Open Access Article
Open Access ArticleSuperhydrophobic bowl-like SERS substrates patterned from CMOS sensors for extracellular vesicle characterization
Sorina
Suarasan
 a,
Juanjuan
Liu
a,
Meruyert
Imanbekova
a,
Tatu
Rojalin
b,
Silvia
Hilt
c,
John C.
Voss
a,
Juanjuan
Liu
a,
Meruyert
Imanbekova
a,
Tatu
Rojalin
b,
Silvia
Hilt
c,
John C.
Voss
 c and
Sebastian
Wachsmann-Hogiu
c and
Sebastian
Wachsmann-Hogiu
 *a
*a
aDepartment of Bioengineering, McGill University, Montreal, Quebec H3A 0E9, Canada. E-mail: sebastian.wachsmannhogiu@mcgill.ca
bDepartment of Biomedical Engineering, University of California, Davis, CA 95616, USA
cDepartment of Biochemistry & Molecular Medicine, University of California, Davis, CA 95616, USA
First published on 2nd September 2020
Abstract
Using a regular CMOS sensor as a template, we are able to fabricate a simple but highly effective superhydrophobic SERS substrate. Specifically, we decorated the microlens layer of the sensor with 7 μm polystyrene beads to obtain a PDMS patterned replica. The process resulted in a uniform pattern of voids in the PDMS (denoted nanobowls) that are intercalated with a few larger voids (denoted here microbowls). The voids act as superhydrophobic substrates with analyte concentration capabilities in bigger bowl-like structures. Silver nanoparticles were directly grown on the patterned PDMS substrate inside both the nano- and microbowls, and serve as strong electromagnetic field enhancers for the SERS substrate. After systematic characterization of the fabricated SERS substrate by atomic force microscopy and scanning electron microscopy, we demonstrated its SERS performance using 4-aminothiophenol as a reporter molecule. Finally, we employed this innovative substrate to concentrate and analyze extracellular vesicles (EVs) isolated from an MC65 neural cell line in an ultralow sample volume. This substrate can be further exploited for the investigation of various EV biomarkers for early diagnosis of different diseases using liquid biopsy.
Introduction
Surface Enhanced Raman Spectroscopy (SERS) is a powerful approach for the detection of extremely low analyte concentrations in various biological samples.1–4 As with Raman spectroscopy, SERS is capable of accurately recording a unique fingerprint and identifying the molecular composition of samples of interest in real-time, in a non-invasive manner.5,6 SERS is in fact a surface-sensitive technique that relies on the amplification of Raman scattering of molecules that reside near (∼within 5–10 nm) or are adsorbed on a plasmonic active surface.7–9 Therefore, it is a reliable label-free method able to provide the spectral data of molecules.10,11An innovative approach is to combine the properties of a good SERS platform with the capabilities of a superhydrophobic substrate. A superhydrophobic substrate presents a surface that consists of unique micrometric patterns able to trap tiny air pockets underneath the liquid droplet to induce a large contact angle.12,13 This is called the Cassie–Baxter state and is characteristic of water droplets on superhydrophobic surfaces, defined by a solid–water contact angle larger than 150° and a rolling angle smaller than 10°.14 Liquid state samples are consequently repelled by forcing the droplet to preserve its original shape rather than spread all over the surface. Hence, this special substrate is able to concentrate analytes in a small area, dependent only on the initial droplet size.15,16 In order to generate an intense electromagnetic field, sufficient for high Raman signal enhancement, on a superhydrophobic substrate, the easiest method is to create hot-spots between noble metal nanoparticles deposited on solid substrates.
Considering that SERS can be used to differentiate between healthy or diseased cells as well as extracellular vesicles (EVs) isolated from these cells,17–19 SERS is more and more employed as a diagnostic tool for early-stage disease detection. EVs are membrane vesicles secreted by all kinds of cells and fall into three major categories: exosomes, microvesicles and apoptotic bodies.20,21 EVs carry RNAs, proteins, and lipids and have a complex function that includes mainly cell to cell communication.22–24 In fact, they possess unique characteristics reflecting the cells of origin and can transport a complex biologically active cargo.25 Also they can be found in different body fluids like blood, saliva, and urine, and are therefore considered as liquid biopsy sources for high efficiency diagnosis of incipient stage diseases.26–28 Unfortunately, conventional EV isolation and purification methods rely on tedious and multistep processes that lead to a very low yield of EVs. This complicates the task of downstream analysis, which usually requires large concentrations of EVs. To overcome this, highly sensitive approaches such as SERS need to be complemented by enrichment methods to increase the concentration of EVs. One way to achieve this is by using superhydrophobic substrates that are both able to concentrate the EVs and provide the material roughness needed for SERS enhancement.
