM or Hex(4)-HexNAc-Fuc-Cer 36:1-O2, m/z 1563.9), and phosphatidylethanolamine (PE 36:6, m/z 736.5; PE O-36:6, m/z 744.5; PE O-37:1, m/z 746.6; PE 37:6, m/z 750.5; PE 38:7, m/z 762.5; PE O-36:3, m/z 766.5; PE O-37:1, m/z 768.6; PE 40:7, m/z 790.5; PE 39:0, m/z 790.6; PE 40:6, m/z 792.6; PE 37:3, m/z 794.5; PE O-39:2, m/z 794.6).
The membrane lipid PI plays an important role in the signal transduction.67 Although experiments performed in a different region of the brain (prefrontal cortex, PFC, and hippocampus), Oliveira et al. also reported an increase of PI in the rat brain due to chronic stress.20 The increase of other phospholipids such as PE and PS reported in our study are supported by other literature observations using neurodegenerative disease models.68 In addition to cholesterol and phospholipids, sphingolipids are the most common membrane lipids in the brain.67,69 Sphingolipids such as gangliosides are implicated in brain development, memory formation as well as synaptic transmission.70 Our results show a decrease in the ganglioside concentration in PTSD samples, which is supported by the observations by Martín et al. and Kracun et al. using the brain tissue of human subjects.71,72 Further study by Oliveira et al. showed that the alteration in lipid levels in the brain is area dependent. They observed an increase of the PI in the hippocampus and a decrease in PE in the PFC, but no changes in the phospholipid levels in the amygdala or cerebellum.73 These findings suggest that lipidome analysis should be performed in specific areas of the brain for meaningful comparisons. Although the lipid distribution in the brain is dynamic and complex, it is suggested to play some role in depression and anxiety disorder.42 The knowledge gained from this study may provide lipid-based targets for disease prevention and treatment.
4. Conclusion
In this study, we have evaluated the application of Raman spectroscopy and imaging for the quantitative analysis of lipid concentrations in paraventricular thalamic nucleus (PVT) of post-traumatic stress disorder (PTSD), and control brain tissues. Raman spectroscopy provided a new tool to non-invasively monitor the lipid changes in the brain tissue. Combining with histology and MALDI mass spectrometry imaging, we have performed a parallel study with Raman imaging and multivariate data analysis to validate the distinguishing of different lipids in brain tissue. Furthermore, we used a direct classical least squares analysis approach for rendering Raman maps and imaging the lipid concentration in the PVT region. By means of this technique and various image processing methods, we have demonstrated the relative alteration of lipids in the PVT region before and after inducing PTSD. Our results show a relative increase in the concentration of phosphatidylinositol (28%), phosphatidylserine (43%), phosphatidylethanolamine (11%), cholesteryl palmitate (4%), cholesterol (27%), sphingomyelin (37%) and sulfatide (58%) in the PTSD sample. Meanwhile, a relative decrease in the concentration of glyceryl tripalmitate (71%) and ganglioside (32%) was observed. However, the relative concentration of galactocerebroside and phosphatidic acid did not change noticeably in the PTSD samples compared to the control. The higher relative concentration of cholesterol and cholesteryl palmitate is directly related to the level of corticosterone.Changes in the concentration of phospholipids and sphingolipids are associated with inflammatory processes in the brain by changing the level of phospholipase A2 (PLA2). Similar Raman imaging methods can be applied to other regions of brain and other types of brain disorders. The Raman scattering-based label-free method could open new ways to perform lipidomic studies on cells and tissue with high spatial resolution for fast and non-destructive analysis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Raman experiments were performed at LSU's Shared Instrumentation Facility (SIF). Mass spectrometry experiments were performed at the LSU Mass Spectrometry Facility (MSF). MRG acknowledges LSU start-up fund and Louisiana Board of Regents Support Fund (RCS award contract number: LEQSF(2017-20)-RD-A-04). AC is supported by LSU Economic Development Assistantships (EDA). The authors thank Dr Olalekan Ogundele for sample preparation of rat brains and Dr David Burk for H&E staining and imaging of brain tissues.References
- D. Piomelli, G. Astarita and R. Rapaka, Nat. Rev. Neurosci., 2007, 8, 743–754 CrossRef CAS.
- J. K. Prasain, L. Wilson, H. D. Hoang, R. Moore and M. A. Miller, Metabolites, 2015, 5, 677–696 CrossRef CAS.
- A. Naudí, R. Cabré, M. Jové, V. Ayala, H. Gonzalo, M. Portero-Otín, I. Ferrer and R. Pamplona, in International review of neurobiology, Elsevier, 2015, vol. 122, pp. 133–189 Search PubMed.
- D. Cheng, A. M. Jenner, G. Shui, W. F. Cheong, T. W. Mitchell, J. R. Nealon, W. S. Kim, H. McCann, M. R. Wenk and G. M. Halliday, PLoS One, 2011, 6, e17299 CrossRef CAS.
- P. L. Wood, M. D. Filiou, D. M. Otte, A. Zimmer and C. W. Turck, Schizophr. Res., 2014, 159, 365–369 CrossRef.
- J. A. Hamilton, C. J. Hillard, A. A. Spector and P. A. Watkins, J. Mol. Neurosci., 2007, 33, 2–11 CrossRef CAS.
- E. H. Seeley and R. M. Caprioli, Trends Biotechnol., 2011, 29, 136–143 CrossRef CAS.
- B. Brügger, Annu. Rev. Biochem., 2014, 83, 79–98 CrossRef.
- J. Soltwisch, H. Kettling, S. Vens-Cappell, M. Wiegelmann, J. Müthing and K. Dreisewerd, Science, 2015, 348, 211–215 CrossRef CAS.
- M. Jermyn, K. Mok, J. Mercier, J. Desroches, J. Pichette, K. Saint-Arnaud, L. Bernstein, M.-C. Guiot, K. Petrecca and F. Leblond, Sci. Transl. Med., 2015, 7, 274ra219 Search PubMed.
- J. M. Surmacki, L. Ansel-Bollepalli, F. Pischiutta, E. R. Zanier, A. Ercole and S. E. Bohndiek, Analyst, 2017, 142, 132–139 RSC.
- S. Ohira, H. Tanaka, Y. Harada, T. Minamikawa, Y. Kumamoto, S. Matoba, H. Yaku and T. Takamatsu, Sci. Rep., 2017, 7, 42401 CrossRef CAS.
- H. Yang, C. Zhao, R. Li, C. Shen, X. Cai, L. Sun, C. Luo and Y. Yin, Analyst, 2018, 143, 2235–2242 RSC.
- M. Haifler, I. Pence, Y. Sun, A. Kutikov, R. G. Uzzo, A. Mahadevan-Jansen and C. A. Patil, J. Biophotonics, 2018, 11, e201700188 CrossRef.
- C. J. Saatkamp, M. L. de Almeida, J. A. M. Bispo, A. L. B. Pinheiro, A. B. Fernandes and L. Silveira, J. Biomed. Opt., 2016, 21, 037001 CrossRef.
- K. Czamara, K. Majzner, M. Z. Pacia, K. Kochan, A. Kaczor and M. Baranska, J. Raman Spectrosc., 2015, 46, 4–20 CrossRef CAS.
- H. Wu, J. V. Volponi, A. E. Oliver, A. N. Parikh, B. A. Simmons and S. Singh, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3809–3814 CrossRef CAS.
- S. Huang, R. Pandey, I. Barman, J. Kong and M. Dresselhaus, ACS Photonics, 2018, 5, 2978–2982 CrossRef.
- A. Rygula, K. Majzner, K. M. Marzec, A. Kaczor, M. Pilarczyk and M. Baranska, J. Raman Spectrosc., 2013, 44, 1061–1076 CrossRef CAS.
- T. G. Oliveira, R. B. Chan, F. V. Bravo, A. Miranda, R. R. Silva, B. Zhou, F. Marques, V. Pinto, J. J. Cerqueira and G. Di Paolo, Mol. Psychiatry, 2016, 21, 80 CrossRef CAS.