A previous study reported SERS analysis of EVs concentrated using an array of silicon micropillars coated with a thin film of Teflon like polymer to ensure hydrophobicity.17 Sivashanmugan et al.29 developed a plasmonic SERS substrate using silver nanocubes deposited on a gold nanorod array substrate for investigation of exosomes at concentrations 104–105 times lower than those in blood samples. Recently, Yan et al.30 used SERS to analyse exosomes isolated from different biological sources deposited onto a hybrid substrate of a quasi-periodic array of gold pyramids covered with graphene. Lee et al.31 also reported SERS analysis of exosomes after capturing them inside a reduced area of SERS active 3D nanobowls. They proved by simulations that the few trapped EVs are exposed to a quite uniform density of hot-spots inside the Ag coated bowl structures. However, all these studies employ complex methods for substrate fabrication and use large volumes of EV samples for SERS analysis.
In this study, we propose a simple, inexpensive, and highly effective substrate capable of concentrating EVs from at least 10 times lower volumes than previously reported while optimizing SERS enhancement. This substrate may be implemented in the rapid diagnosis of early-stage diseases using non-invasive liquid biopsy.
Here, we used a regular CMOS sensor as a template for our substrate fabrication. CMOS chips are ideal candidates for miniaturization of point of care devices for sample identification, imaging, signal processing, and readout.32–35 However, we leverage here only the existing patterned surface of the CMOS chip decorated with a few, larger 7 μm polystyrene beads. By pouring PDMS on top of this surface, we fabricate, via soft lithography, an innovative PDMS substrate consisting of nano- and microbowl structures. These structures exhibit superhydrophobic properties that enable us to concentrate and trap EVs in bowls.
The in situ grown silver nanoparticles (AgNPs) inside these bowls provide strong electromagnetic field enhancement to the analyte signal. The substrate was further characterized using scanning electron microscopy (SEM) and atomic force microscopy (AFM). The SERS properties and performance of this substrate were then evaluated using 4-aminothiophenol (4-ATP) as a reporter molecule. Subsequently, we analysed EVs isolated by ultracentrifugation from a MC65 neural cell line and were able to record high quality Raman and SERS fingerprint spectra of EV components.
Thus, due to the ability to concentrate the analyte via superhydrophobicity and to achieve SERS enhancement via metallic nanoparticles, this innovative substrate provides an ideal platform to investigate EVs with sample volumes as low as 0.5 μl.
Experimental
Materials
Silver nitrate (>99%), sodium citrate dihydrate (>99%), L-ascorbic acid, and 4-aminothiophenol (97%) were purchased from Sigma-Aldrich. The PDMS (polydimethylsiloxane) Sylgard 184 silicone elastomer kit was purchased from Dow Silicones Corporation. Polystyrene beads with 7 μm diameter were purchased from Corpuscular, Inc. and diluted in Milli-Q water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 v/v dilution) before use. The CMOS sensors (model OV5647) used in this report were purchased from OmniVision Technologies.
1000 v/v dilution) before use. The CMOS sensors (model OV5647) used in this report were purchased from OmniVision Technologies.
Substrate fabrication
To fabricate the SERS substrate, we used a regular CMOS sensor as a template. The original lens and IR filter of the chip were removed to reveal the microlens sensor array of 3670 μm × 2740 μm, containing approximately 5 million lenses that can be replicated into nanobowls. Then, we dropped a 10 μl solution containing ∼1400, 7 μm polystyrene beads on top of the microlens layer of a CMOS chip and placed it in an oven at 60 °C to allow water evaporation. In the meantime, we prepared a PDMS polymer by mixing the elastomer and the initiator from the kit in a ratio of 10:1. PDMS was carefully poured onto the chip surface and cured for 2 hours at 60 °C. After curing, PDMS was peeled off from the chip and used as a Raman substrate. To obtain a SERS substrate, we grew Ag nanoparticles directly onto the patterned PDMS surface. We obtained an efficient SERS substrate by growing in situ AgNPs directly on the patterned PDMS substrate using a method adapted from a previously reported protocol.36 Specifically, we dropped successively 20 μl volumes of 25 mM ascorbic acid (AA), 25 mM sodium citrate and 25 mM silver nitrate (AgNO3) directly onto the substrate surface, gently mixed the solutions by pipetting, and allowed the mixture to react for 5 minutes. Then, we discarded the colloidal AgNPs from the surface and rinsed the substrate with Milli-Q water.EVs isolation and purification
Cell culture supernatant (approximately 100 ml) from the MC-65 neural cell line was collected and centrifuged at 300 × g for 10 min at 4 °C to clear out potentially remaining whole cells. The supernatant was then centrifuged at 2000g for 15 min at 4 °C to pellet dead cells and cell fragments and debris. This step was followed by a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g spin for 30 min at 4 °C to remove larger microvesicles. All the low-speed spins (300–10
000g spin for 30 min at 4 °C to remove larger microvesicles. All the low-speed spins (300–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) were performed using a Beckman Coulter Microfuge 20R centrifuge with a FA361.5 Biosafe rotor. Next, the samples were ultracentrifuged (UC) two times at 120
000g) were performed using a Beckman Coulter Microfuge 20R centrifuge with a FA361.5 Biosafe rotor. Next, the samples were ultracentrifuged (UC) two times at 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C to pellet extracellular vesicles, dispersing in ultrapure water between spins. UC was performed using a Beckman Optima TLX Ultracentrifuge with an SW 28 swinging bucket rotor. The resulting pellets were finally resuspended in up to 100 μl of ultrapure water and stored at −80 °C until use. It is noted that the samples were aliquoted (20 μl) to reduce freeze–thaw cycling.