- K. Kochan, E. Maslak, C. Krafft, R. Kostogrys, S. Chlopicki and M. Baranska, J. Biophotonics, 2015, 8, 597–609 CrossRef CAS.
- I. P. Santos, P. J. Caspers, T. C. Bakker Schut, R. van Doorn, V. Noordhoek Hegt, S. Koljenović and G. J. Puppels, Anal. Chem., 2016, 88, 7683–7688 CrossRef CAS.
- M. Köhler, S. Machill, R. Salzer and C. Krafft, Anal. Bioanal. Chem., 2009, 393, 1513–1520 CrossRef.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- M. J. Bovin, B. P. Marx, F. W. Weathers, M. W. Gallagher, P. Rodriguez, P. P. Schnurr and T. M. Keane, Psychol. Assess., 2016, 28, 1379 Search PubMed.
- A. De Jongh, P. A. Resick, L. A. Zoellner, A. Van Minnen, C. W. Lee, C. M. Monson, E. B. Foa, K. Wheeler, E. t. Broeke and N. Feeny, Depression Anxiety, 2016, 33, 359–369 CrossRef.
- P. J. Ebenezer, C. B. Wilson, L. D. Wilson, A. R. Nair and J. Francis, PLoS One, 2016, 11, e0160923 CrossRef.
- G. Ronzoni, A. del Arco, F. Mora and G. Segovia, Psychoneuroendocrinology, 2016, 70, 1–9 CrossRef CAS.
- S. Seetharaman, M. Fleshner, C. R. Park and D. M. Diamond, Brain Behav., 2016, 6, e00458 Search PubMed.
- J. Deslauriers, M. Toth, A. Der-Avakian and V. B. Risbrough, Biol. Psychiatry, 2018, 83, 895–907 CrossRef.
- H. Manjoch, E. Vainer, M. Matar, G. Ifergane, J. Zohar, Z. Kaplan and H. Cohen, Behav. Brain Res., 2016, 306, 91–105 CrossRef.
- Y. Levkovitz, D. Fenchel, Z. Kaplan, J. Zohar and H. Cohen, Eur. Neuropsychopharmacol., 2015, 25, 124–132 CrossRef CAS.
- L. M. Shin, S. L. Rauch and R. K. Pitman, Ann. N. Y. Acad. Sci., 2006, 1071, 67–79 CrossRef.
- J. P. Herman and J. G. Tasker, Front. Endocrinol., 2016, 7, 137 Search PubMed.
- S. J. van Rooij, J. S. Stevens, T. D. Ely, R. Hinrichs, V. Michopoulos, S. J. Winters, Y. E. Ogbonmwan, J. Shin, N. R. Nugent and L. A. Hudak, Biol. Psychiatry, 2018, 84, 106–115 CrossRef.
- M. D. Nelson and A. M. Tumpap, CNS Spectr., 2017, 22, 363–372 CrossRef.
- M. A. Penzo, V. Robert, J. Tucciarone, D. De Bundel, M. Wang, L. Van Aelst, M. Darvas, L. F. Parada, R. D. Palmiter and M. He, Nature, 2015, 519, 455 CrossRef CAS.
- D. T. Hsu, G. J. Kirouac, J.-K. Zubieta and S. Bhatnagar, Front. Behav. Neurosci., 2014, 8, 73 Search PubMed.
- G. J. Kirouac, Neurosci. Biobehav. Rev., 2015, 56, 315–329 CrossRef.
- E. Z. Millan, Z. Ong and G. P. McNally, in Progress in Brain Research, ed. T. Calvey and W. M. U. Daniels, Elsevier, 2017, vol. 235, pp. 113–137 Search PubMed.
- K. Zhou and Y. Zhu, Pharmacol. Res., 2019, 142, 70–76 CrossRef.
- C. P. Müller, M. Reichel, C. Mühle, C. Rhein, E. Gulbins and J. Kornhuber, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2015, 1851, 1052–1065 CrossRef.