000g for 70 min at 4 °C to pellet extracellular vesicles, dispersing in ultrapure water between spins. UC was performed using a Beckman Optima TLX Ultracentrifuge with an SW 28 swinging bucket rotor. The resulting pellets were finally resuspended in up to 100 μl of ultrapure water and stored at −80 °C until use. It is noted that the samples were aliquoted (20 μl) to reduce freeze–thaw cycling.
Characterization methods
Scanning electron microscopy (SEM) images were recorded using a Quanta 450 FEG electron microscope operating at an accelerating voltage of 10 kV and magnification of up to ∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in a high vacuum environment.
000 in a high vacuum environment.
Atomic force microscopy (AFM) images of the CMOS sensor and the patterned PDMS substrate were performed using a Bruker MultiMode8 system in contact mode.
Nanoparticle Tracking Analysis (NTA) was carried out using a NanoSight model LM10 (Malvern Panalytical Ltd, UK), equipped with a blue (405 nm) laser and an sCMOS camera. The isolated EVs were thawed to room temperature and typically diluted 500-fold in filtered ultrapure water. Filtered ultrapure water (typically ∼2 ml) was also used to thoroughly flush the NTA tubing to confirm the background to be free of any nanoparticle contamination prior to the next sample addition. 1 ml of each diluted sample was loaded into a single-use syringe and the syringe was placed on an automated syringe pump (Harvard Bioscience, MA, USA) for injection. Three consecutive 30 s videos of each sample under flow conditions with at least 130 particles per frame during each run were recorded at camera level 12. The data were analysed using NanoSight NTA 3.1 software with the detection threshold set to 5 and screen gain 10 to track a statistically relevant number of particles, concurrently minimizing the distorting background artefacts.
Raman and SERS measurements were performed with a WITec Alpha300R Confocal Raman Microscopy system, equipped with a 633 nm laser line and a maximum power of 5 mW at the sample. Maximum laser power was used for spontaneous Raman measurements and reduced laser power was employed for SERS measurements. Single spectra were recorded under a 50× objective (NA 0.8, WD 0.58 mm) and SERS mapping over a 30 × 30 μm area was recorded under a 100× objective (NA 0.9, WD 0.31 mm) corresponding to a spatial resolution (pixel size) of 0.858 μm. This is smaller than the size of the nanobowls (1.38 μm) and the size of the microbowls (up to 7 μm), indicating that we investigate samples within nano- and microbowls.
For EVs characterization, the exposure time for obtaining Raman and SERS spectra was 1 min and 10 s, respectively. For data analysis we used the WITec Project FIVE 5.1.
Results and discussion
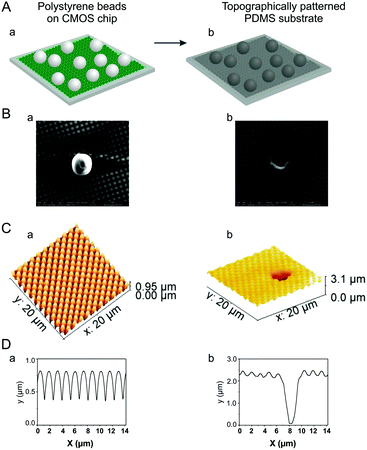
Fig. 1A schematically illustrates the fabrication of the substrate employed for the analysis of EVs. We used an inexpensive, commercial CMOS sensor as a template for substrate fabrication. The CMOS sensor presents a layer of a large area (3670 μm × 2740 μm) with perfectly arranged, close-packed microlenses arranged in a square pattern, with a diameter of 1.4 μm, for a total of approximately 5 million microlenses. This surface was used as a template for the PDMS-based superhydrophobic substrate. PDMS is an inexpensive hydrophobic elastomer highly employed to confer water-resistance to solid surfaces, for the fabrication of flexible substrates and pattern replication since it can easily adjust to every surface irregularity to assist the formation of superhydrophobic surfaces. When used as a mold for PDMS patterned replica, the CMOS surface produces a superhydrophobic substrate that is able to concentrate EVs by preventing the spreading of the EVs solution droplet. The droplet touches the superhydrophobic substrate over a relatively small area and, as the water evaporates, the dried EVs are restricted to a small area on the substrate. In addition, by depositing sparse 7 μm polystyrene beads on top of the sensor surface (Fig. 1A.a and B.a), we are able to obtain a patterned PDMS replica with bowl-like structures representing both CMOS lenses and polystyrene beads. For an easier description, we will further denote the bowls resulting from sensor array structures as nanobowls, and the bigger bowls originating from the polystyrene beads as microbowls.The morphological features of these nano- and microbowl structures are presented in Fig. 1B–D. Since there are only a few microbowls located at a relatively high distance from each other on the substrate, the superhydrophobicity of the substrate will not be disrupted. Additionally, the microbowls improve the concentration of the EVs by trapping them inside in a small area. Due to this property conferred by the unique nanogeometry, the substrate is suitable for the detection of EVs and can be explored as a Raman/SERS substrate.
However, to build an effective SERS substrate, a careful choice of plasmonic materials and structures is needed. Noble metals such as gold and silver, in particular, at the nanometre scale are most commonly used for plasmonic substrate fabrication as they can not only induce a strong electromagnetic field near the surface but are also chemically inert and confer biocompatibility to the substrate.37,38
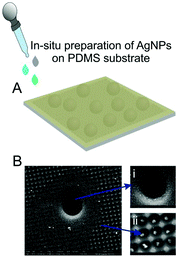
To produce a good SERS substrate, the patterned PDMS was coated with AgNPs via in situ synthesis as described in the Experimental section. In this way, we obtain a large area of uniformly distributed NPs with a high density of plasmonic hot-spots. As shown in Fig. 2B in the SEM images, in situ synthesized AgNPs indeed reside not only in the microbowls (inset (i)) but also in the nanobowls (inset (ii)). Once the analyte EVs come into proximity with these plasmonic active NPs, their Raman signal will be increased as a result of both the enhanced electric-field created by NPs, and the enhanced concentration due to superhydrophobicity.
To better illustrate the uniformity and periodicity of the nanopatterns in the obtained PDMS substrate, we recorded AFM images. Fig. 1C.a shows the AFM image from the sensor surface used as a mold for our substrate. This surface presents an array of perfectly arranged lenses with a 1.4 μm diameter and 0.8 μm depth between them, as measured from the profile shown in Fig. 1D.a. Using this surface, we obtained a large substrate area with perfectly arranged nanobowls with a diameter of ∼1.38 μm and a depth of 440 nm, as measured from the corresponding profile from Fig. 1D.b. The uniformity is determined by the CMOS sensor surface that is precisely copied as the liquid PDMS is able to fill all the spaces between the closely packed microlenses. On the other hand, from spot to spot, the uniform structure of the substrate is disrupted by bigger bowls, as shown in Fig. 1C.b, resulting from the reproduction of the substrate with polystyrene beads on top. Here, due to system limitations of the AFM, we used 3 μm beads to illustrate the size and distribution of particles on the substrate. The bigger bowls formed by these beads exhibit a diameter of 2.7 μm and a depth of 2.75 μm, as measured from the corresponding profile in Fig. 1D.b. By translating this information to the 7 μm beads, we can conclude that we can obtain bowls of approximately 6 μm in diameter that can facilitate the concentration of EVs.
To evaluate the superhydrophobicity, a water droplet was cast on top of the fabricated substrate (Fig. 3A.a). The droplet is nearly spherical in shape due to the repulsive effect of the micropatterned substrate that imposes a large contact angle, indicating a Cassie–Baxter superhydrophobicity regime.39 The superhydrophobic effect of the SERS substrate remains unchanged even after AgNPs are grown inside the nano- and microbowls (Fig. 3A.b). It is important to mention that the size of the droplet footprint after water evaporation is identical to the initial size of the droplet which means that the EVs of interest will be concentrated exclusively onto the restricted area determined by the sample droplet size.