- G. Z. Réus, H. M. Abelaira, A. L. Maciel, M. A. B. dos Santos, A. S. Carlessi, A. V. Steckert, G. K. Ferreira, S. D. De Prá, E. L. Streck, D. S. Macêdo and J. Quevedo, Metab. Brain Dis., 2015, 30, 545–553 CrossRef.
- T. G. Oliveira, R. B. Chan, F. V. Bravo, A. Miranda, R. R. Silva, B. Zhou, F. Marques, V. Pinto, J. J. Cerqueira, G. Di Paolo and N. Sousa, Mol. Psychiatry, 2015, 21, 80 CrossRef.
- P. R. Zoladz, M. Fleshner and D. M. Diamond, Psychoneuroendocrinology, 2012, 37, 1531–1545 CrossRef CAS.
- O. M. Ogundele, P. J. Ebenezer, C. C. Lee and J. Francis, Neuroscience, 2017, 353, 147–165 CrossRef CAS.
- C. B. Wilson, P. J. Ebenezer, L. D. McLaughlin and J. Francis, PLoS One, 2014, 9, e89104 CrossRef.
- C. B. Wilson, L. D. McLaughlin, P. J. Ebenezer, A. R. Nair, R. Dange, J. G. Harre, T. L. Shaak, D. M. Diamond and J. Francis, Front. Behav. Neurosci., 2014, 8, 256 Search PubMed.
- R. M. Spiers, J. Marzi, E. M. Brauchle, S. E. Cross, R. H. Vaughan, P. A. Bateman, S. J. Hughes, K. Schenke-Layland and P. R. Johnson, Acta Biomater., 2019, 99, 269–283 CrossRef CAS.
- A. Ditta, H. Nawaz, T. Mahmood, M. Majeed, M. Tahir, N. Rashid, M. Muddassar, A. Al-Saadi and H. Byrne, Spectrochim. Acta, Part A, 2019, 221, 117173 CrossRef CAS.
- M. Karas, H. Ehring, E. Nordhoff, B. Stahl, K. Strupat, F. Hillenkamp, M. Grehl and B. Krebs, Org. Mass Spectrom., 1993, 28, 1476–1481 CrossRef CAS.
- M. T. Bokhart, M. Nazari, K. P. Garrard and D. C. Muddiman, J. Am. Soc. Mass Spectrom., 2017, 29, 8–16 CrossRef.
- S. E. Lakhan and K. F. Vieira, Nutr. J., 2008, 7, 2 CrossRef.
- C. Krafft, L. Neudert, T. Simat and R. Salzer, Spectrochim. Acta, Part A, 2005, 61, 1529–1535 CrossRef.
- B. Schrader, Raman, infrared atlas of organic compounds, VCH-Verlag-Ges., 1989 Search PubMed.
- R. L. Frost, M. L. Weier, K. L. Erickson, O. Carmody and S. J. Mills, J. Raman Spectrosc., 2004, 35, 1047–1055 CrossRef CAS.
- K. Ben Mabrouk, T. H. Kauffmann, H. Aroui and M. D. Fontana, J. Raman Spectrosc., 2013, 44, 1603–1608 CrossRef CAS.
- L. Zhang, M. J. Henson and S. S. Sekulic, Anal. Chim. Acta, 2005, 545, 262–278 CrossRef CAS.
- S. E. Bohndiek, A. Wagadarikar, C. L. Zavaleta, D. Van de Sompel, E. Garai, J. V. Jokerst, S. Yazdanfar and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12408–12413 CrossRef CAS.
- D. Danan and H. Cohen, Eur. Neuropsychopharmacol., 2018, 28, S36 CrossRef.
- M. Noponen, M. Sanfilipo, K. Samanich, H. Ryer, G. Ko, B. Angrist, A. Wolkin, E. Duncan and J. Rotrosen, Biol. Psychiatry, 1993, 34, 641–649 CrossRef CAS.
- T. Cajka and O. Fiehn, TrAC, Trends Anal. Chem., 2014, 61, 192–206 CrossRef CAS.