Next, to evaluate the SERS performance of the substrate obtained as described above, 4-ATP was used as a reporter molecule. This molecule has high affinity for binding with AgNPs via the thiol group and forms a uniform layer on the test surface. 10−4 M 4-ATP dissolved in ethanol solution was dropped on the SERS substrate and incubated for 30 minutes to interact with the AgNPs inside the bowls. Subsequently, the surface was thoroughly washed with water such that only molecules bound to AgNPs could be detected.
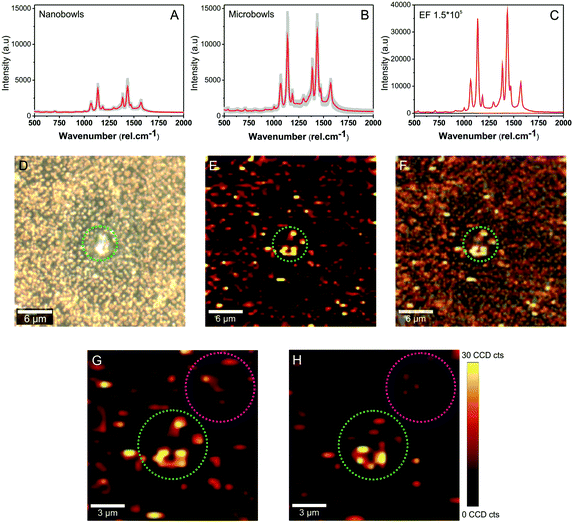
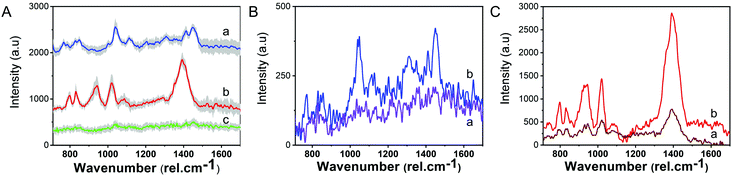
As shown in Fig. 4A and B, high-quality spectra were obtained under 633 nm laser irradiation. The characteristic SERS fingerprint of 4-ATP was obtained in both nano- and microbowls from the substrate. A particularly large enhancement obtained in the microbowls (Fig. 4B) clearly confirms that the analyte is trapped and concentrated inside these structures. The data shown in the figure presents the average spectra (red spectra) and standard deviation (grey area) recorded from more than 10 different spots and show the characteristic peaks of the benzene and amine group. More specifically, they exhibit a typical SERS response of 4-ATP molecules with characteristic bands located at 1075, 1140, 1190, 1303, 1390, 1440 and 1575 cm−1, previously assigned to a1 and b2 vibration modes.40,41
To calculate the substrate SERS enhancement factor (EF) in microbowls, we measured under similar experimental conditions the Raman and SERS spectra of different concentrations of 4-ATP molecules. Note that a higher laser power is required to acquire Raman spectra. Fig. 4C illustrates the obtained spectra. Based on these data, quantitative assessment of the signal enhancement factor (EF) was done and it was determined to be 1.55 × 105.
To further evaluate the distribution of hot-spots that are responsible for the enhancement, SERS maps were recorded by scanning a 30 × 30 μm area including the microbowl structure. Fig. 4D shows the optical image of the SERS substrate and Fig. 4E shows the SERS map that resulted after plotting the intensity of the 1440 cm−1 band of 4-ATP over the scanned region. The intensity distribution of the SERS signal matches the intensity observed in the optical image (Fig. 4F), indicating that AgNPs indeed enhance the Raman signal of 4-ATP molecules. In addition, the SERS map reveals that compared with nanobowls, higher SERS intensities are recorded inside the microbowls, suggesting a higher accumulation of AgNPs and/or a higher concentration of the analyte. Moreover, a detailed analysis of the enlarged areas shown in Fig. 4G and H demonstrates that a strong SERS intensity can be recorded from a large volume of microbowls, further confirming the higher level of analyte entrapment in the bigger bowls of the substrate. Indeed, the green circle in Fig. 4G, corresponding to a microbowl structure, shows a 14 times higher intensity signal compared with the same surface area decorated with nanobowls (pink circle in Fig. 4G). Moreover, as shown in Fig. 4H, this trend can also be observed when we change the focus of the laser beam towards the bottom of the bowls (green and pink circles). However, when we collect individual spectra from the nano- and microbowls, we get approximately 3 times higher intensity for the microbowls (Fig. 4A and B). This is a consequence of surface morphology that helps to concentrate/grow more AgNPs inside larger bowls. In this way, additional hot-spots are formed in the microbowls which can capture and subsequently amplify the signal of analyte molecules.
This interesting result confirms once again not only the growth of AgNPs in the fabricated voids but also the higher molecule concentration in the microbowls.
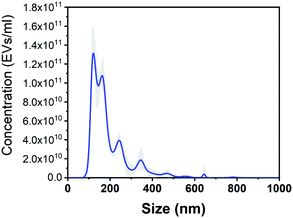
To demonstrate the ability of the substrate to be used for the detection of biological samples of significant interest, we focused next on the concentration and characterization of EVs. EVs were isolated from the MC65 neuronal cell line and isolated by ultracentrifugation, as described in the Experimental section. As determined by the NTA analysis (Fig. 5), EVs have sizes ranging mainly from 100 nm to 200 nm and a concentration of 1.4 × 1011 EVs per millilitre.
Fig. 6A.a presents the Raman spectrum of EVs recorded under 633 nm laser excitation. The Raman spectra exhibit the specific peaks of EVs known to arise from their primary chemical constituents: protein, lipids, and nucleic acids. Specifically, peaks located at 830, 1038 and 1443 cm−1 are attributed to the vibrations from lipids,17,31,42 the peaks arising at 820, 920 and 1211 cm−1 are coming from proteins,10,31,43 while the 786 and 852 cm−1 bands originate from nucleic acid and tyrosine ring breathing modes of the DNA/RNA bases.31,44,45
Using the described SERS substrate, we recorded SERS signals from EVs (Fig. 6A, curve b) using ∼1 mW laser power. SERS spectra acquired exhibit similar features; however, the peaks show higher intensities and are in some cases shifted with respect to the Raman position due to their interaction with the plasmonic substrate. To explain this result, we hypothesise that EVs come in contact with the hot-spots created between AgNPs attached to the substrate in different orientations leading to selective enhancement of specific Raman vibrations of molecules or molecular groups in closer vicinity to the hot-spots.42 We note that no Raman signal could be achieved using the same parameters (Fig. 6A, curve c), which confirm the great enhancement capacity of our SERS substrate.
Another important characteristic of a good, reliable SERS biosensing platform is the reproducibility of the signal in different regions of the substrate. Fig. 6A presents an average collection of individual SERS spectra of EVs from more than 10 different spots within the substrate. The standard deviation is shown by the grey area in the spectra and the coefficients of variation are 10.67% and 23.87% for the 1390 cm−1 peak of EVs in nanobowls and microbowls, respectively).
As mentioned before for the 4-ATP reporter molecule, the enhancement factor in the microbowls is approximately 3 times higher than that in the nanobowls. Higher enhancement can also be observed for EVs in the microbowls, as shown in Fig. 6B and C. Better EVs spectra are recorded from the inside of the microbowls using both Raman and SERS substrates. In the case of the Raman substrate, the enhancement is coming from the EVs concentration on a restricted area of the substrate and a higher entrapment in the bigger bowls. On the other hand, in the case of SERS, the topography concentration effect is cumulated with an additional SERS effect from plasmonic active hot-spots resulting in up to ∼6 times higher amplification of the EV signal (Fig. 6C).
Therefore, our SERS substrate is a good candidate for the analysis of EVs as potential disease biomarkers.
Conclusions
In this study, we successfully combined the ability of our superhydrophobic substrate to concentrate the analyte with the capability of in situ AgNPs growing to obtain a highly efficient SERS substrate for the characterization of analytes at low concentrations. We used a polystyrene bead-decorated CMOS sensor as a template for a PDMS replica capable of concentrating analytes inside the patterned bowls. In these structures, we grew AgNPs that are able to concentrate light into intense electromagnetic fields. We have demonstrated the performance of this SERS substrate using a common Raman reporter molecule, namely 4-ATP, which attaches onto the Ag nanoparticles via the thiol group and forms a monolayer that allows for the quantification of the enhancement ability and reproducibility of the substrate. Microbowls from superhydrophobic SERS substrates act as capturing and concentrating structures for the analyte. They also highly enhance the Raman signal of the analyte due to the hot-spots created at the bottom of the microbowls by the AgNPs. Finally, this substrate was employed to analyse very small volumes of EVs isolated from the MC65 neuronal cell line. This SERS substrate could be further implemented for portable and inexpensive point-of-care diagnostic applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), Discovery Grant RGPIN-2018-05675 (to S. W.-H.). M. I. acknowledges the Bolashak International Scholarship for PhD studies. The authors also thank the Faculty of Engineering at McGill University. J. L. acknowledges the McGill Engineering Doctoral Award (MEDA). T. R. gratefully acknowledges the Sigrid Juselius Foundation, Helsinki, Finland. We also thank Dr Ayyappasamy Sudalaiyadum Perumal for recording EM images shown in Fig. 1 and 2.Notes and references
- A. Merdalimova, V. Chernyshev, D. Nozdriukhin, P. Rudakovskaya, D. Gorin and A. Yashchenok, Appl. Sci., 2019, 9, 1135 CrossRef CAS.