- C. Zhu, A. Dane, G. Spijksma, M. Wang, J. Van der Greef, G. Luo, T. Hankemeier and R. J. Vreeken, J. Chromatogr., A, 2012, 1220, 26–34 CrossRef CAS.
- H. Nygren, P. Pöhö, T. Seppänen-Laakso, U. Lahtinen, M. Oresic and T. Hyötyläinen, LC·GC Eur., 2013, 26, 142–148 CAS.
- I. Kaya, W. Michno, D. Brinet, Y. Iacone, G. Zanni, K. Blennow, H. Zetterberg and J. r. Hanrieder, Anal. Chem., 2017, 89, 4685–4694 CrossRef CAS.
- C. G. Pick, The Consequences of Exposure to Mission-Related Shock Waves Upon Cognitive Potential, TEL-AVIV UNIV, Israel, 2010 Search PubMed.
- S. N. Jackson, H.-Y. J. Wang and A. S. Woods, Anal. Chem., 2005, 77, 4523–4527 CrossRef CAS.
- F. Mesa-Herrera, L. Taoro-González, C. Valdés-Baizabal, M. Diaz and R. Marín, Int. J. Mol. Sci., 2019, 20, 3810 CrossRef CAS.
- M. Jain, S. Ngoy, S. A. Sheth, R. A. Swanson, E. P. Rhee, R. Liao, C. B. Clish, V. K. Mootha and R. Nilsson, Am. J. Physiol.: Endocrinol. Metab., 2014, 306, E854–E868 CrossRef CAS.
- S. Sonnino and V. Chigorno, Biochim. Biophys. Acta, Rev. Biomembr., 2000, 1469, 63–77 CrossRef CAS.
- V. Martín, N. Fabelo, G. Santpere, B. Puig, R. Marín, I. Ferrer and M. Díaz, J. Alzheimer's Dis., 2010, 19, 489–502 Search PubMed.
- I. Kracun, H. Rosner, V. Drnovsek, Z. Vukelic, C. Cosovic, M. Trbojevic-Cepe and M. Kubat, Neurochem. Int., 1992, 20, 421–431 CrossRef CAS.
- T. G. Oliveira, R. B. Chan, F. V. Bravo, A. Miranda, R. R. Silva, B. Zhou, F. Marques, V. Pinto, J. J. Cerqueira and G. Di Paolo, Mol. Psychiatry, 2016, 21, 80–88 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Schematic showing the design of the experiment; paraventricular nucleus of the thalamus (PVT) region inside rat brain; the method used for quantification of Raman maps by means of brightness intensity values; different preprocessing methods performed on control and PTSD datasets; average spectra of control and PTSD samples; full range Raman spectra of all the lipids used in this study; comparison of cholesterol (FC) and cholesteryl palmitate (or cholesterol ester, CE) chemical structures; comparison of glyceryl palmitate (or triacylglyceride, TAG) and phosphatidic acid (PA) chemical structures; chemical structure of different phospholipids (PE, PI, PC, PS); comparison of the Raman spectra obtained from sphingolipids; Raman spectra of sphingolipids in the fingerprint region; PCA analysis of the most common lipids in brain tissue for (a) raw data, (b) normalized data, (c) smoothed data and (d, e) 2nd derivative data; Raman maps of phosphatidylserine, phosphatidylinositol, phosphatidylethanolamine and cholesteryl palmitate in the PVT region of the rat brain; Raman maps of cholesterol, galactocerebroside, glyceryl tripalmitate and phosphatidic acid in the PVT region of the rat brain; Raman maps of sphingomyelin, sulfatide and ganglioside in the PVT region of the rat brain; pixel value distributions of different lipids in control and PTSD samples; comparison of positive and negative ion mode spectra; MALDI signal obtained from rat brain tissue in both positive and negative modes; MALDI images acquired from the PVT region of rat brain for control and PTSD; peak positions of the Raman spectrum for control and PTSD samples; ANOVA with Levene test for homoscedasticity for the Raman spectra of the lipids. See DOI: 10.1039/d0an01615b |
| This journal is © The Royal Society of Chemistry 2021 |