- Y. Zhang, X. Mi, X. Tan and R. Xiang, Theranostics, 2019, 9, 491–525 CrossRef CAS.
- S. Yang, X. Dai, B. B. Stogin and T.-S. Wong, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 268–273 CrossRef CAS.
- A. M. Giovannozzi, F. Rolle, M. Sega, M. C. Abete, D. Marchis and A. M. Rossi, Food Chem., 2014, 159, 250–256 CrossRef CAS.
- L. Guerrini and R. A. Alvarez-Puebla, Cancers, 2019, 11, 748 CrossRef CAS.
- D. Cialla-May, X.-S. Zheng, K. Weber and J. Popp, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- G. Demirel, H. Usta, M. Yilmaz, M. Celik, H. A. Alidagi and F. Buyukserin, J. Mater. Chem. C, 2018, 6, 5314–5335 RSC.
- R. Pilot, R. Signorini, C. Durante, L. Orian, M. Bhamidipati and L. Fabris, Biosensors, 2019, 9, 57 CrossRef CAS.
- J. Liu, M. Jalali, S. Mahshid and S. Wachsmann-Hogiu, Analyst, 2020, 145, 364–384 RSC.
- J. C. Fraire, S. Stremersch, D. Bouckaert, T. Monteyne, T. De Beer, P. Wuytens, R. De Rycke, A. G. Skirtach, K. Raemdonck, S. De Smedt and K. Braeckmans, ACS Appl. Mater. Interfaces, 2019, 11, 39424–39435 CrossRef CAS.
- I. Bruzas, W. Lum, Z. Gorunmez and L. Sagle, Analyst, 2018, 143, 3990–4008 RSC.
- F. Gentile, M. L. Coluccio, A. Accardo, M. Asande, G. Cojoc, F. Mecarini, G. Das, C. Liberale, F. De Angelis, P. Candeloro, P. Decuzzi and E. Di Fabrizio, Microelectron. Eng., 2011, 88, 1749–1752 CrossRef CAS.
- J. Zhi and L.-Z. Zhang, Sci. Rep., 2017, 7, 1–12 CrossRef CAS.
- H. M. Ali, M. A. Qasim, S. Malik and G. Murtaza, Heat Transf. – Models Methods Appl., 2018, 283 Search PubMed.
- A. Accardo, L. Tirinato, D. Altamura, T. Sibillano, C. Giannini, C. Riekel and E. Di Fabrizio, Nanoscale, 2013, 5, 2295–2299 RSC.
- X. Ma, L. Jiang, X. Li, B. Li, J. Huang, J. Sun, Z. Wang, Z. Xu, L. Qu, Y. Lu and T. Cui, Microsyst. Nanoeng., 2019, 5, 1–10 CrossRef CAS.
- L. Tirinato, F. Gentile, D. Di Mascolo, M. L. Coluccio, G. Das, C. Liberale, S. A. Pullano, G. Perozziello, M. Francardi, A. Accardo, F. De Angelis, P. Candeloro and E. Di Fabrizio, Microelectron. Eng., 2012, 97, 337–340 CrossRef CAS.
- J. Park, M. Hwang, B. Choi, H. Jeong, J. Jung, H. K. Kim, S. Hong, J. Park and Y. Choi, Anal. Chem., 2017, 89, 6695–6701 CrossRef CAS.
- T.-D. Li, R. Zhang, H. Chen, Z.-P. Huang, X. Ye, H. Wang, A.-M. Deng and J.-L. Kong, Chem. Sci., 2018, 9, 5372–5382 RSC.
- C.-Y. Kao and E. T. Papoutsakis, Curr. Opin. Biotechnol., 2019, 60, 89–98 CrossRef CAS.
- D. E. Murphy, O. G. de Jong, M. Brouwer, M. J. Wood, G. Lavieu, R. M. Schiffelers and P. Vader, Exp. Mol. Med., 2019, 51, 1–12 CAS.
- S. Saeedi, S. Israel, C. Nagy and G. Turecki, Transl. Psychiatry, 2019, 9, 1–11 CrossRef.
- G. K. Patel, H. Zubair, M. A. Khan, S. K. Srivastava, A. Ahmad, M. C. Patton, S. Singh, M. Khushman and A. P. Singh, in Diagnostic and Therapeutic Applications of Exosomes in Cancer, ed. M. Amiji and R. Ramesh, Academic Press, 2018, pp. 261–283 Search PubMed.
- J. Daßler-Plenker, V. Küttner and M. Egeblad, Biochim. Biophys. Acta, Rev. Cancer, 2020, 188340 CrossRef.
- Y. Zhang, Y. Liu, H. Liu and W. H. Tang, Cell Biosci., 2019, 9, 19 CrossRef.
- J. Carmicheal, C. Hayashi, X. Huang, L. Liu, Y. Lu, A. Krasnoslobodtsev, A. Lushnikov, P. G. Kshirsagar, A. Patel, M. Jain, Y. L. Lyubchenko, Y. Lu, S. K. Batra and S. Kaur, Nanomedicine, 2019, 16, 88–96 CrossRef CAS.
- L. Console, M. Scalise and C. Indiveri, Clin. Chim. Acta, 2019, 488, 165–171 CrossRef CAS.
- S. H. Jalalian, M. Ramezani, S. A. Jalalian, K. Abnous and S. M. Taghdisi, Anal. Biochem., 2019, 571, 1–13 CrossRef CAS.
- K. Sivashanmugan, W.-L. Huang, C.-H. Lin, J.-D. Liao, C.-C. Lin, W.-C. Su and T.-C. Wen, J. Taiwan Inst. Chem. Eng., 2017, 80, 149–155 CrossRef CAS.
- Z. Yan, S. Dutta, Z. Liu, X. Yu, N. Mesgarzadeh, F. Ji, G. Bitan and Y.-H. Xie, ACS Sens., 2019, 4, 488–497 CrossRef CAS.
- C. Lee, R. P. Carney, S. Hazari, Z. J. Smith, A. Knudson, C. S. Robertson, K. S. Lam and S. Wachsmann-Hogiu, Nanoscale, 2015, 7, 9290–9297 RSC.
- S. B. Kim, H. Bae, K. Koo, M. R. Dokmeci, A. Ozcan and A. Khademhosseini, J. Lab. Autom., 2012, 17, 43–49 CrossRef.
- K.-M. Lei, P.-I. Mak, M.-K. Law and R. P. Martins, Lab Chip, 2016, 16, 3664–3681 RSC.
- Y. H. Ghallab and I. Yehea, CMOS Circuits and Systems for Lab-on-a-Chip Applications, Intech Open, 2016, p. 17 Search PubMed.
- S. Carrara, Bio/CMOS Interfaces and Co-Design, Springer, 2013 Search PubMed.
- B. Fortuni, T. Inose, S. Uezono, S. Toyouchi, K. Umemoto, S. Sekine, Y. Fujita, M. Ricci, G. Lu, A. Masuhara, J. A. Hutchison, L. Latterini and H. Uji-i, Chem. Commun., 2017, 53, 11298–11301 RSC.
- P. A. Mosier-Boss, Nanomaterials, 2017, 6, 142 CrossRef.
- T. Xia, H. Luo, S. Wang, J. Liu, G. Yu and R. Wang, CrystEngComm, 2015, 17, 4200–4204 RSC.
- J. Jeevahan, M. Chandrasekaran, G. Britto Joseph, R. B. Durairaj and G. Mageshwaran, J. Coat. Technol. Res., 2018, 15, 231–250 CrossRef CAS.
- D. Zhu, Z. Wang, S. Zong, H. Chen, X. Wu, Y. Pei, P. Chen, X. Ma and Y. Cui, Nanoscale, 2014, 6, 8155–8161 RSC.
- X. Hu, T. Wang, L. Wang and S. Dong, J. Phys. Chem. C, 2007, 111, 6962–6969 CrossRef CAS.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Faraday Discuss., 2004, 126, 141–157 RSC , discussion 169-183.
- P. Zhang, L. Wang, Y. Fang, D. Zheng, T. Lin and H. Wang, Mol. Basel Switz., 2019, 16, 2947 Search PubMed.
- G. Shetty, C. Kendall, N. Shepherd, N. Stone and H. Barr, Br. J. Cancer, 2006, 94, 1460–1464 CrossRef CAS.
- J. W. Chan, D. S. Taylor, T. Zwerdling, S. M. Lane, K. Ihara and T. Huser, Biophys. J., 2006, 90, 648–656 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |