Pyridinium-substituted tetraphenylethylene salt-based photosensitizers by varying counter anions: a highly efficient photodynamic therapy for cancer cell ablation and bacterial inactivation†
Wei
Xiong
a,
Lingyun
Wang
 *a,
Xiaoli
Chen
a,
Hao
Tang
*a,
Xiaoli
Chen
a,
Hao
Tang
 a,
Derong
Cao
a,
Derong
Cao
 a,
Guozhen
Zhang
a,
Guozhen
Zhang
 *b and
Wei
Chen
*b and
Wei
Chen
 *c
*c
aSchool of Chemistry and Chemical Engineering, South China University of Technology, Guangzhou, 510640, China. E-mail: lingyun@scut.edu.cn
bHefei National Laboratory for Physical Sciences at the Microscale, Collaborative Innovation Center of Chemistry for Energy Materials, CAS Center for Excellence in Nanoscience, School of Chemistry and Materials Science, University of Science and Technology of China, Hefei, Anhui 230026, P. R. China. E-mail: guozhen@ustc.edu.cn
cDepartment of Physics, The University of Texas at Arlington, Arlington, Texas 76019, USA. E-mail: weichen@uta.edu
First published on 12th May 2020
Abstract
Cancer and bacterial infection seriously threaten the health of human beings. The development of an image-guided photosensitizer with a “Two-in-One” function that can be simultaneously used for both efficient cancer cell ablation and rapid bacterial inactivation is highly in demand. In this project, we designed and prepared two aggregation-induced emission luminogens (AIEgens) (called TPEPy-I and TPEPy-PF6) with a strong electron push–pull effect. They have a near-infrared (NIR) emission, a high 1O2 quantum yield up to 0.93 and a fluorescence turn-on effect in mitochondria. Upon white light irradiation, the two mitochondria-targeting AIEgens exhibit a highly efficient photodynamic ablation of HeLa cells as well as excellent photodynamic inactivation of both Gram-positive S. aureus and Gram-negative E. coli. The time-dependent density functional theory (TD-DFT) results indicate that compared to TPEPy-PF6, TPEPy-I can easily produce the triplet state that is a prerequisite for 1O2 formation. Moreover, the positive effect of iodide anions gives TPEPy-I a higher photodynamic efficacy in cancer cell ablation and bacterial inactivation as compared with TPEPy-PF6.
1. Introduction
Cancer and bacterial infectious diseases threaten human health. Especially, cancer-associated bacteria seriously reduce the efficiency of cancer treatments.1 It is important and urgent to develop an effective method enabling cancer therapy and killing of pathogenic bacteria simultaneously. Photodynamic therapy (PDT) has emerged as a promising method to combat cancer and pathogenic bacteria due to its distinct advantages, such as noninvasiveness, negligible drug resistance, localized treatment, high spatiotemporal precision and low side effects.2 For instance, Photofrin is the first FDA-approved photosensitizer (PS) and is still widely employed in clinical cancer treatment. Its mechanism is associated with the generation of destructive singlet oxygen (1O2) or other reactive oxygen species (ROS) under light irradiation.3 Specifically, using an emissive PS to realize image-guided PDT is important and attractive, since it conveniently offers diagnosis and treatment integration. However, traditional PSs suffer from quenched fluorescence and reduced ROS production in the aggregated state because of π–π stacking and the resulting aggregation-caused quenching (ACQ) effect, which makes the image-guided PDT unsatisfactory.In contrast, aggregation-induced-emission PSs (AIE-PSs) show enhanced fluorescence and efficient photosensitizing characteristics in aggregate states, which can avoid the ACQ effect of PSs.4–16 In addition, some groups revealed that AIE luminogens (AIEgens) with an electron push–pull effect would favor 1O2 generation.17 To date, AIE-PSs have been widely employed in imaging-guided cancer cell ablation, and bacterial detection and inactivation.18–20 However, some AIE-PSs are ineffective in PDT mediated killing of Gram-negative bacteria.20f,g Particularly in solution, Gram-negative bacteria are protected from extracellularly produced singlet oxygen.21 More importantly, porins at the outer membranes of Gram-negative bacteria can work as ‘molecular sieves’, which hinder PSs of large size from going through the porin channels. These synergistic effects make PSs fail to kill Gram-negative species. Furthermore, bacterial infection as a cause of cancer has been studied, where the induction of chronic inflammation and the production of carcinogenic bacterial metabolites are regarded as possible mechanisms.22 The most specific example of the inflammatory mechanism of carcinogenesis is Helicobacter pylori infection. In addition, skin cancers are easily induced by bacterial infections. Therefore, the development of efficient AIE-PSs that can simultaneously kill cancer cells and bacteria would be appealing.
Recently, Chen's group reported a powerful combination of copper-cysteamine nanoparticles with potassium iodide to inactivate both Gram-positive MRSA and Gram-negative E. coli.23 In that case, iodide anions showed a positive effect by the formation of triiodide ions to enhance the bactericidal ability. Inspired by this, a cationic AIE-PS containing an iodide anion (TPEPy-I, Scheme 1) was designed and synthesized by a simple and straightforward synthetic protocol. For comparison, another AIE-PS containing hexafluorophosphate (TPEPy-PF6, Scheme 1) was introduced for the same study. Both AIE-PSs have some interesting features, such as: (1) the AIEgens comprise a TPE segment (working as a donor), a thiophene fragment (π bridge), and a cationic pyridinium moiety (acceptor), enabling a broad absorption in the visible range and 1O2 formation through a strong charge transfer. (2) Mitochondrion is an important site for energy conversion and the main source of cellular ROS, which plays a crucial role in mediating cell apoptosis. The presence of cationic pyridinium endows the two AIEgens with a function of targeting mitochondria that may improve PDT performance and imaging qualities. Meanwhile, the cationic pyridinium may help AIEgens penetrate the bacterial membranes by electrostatic interaction and improve their water solubility. (3) The D–π–A structure endows AIE-PSs with NIR emissions, which is highly desirable to achieve image-guided PDT with a low background noise. Our observations indicate that both TPEPy-I and TPEPy-PF6 have the “two in one” functions that can be used simultaneously for cancer cell ablation and bacterial inactivation. However, TPEPy-I is more efficient as an AIE-PS than TPEPy-PF6, in which the iodide ions may play some important roles in the photodynamic performance.
2. Results and discussion
2.1 Molecular design and synthesis
As shown in Scheme S1 (ESI†), compound 2 was prepared by a Suzuki reaction between TPE derivative 1 and 5-formyl-2-thiopheneboronic acid. TPEPy-I was obtained by the Knoevenagel reaction between 2 and pyridinium salt. The anion exchange between TPEPy-I and hexafluorophosphate generated TPEPy-PF6, which was expected to avoid the fluorescence quenching effect of iodine atoms. The combination of a strong electron donor–acceptor (D–A) interaction with extended π-conjugation could facilitate intramolecular charge transfer (ICT), therefore resulting in low electronic bandgaps, broad absorption and long emission wavelengths. The two target products were characterized by NMR and HRMS (see Fig. S1–S6 in the ESI† for the details).2.2 Solvatochromism and AIE properties
As shown in Fig. S7 and S8 (ESI†), TPEPy-I and TPEPy-PF6 show obvious solvatochromism, where their absorption and emission spectra are remarkably affected by the polarity of solvents. For example, TPEPy-I in THF has a broad absorption from 350 to 550 nm with the absorption maximum at 453 nm (with a 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 molar extinction coefficient) and an emission at 744 nm with a large Stokes shift of 191 nm. Similarly, TPEPy-PF6 in THF show a large Stokes shift of 169 nm with a maximum absorption at 464 nm (with a 44
000 M−1 cm−1 molar extinction coefficient) and an emission at 744 nm with a large Stokes shift of 191 nm. Similarly, TPEPy-PF6 in THF show a large Stokes shift of 169 nm with a maximum absorption at 464 nm (with a 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 molar extinction coefficient) and an emission maximum at 733 nm. In addition, the emission intensity of the two AIEgens is greatly reduced or even quenched in polar solvents such as DMSO and DMF, suggesting a strong ICT effect in polar media. As compared with TPEPy-PF6, the heavy atom effect of TPEPy-I slightly weakens the emission intensity. The Stokes shift for TPEPy-I and TPEPy-PF6 is much larger than that of the commercial NIR fluorophores (less than 50 nm),19 which can avoid light interference from the excitation light and the self-absorption of emission during biomedical imaging.
000 M−1 cm−1 molar extinction coefficient) and an emission maximum at 733 nm. In addition, the emission intensity of the two AIEgens is greatly reduced or even quenched in polar solvents such as DMSO and DMF, suggesting a strong ICT effect in polar media. As compared with TPEPy-PF6, the heavy atom effect of TPEPy-I slightly weakens the emission intensity. The Stokes shift for TPEPy-I and TPEPy-PF6 is much larger than that of the commercial NIR fluorophores (less than 50 nm),19 which can avoid light interference from the excitation light and the self-absorption of emission during biomedical imaging.
The investigation of the AIE feature in DMSO/water mixtures with different water fractions was carried out and it was found that their emissions were almost totally quenched in DMSO. Their photoluminescence (PL) intensities are gradually enhanced with increasing the fraction of water (Fig. S9, ESI†). The strongest PL intensity was observed at a 90% fraction of water, in which the PL intensities of TPEPy-I and TPEPy-PF6 were enhanced to about 6.5- and 5.9-fold, respectively, as compared with that of the DMSO solutions. DLS revealed that the average hydrodynamic diameters of nanoaggregates that formed in DMSO/water (1/9, v/v) are around 179 and 301 nm for TPEPy-I and TPEPy-PF6, respectively (Fig. S10, ESI†). Meanwhile, TPEPy-I and TPEPy-PF6 also have strong fluorescence in the solid state, and their powder samples show emission peaks at 720 and 718 nm, respectively, (Fig. S11a, ESI†). The emission spectra of TPEPy-I and TPEPy-PF6 solids in PMMA films are shown in Fig. S11b (ESI†) and their absolute PL quantum yields were found to be 16% and 45%, respectively. All these results demonstrate that both TPEPy-I and TPEPy-PF6 are typical AIE-active molecules.
2.3 ROS and singlet oxygen generation
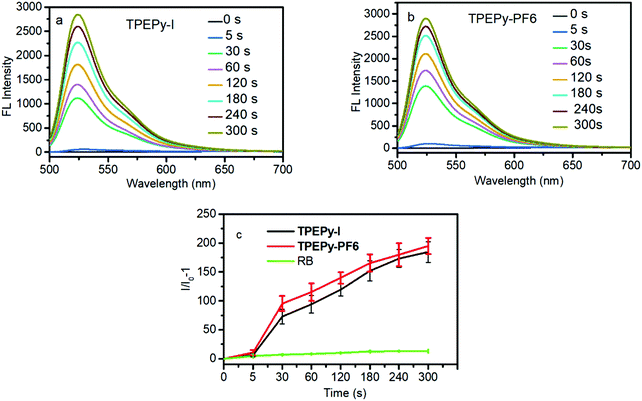
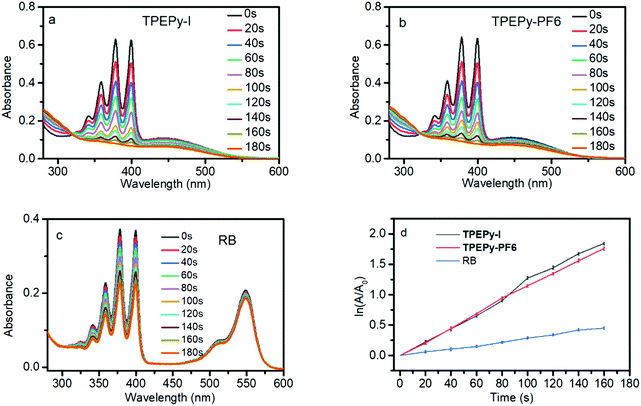
Because the two AIEgens have a strong absorption in the visible light region, they can be activated by visible light for PDT. The ROS generation was evaluated under white light excitation using a commercial ROS indicator, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA), which is capable of emitting a green fluorescence at around 525 nm when oxidized by ROS. As shown in Fig. 1, for the mixture of each AIEgen and H2DCF-DA in aqueous solution, the emission at 525 nm was gradually intensified with increasing irradiation time duration. The 525 nm emission reaches its maximum value after 5 min activation with a 193-fold fluorescence enhancement, suggesting that the two AIEgens have an extremely high capability for ROS generation. Such a change, however, was not observed for each AIEgen or H2DCF-DA alone under the same conditions.It is well known that many types of ROS including type I (radicals or radical ions such as O2˙−, OH˙, and O22−) and type II (1O2) are involved in the PDT process.241O2 is generally identified as the primary species for PDT due to its higher diffusion and stronger interaction with tissues than free radicals.25 Thus, we measured 1O2 production by TPEPy-I and TPEPy-PF6 upon white light irradiation using 9,10-anthracenediylbis(methylene)dimalonic acid (ABDA) as a 1O2 probe. ABDA can selectively undergo oxidation by 1O2 to yield endoperoxid, resulting in a corresponding decrease in its absorbance. As shown in Fig. 2a and b, the characteristic absorption peaks of ABDA decrease promptly with prolonged light irradiation. After 3 minutes exposure to white light, only 40% ABDA degraded in the presence of Rose Bengal (the most widely used PS in PDT), compared to 87% and 85% ABDA consumption in the presence of TPEPy-I and TPEPy-PF6 aggregates, respectively (Fig. 2c and d). When Rose Bengal (RB) was employed as the standard PS (0.75 in water), the 1O2 quantum yields of TPEPy-I and TPEPy-PF6 were calculated to be 0.89 and 0.93, respectively, indicating a high 1O2 generation efficiency by the two AIEgens (Fig. S12, ESI†). Their 1O2 efficiencies are even higher than those of clinically used PSs such as Photofrin (0.28) or Laserphyrin (0.48).26
We further detected ROS formation by TPEPy-I and TPEPy-PF6 inside HeLa cells upon white-light activation using H2DCF-DA. As shown in Fig. S13 (ESI†), an obvious green fluorescence signal was observed inside the cells from H2DCF-DA when it was incubated into the cells with either of the two AIEgens, indicating the formation of ROS inside the cells. The superior ROS and 1O2 generation efficacy suggests that the two AIEgens are ideal PS candidates for PDT applications.
2.4 TD-DFT calculation
As we know, there are inherent relationships between ROS generation and intersystem crossing (ISC). ISC between the singlet and triplet excited states of molecules is a prerequisite for 1O2 generation. It has been recognized that small singlet–triplet gaps appreciably boost ISC rates. The ISC rate (kISC) can be estimated from an empirical formula based on perturbation theory: kISC ∝ |〈Sm|HSO|Tn〉|2/(ΔES–T)2. HSO is the spin–orbit perturbation Hamiltonian and 〈Sm|HSO|Tn〉 is the spin–orbit matrix element (SOCME) between the mth singlet excited state and the adjacent nth triplet excited state. ΔES–T is the singlet–triplet energy gap of the ISC channel of interest, which can play a crucial role in enhancing the ISC rate, as well as the high 1O2 generation efficiency. To gain a mechanistic understanding of the extremely high 1O2 quantum yield of the two AIEgens, we carried out time-dependent density functional theory (TD-DFT) calculations to optimize their excited states structures and calculate ΔES–T using the Gaussian 09 program and compute SOCME under scalar relativistic effects at the level of the zero-order regular approximation (ZORA) as implemented in the ORCA 4.2.1 program (more detail is provided in the ESI†). The conductor-like polarizable continuum model (C-PCM) for water was employed to account for the implicit solvent effect of water in experiments.We firstly found through natural transition orbital (NTO) analysis (Fig. S14, ESI†) that the lowest singlet excited state (S1) of TPEPy+ is mainly a local transition (LT) in the acceptor with a minor participation in the donor. It has a strong light adsorption, indicated by an oscillator strength (f) of 1.94. Similarly, the lowest spin-forbidden triplet state (T1) is also a LT in the acceptor part. Considering Kasha's rule, we assume that all types of transitions (fluorescence, thermal relaxation, and ISC) after excitation of this molecule will start from S1. We found that the third lowest triplet state (T3) is the most adjacent triplet state to S1. They can form effective ISC channels, thanks to the small ΔES–T (S1 → T3: −0.22 eV), albeit the SOCME is small (S1 → T3: 0.33 cm−1). The intrinsically small spin–orbit coupling is typical for pure organic chromophores. To further strengthen the ISC, we replace PF6− with an iodide ion (I−), based on well-accepted experience that halogen atoms can effectively boost ISC of organic molecules because of their prominent relativistic effect. I− and TPEPy+ form a weakly bound complex, with I− being ca. 3.7 Å away from the center of the pyridine ring. As clearly shown in Table 1, the introduction of I− greatly increases the SOCME of the S1 → T3 ISC channel by two orders of magnitude. This is because of the contribution of the 5p orbital of I to S1 and T3 (Fig. S14, ESI†), which enables efficient spin–orbit coupling. The reduction of ΔES–T also helps to boost the ISC from S1 to T3. Furthermore, the T1–S0 gaps of both TPEPy-PF6 and TPEPy-I are sufficiently large (1.17 eV and 1.62 eV) to supply the energy needed to activate 3O2, because previous studies revealed that the molecule should have a T1 state with an energy higher than 0.98 eV to convert ground-state molecular oxygen into excited state singlet oxygen.27 Therefore, compared to TPEPy-PF6, TPEPy-I can more easily generate a triplet state that is a prerequisite for 3O2 sensitization to produce 1O2.
| Species | Transition | ΔE(S–T)/eV | SOCME/cm−1 |
|---|---|---|---|
| Note: for simplicity, TPEPy-PF6 is modeled by its cation part, i.e.TPEPy+, in TD-DFT calculations. All energies are computed using the ωB97XD functional in conjunction with the TZVP basis set for all light elements and SDD pseudopotential for iodine. The implicit solvent effect of water is treated by the C-PCM solvation model. | |||
| TPEPy + | S1 → T1 | 1.27 | 0.04 |
| S1 → T2 | 0.76 | 0.32 | |
| S1 → T3 | −0.22 | 0.33 | |
| S0 → T1 | 1.17 | 0.19 | |
| TPEPy-I | S1 → T1 | 1.30 | 5.45 |
| S1 → T2 | 0.47 | 5.21 | |
| S1 → T3 | −0.03 | 33.18 | |
| S0 → T1 | 1.62 | 0.98 | |
2.5 Mitochondria-specific targeting bioimaging
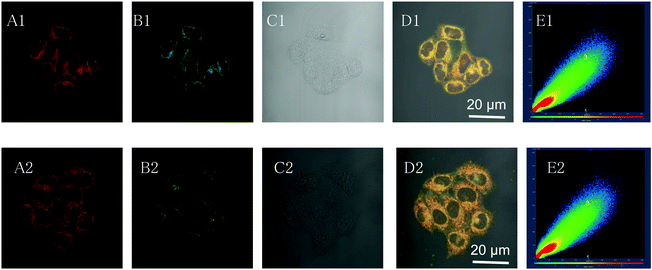
The intracellular distribution of TPEPy-I and TPEPy-PF6 in living HeLa cells was studied by using confocal laser scanning microscopy (CLSM). After treatment with TPEPy-I and TPEPy-PF6 for 1 h, HeLa cells exhibited a strong red fluorescence in the cytoplasm, suggesting a fast permeability of AIEgens into living cells. To realize the specificity of the AIEgens for cell imaging, co-localization experiments were then carried out with MitoTracker Deep Red, which is a commercial probe for mitochondrial imaging. It is interesting to find that TPEPy-I and TPEPy-PF6 can specifically stain the mitochondria. The perfect overlapping of their images with that of MitoTracker Deep Red gives rise to high overlap coefficiencies (0.97) (Fig. 3), indicating their superior specificities for staining mitochondria. The excellent mitochondria-targeting specificity could be attributed to the suitable lipophilicity and cationic property of AIEgens.282.6 PDT in cancer cell ablation
TPEPy-I and TPEPy-PF6 exhibit a negligible dark cytotoxicity at concentrations up to 50 μM with 90% viability of HeLa cells (Fig. S15, ESI†), which means they have a good biocompatibility.In contrast with their low dark cytotoxicity, TPEPy-I and TPEPy-PF6 exhibit a high phototoxicity (Fig. 4). For example, the cell viabilities are found to be 88.8% vs. 93.6%, 70.4% vs. 92.6%, and 58.2% vs. 66.6% at 0.05, 0.1 and 0.5 μM for TPEPy-I and TPEPy-PF6, respectively. This indicates that TPEPy-I is more efficient in cancer cell ablation than TPEPy-PF6 at low concentrations less than 0.5 μM. As the concentration of the two AIEgens reaches 1 μM, there is no actual difference between them; both are effective in cancer cell ablation with the cell viability lower than 5% upon white light irradiation. The dependence of cell viability on the PS dose is in line with the characteristics of PDT. The high potency at low concentration indicates that TPEPy-I and TPEPy-PF6 outperform most conventional PSs, such as porphyrin, chlorin, BODIPY, or their respective derivatives.2 As we know, mitochondrion is the main organelle targeted by PDT, and the mitochondria-specific targeting capability29 and rapid 1O2 generation of TPEPy-I and TPEPy-PF6 make them ideal for photodynamic applications, which cause damage in situ to exert excellent therapeutic efficiency.
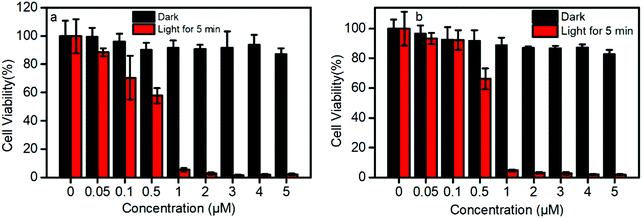 | ||
| Fig. 4 Effect of (a) TPEPy-I and (b) TPEPy-PF6 with and without room-light irradiation on cell proliferation of HeLa cells evaluated by the MTT assay. Light power: 30 mW cm−2. | ||
2.7 Observation of cell death and morphological changes during PDT
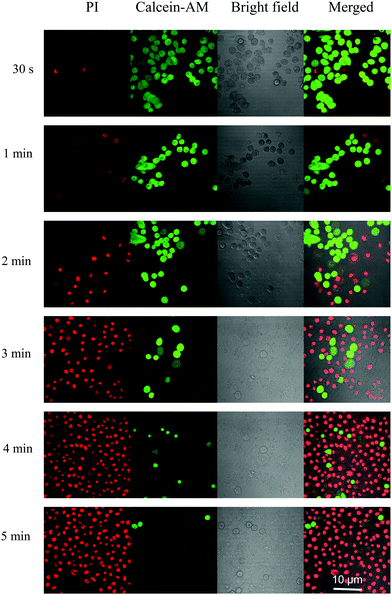
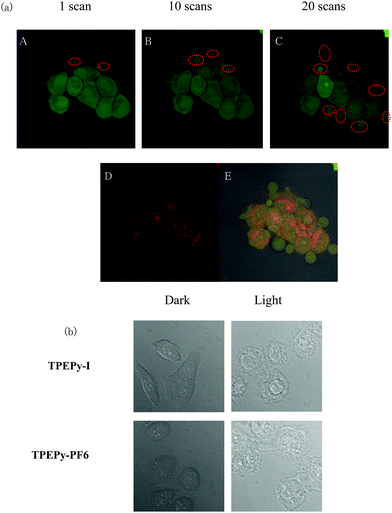
The live- and dead-cell staining experiments were used to further justify the photodynamic efficacy of TPEPy-I and TPEPy-PF6. The representative images are presented in Fig. 5 and Fig. S16 (ESI†), where live and dead cells were stained by calcein AM (green fluorescence) and propidium iodide (red fluorescence), respectively. Under white light irradiation for different time durations, the population of dead cells, as indicated by red fluorescence, increased while the live cell population as indicated by green emission decreased. More cells were destroyed in both cases with the increase of the irradiation duration. After white light irradiation for 5 min, almost all HeLa cells were killed in both cases, indicating the high PDT efficacy of TPEPy-I and TPEPy-PF6.In addition, we found that laser irradiation induced a dramatic change in the cell morphology. Interestingly, bubble generation was present during the CLSM observations, where calcein AM was used to detect cell survival for labeling. Before irradiation, regular and normal cell morphology was observed. However, upon laser stimulation, swelling and blebbing appeared on the cells. More bubbles formed with increasing laser duration (Fig. 6a). As we know, cell morphological changes, such as blebbing, are a sign of cell death.30 When the cells were irradiated by laser for desired time (20 scanning cycles) and then left for 5 min for PI staining, the red emission from PI in the cell nuclei was obviously present due to the PI intercalating with the DNA from the nuclei. These changes can be attributed to the fact that the ROS generated from the AIEgen considerably disrupted the rigidity and permeability of the plasma membrane, causing cancer cell apoptosis and necrosis. The bright field images also confirmed that the shape of the cell changed dramatically after HeLa cells were stained by AIEgens under white light irradiation (Fig. 6b). The shrinkage and fusion of cell walls as well as the appearance of multiple protuberances were found, providing direct and strong evidence for the toxicity on HeLa cells.
2.8 Bacterial elimination
Since TPEPy-I and TPEPy-PF6 are positively charged, it is expected that they can target bacteria through electrostatic interactions.31 The bacterial staining performance of the two AIEgens on various bacteria was observed by using CLSM. Gram-positive S. aureus and Gram-negative E. coli were selected as representatives, because they are negatively charged and facilitate the positively charged AIEgens to stain bacteria. The bright red fluorescence in S. aureus incubated with TPEPy-I and TPEPy-PF6 was readily observed with high contrast relative to the background (Fig. S17, ESI†), indicating that both AIEgens can bind to Gram-positive bacteria efficiently. However, the two AIEgens failed to stain E. coli, where scarcely any fluorescence signals could be detected under CLSM (Fig. S18a, ESI†). The selective imaging behavior of Gram-positive bacteria verses Gram-negative bacteria could be ascribed to their differences in surface structures and chemical components.As shown in Fig. S18b (ESI†), G+ bacteria only have a cytoplasmic membrane covered by a loose and poriferous cell wall, and a crosslinked and thick peptidoglycan layer about 20–80 nm in size with acidic residues in the outer walls. In contrast, G− bacteria have a thinner peptidoglycan layer, which is embedded in the phospholipid bilayer. Meanwhile, G− bacteria possess an additional outer membrane, which performs a barrier function.32 These differences in the two types of pathogens allow AIEgens to penetrate their cell membrane and thus localize in them in different microenvironments. No fluorescence signals detected under CLSM were observed for E. coli, while S. aureus presented bright red fluorescence with high labeling efficiency, indicating the distinct binding affinities of the two AIEgens to the two pathogens. Lacking the protection of an outer membrane, TPEPy-I and TPEPy-PF6 can readily penetrate the cell membrane and enter the inside of S. aureus. The two AIEgens are restricted effectively by the internal environment, which turns on their emission. For E. coli, it is possible that the outer membrane (phospholipid layer) inhibits the insertion of the two AIEgens or the inserted part is easily removed due to weak interaction between E. coli and the two AIEgens. Several references reported selective imaging of G+ over G− with AIEgens.33 The bacterial imaging results indicate that TPEPy-I and TPEPy-PF6 have the potential to selectively recognize Gram-positive bacteria over Gram-negative ones.
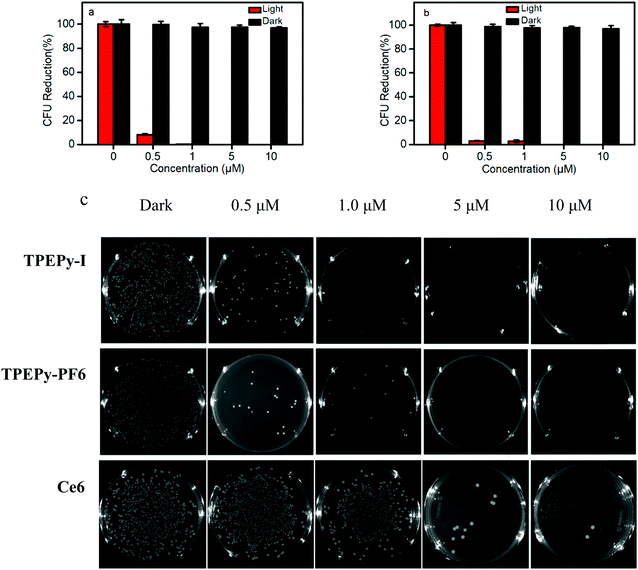
Next, we examined the antibacterial activity of TPEPy-I and TPEPy-PF6 upon white light irradiation. The standard plate colony-counting method was used to determine the percentage of live bacteria. The two AIEgens display a dose-dependent antibacterial efficiency toward both S. aureus and E. coli. Regarding the ability of TPEPy-I and TPEPy-PF6 to kill S. aureus, the inhibition percentage is more than 92% and 97% at 0.5 μM AIEgens, respectively (Fig. 7a and b). When the concentration of the two AIEgens reached 1 μM (Fig. 7c), the inhibition percentage reached nearly 100%. For commercial PSs, the concentration of Toluidine blue O used to kill more than 99.0% S. aureus is up to 80 μM.34 For chlorin e6 (Ce6), the concentration required to kill 99.9% of S. aureus is 40 μM.35 This comparison strongly suggests that TPEPy-I and TPEPy-PF6 are excellent antibacterial PDT agents for S. aureus.
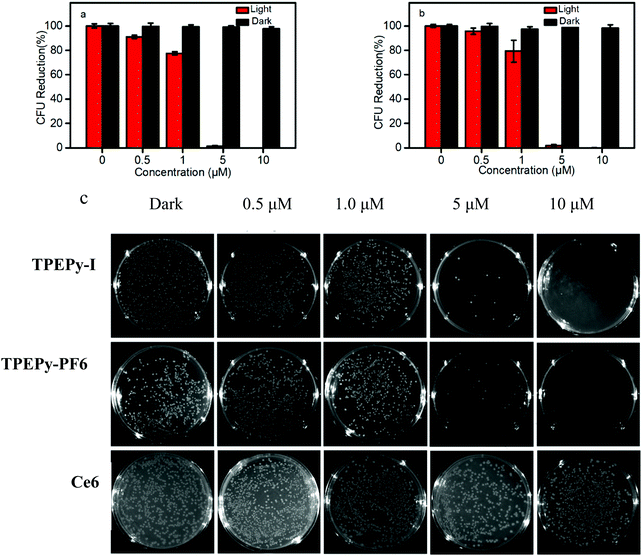
Meanwhile, TPEPy-I exhibits a better antibacterial activity toward E. coli than TPEPy-PF6 at low concentrations less than 1 μM. About 97% E. coli was eliminated at the concentration of 5 μM AIEgens (Fig. 8). The killing effect of TPEPy-I and TPEPy-PF6 on S. aureus is more efficient than that on E. coli because Gram-negative bacteria have an additional protecting layer in the outer membrane. Similar findings have been reported in previous studies.36 Photographs of bacteria cultured on agar plates further confirmed the results mentioned above. Almost no bacterial colony was observed on the agar for S. aureus and E. coli in the presence of 1 and 5 μM AIEgens, respectively, which showed better antibacterial activity than that of commercial Ce6 (Fig. 8c).
Some control experiments were carried out. In the dark or AIEgens alone or light alone, no obvious drop in the survival rate of bacteria was found, which suggests that the potent antimicrobial activity is entirely a consequence of the intrinsic ROS generation induced by AIEgens and white-light irradiation (Fig. S19, ESI†). All results indicate that TPEPy-I and TPEPy-PF6 possess excellent antibacterial activity towards both Gram-positive and Gram-negative bacteria at low PS concentrations compared to reported data (Table S1, ESI†), suggesting that they are excellent broad-spectrum antibacterial agents.
PDT can target both external and internal structures of bacteria, and it does not really require the PSs to enter the bacteria, thus the sterilization mechanism of PDT is different from traditional antibiotics. It has been proposed that the cationic PS can penetrate the outer membrane of Gram-negative bacteria by the “self-promoted uptake pathway” in which the divalent metal cations Ca2+ and Mg2+ are gradually displaced by the PS, and the lipopolysaccharide in the outer membrane permeability barrier is destabilized.37 In our case, the two AIEgens exhibited remarkable 1O2 generation ability (up 93%) under illumination of white light. So, the proposed molecules can provide an efficient bacterial inactivation route toward E. coli. Some white light triggered PDT methods using AIEgens with highly efficient cancer cell ablation and bacterial inactivation have been reported.38
2.9 The mechanism of cancer cell ablation and bacterial elimination
From the results discussed above, TPEPy-I is more efficient in cancer cell ablation and bacterial elimination than TPEPy-PF6. The proposed mechanism can be ascribed to several possibilities. First, TPEPy-I may produce more triplet states in favor of 1O2 formation from the TD-DFT calculations. Second, more ROS can be generated through iodide anions involved a type-I electron-transfer photochemical mechanism. Third, the iodide anion can be oxidized to molecular iodine I2 or I3− by both type-I and type-II ROS.39 The higher generation of toxic species, such as H2O2, triiodide ions, and singlet oxygen from TPEPy-I may be the major killing mechanism for its enhanced cancer cell ablation and bacterial elimination activity. Some groups have reported similar results that the combination of triiodide with H2O2 can promote cell and microbial killing.40The decrease of mitochondrial membrane potential (MMP) is a crucial indicator to assess the dysfunction of mitochondria and it plays an important role in the release of the pro-apoptotic proteins to trigger caspase activation and cell apoptosis.41 The change of mitochondrial membrane potential was determined using JC-10 dye as the indicator, which tends to aggregate with red fluorescence with a high MMP but becomes monomeric with green fluorescence with a low MMP. Therefore, the change of JC-10 fluorescence can be used to assess the status of mitochondria. As shown in Fig. S20 (ESI†), under irradiation from white light, HeLa cells incubated with TPEPy-I and TPEPy-PF6 display enhanced green fluorescence. The ratio of Igreen/Ired in the presence of the two AIEgens was higher than that of light alone, proving that the generated ROS by the two AIEgens injured the cellular mitochondria and resulted in the loss of the MMP.
As we know, cationic fluorescent dyes preferr to stain mitochondria in living cells through electronic interaction due to the negative charge of the mitochondrial inner membrane.42 We assume that TPEPy-I and TPEPy-PF6 could be taken up by cancer cells through endocytosis. Then, the two AIEgens selectively accumulated into the mitochondria, thereby inducing efficient mitochondrial dysfunction and intrinsic cancer cell apoptosis by PDT.
3. Conclusions
In summary, two NIR AIEgens (TPEPy-I and TPEPy-PF6) based on an electron push–pull framework are synthesized and characterized. Due to their AIE backbones with D–A structural units, both the AIEgens show broad absorption in the visible range, while a cationic group is introduced into the molecular design for mitochondria-specific targeting and bacterial membrane anchoring. They can generate ROS efficiently inside cancer cells and bacteria, causing oxidative damage to the mitochondria of cancer cells and cell walls of bacteria. It was demonstrated that low concentrations of AIEgens (1 μM) almost completely kill cancer cells upon white light irradiation. Moreover, benefitting from the enhanced membrane interactions and photosensitizing ability, TPEPy-I and TPEPy-PF6 exhibited efficient antibacterial capability in destroying S. aureus and E. coli at a low concentration (0.5 μM). These results revealed the great potential of these AIEgens with NIR emission, a high yield of ROS generation, and low dark toxicity to serve in image guided PDT.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The support by the China Scholarship Council (201906155012), the National Key Research and Development Program of China (2016YFA0602900), the National Natural Science Foundation of China (21772045), the Natural Science Foundation of Guangdong Province (2016A030311034, 2018B030311008) and Technology Program of Guangzhou (201904010414) is gratefully acknowledged. G. Z. Zhang is grateful to the Supercomputing Center of University of Science and Technology of China for the computing resource. W. Chen would like to acknowledge the distinguished record award in research/creative activity from the University of Texas.References
- B. D. Wallace, H. Wang, K. T. Lane, J. E. Scott, J. Orans, J. S. Koo, M. Venkatesh, C. Jobin, L. A. Yeh, S. Mani and M. R. Redinbo, Science, 2010, 330, 831–835 CrossRef CAS PubMed.
- (a) D. E. J. G. J. Dolmans, D. Fukumura and R. K. Jain, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef CAS PubMed; (b) A. P. Castano, P. Mroz and M. R. Hamblin, Nat. Rev. Cancer, 2006, 6, 535–545 CrossRef CAS PubMed; (c) M. H. Al-Afyouni, T. N. Rohrabaugh, K. F. Al-Afyouni and C. Turro, Chem. Sci., 2018, 9, 6711–6720 RSC; (d) H. Shi, X. Ma, Q. Zhao, B. Liu, Q. Qu, Z. An, Y. Zhao and W. Huang, Adv. Funct. Mater., 2014, 24, 4823–4830 CrossRef CAS; (e) D. Tao, L. Feng, Y. Chao, C. Liang, X. Song, H. Wang, K. Yang and Z. Liu, Adv. Funct. Mater., 2018, 28, 1804901 CrossRef; (f) H. Yuan, H. Chong, B. Wang, C. Zhu, L. Liu, Q. Yang, F. Lv and S. Wang, J. Am. Chem. Soc., 2012, 134, 13184–13187 CrossRef CAS PubMed; (g) Y. Li, Q. Wu, M. Kang, N. Song, D. Wang and B. Z. Tang, Biomaterials, 2020, 232, 119749 CrossRef CAS PubMed; (h) M. Gao and B. Z. Tang, Coord. Chem. Rev., 2020, 402, 213076 CrossRef CAS; (i) C. Sun, H. Zhang, L. Yue, S. Li, Q. Cheng and R. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 22925–22931 CrossRef CAS PubMed; (j) Y. Ding, S. Li, L. Liang, Q. Huang, L. Yuwen, W. Yang, R. Wang and L. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 9980–9987 CrossRef CAS PubMed.
- T. J. Dougherty, C. J. Gomer, B. W. Henderson, G. Jori, D. Kessel, M. Korbelik, J. Moan and Q. Peng, J. Natl. Cancer Inst., 1998, 90, 889–905 CrossRef CAS PubMed.
- (a) X. Zhao, Y. J. Yang, Y. Yu, S. M. Guo, W. X. Wang and S. M. Zhu, Chem. Commun., 2019, 55, 13542–13545 RSC; (b) G. X. Feng and B. Liu, Acc. Chem. Res., 2018, 51, 1404–1414 CrossRef CAS PubMed.
- Z. Y. Liu, H. Zou, Z. Zhao, P. F. Zhang, G. G. Shan, T. K. Kwok. Ryan, J. W. Y. Lam, L. Zheng and B. Z. Tang, ACS Nano, 2019, 13, 11283–11293 CrossRef CAS PubMed.
- N. Alifu, X. Dong, D. Li, X. Sun, A. Zebibula, D. Zhang, G. Zhang and J. Qian, Mater. Chem. Front., 2017, 1, 1746–1753 RSC.
- X. Sun, A. Zebibula, X. Dong, G. Zhang, D. Zhang, J. Qian and S. He, ACS Appl. Mater. Interfaces, 2018, 10, 25037–25046 CrossRef CAS PubMed.
- C. Chen, H. Qu, R. Liu and D. Ding, Adv. Mater., 2019, 1806331 Search PubMed.
- (a) Z. Zheng, T. Zhang, H. Liu, Y. Chen, R. T. K. Kwok, C. Ma, P. Zhang, H. H. Y. Sung, I. D. Williams, J. W. Y. Lam, K. S. Wong and B. Z. Tang, ACS Nano, 2018, 12, 8145–8159 CrossRef CAS PubMed; (b) B. Gu, W. Wu, G. Xu, G. Y. F. Feng, P. H. J. Chong, J. Qu, K. T. Yong and B. Liu, Adv. Mater., 2017, 29, 1701076 CrossRef PubMed; (c) Y. Yang, L. Wang, H. Q. Cao, Q. Li, Y. Li, M. J. Han, H. Wang and J. B. Li, Nano Lett., 2019, 19, 1821–1826 CrossRef CAS PubMed.
- (a) J. Qi, C. Chen, D. Ding and B. Z. Tang, Adv. Healthcare Mater., 2018, 7, 1800477 CrossRef PubMed; (b) C. Chen, X. Ni, S. R. Jia, Y. Liang, X. L. Wu, D. L. Kong and D. Ding, Adv. Mater., 2019, 31, 1904914 CrossRef CAS PubMed.
- H. Gao, X. Zhang, C. Chen, K. Li and D. Ding, Adv. Biosyst., 2018, 2, 1800074 CrossRef.
- X. Ni, X. Zhang, X. Duan, H. Zheng, X. Xue and D. Ding, Nano Lett., 2019, 19, 318–330 CrossRef CAS PubMed.
- D. Wang, M. M. S. Lee, W. Xu, G. Shan, X. Zheng, R. T. K. Kwok, J. W. Y. Lam, X. Hu and B. Z. Tang, Angew. Chem., Int. Ed., 2019, 58, 5628–5632 CrossRef CAS PubMed.
- W. Wu, D. Mao, S. Xu, S. Ji, F. Hu, D. Ding, D. Kong and B. Liu, Mater. Horiz., 2017, 4, 1110–1114 RSC.
- F. Hu, D. Mao, K. Kenry, X. Cai, W. Wu, D. Kong and B. Liu, Angew. Chem., Int. Ed., 2018, 57, 10182–10186 CrossRef CAS PubMed.
- S. Wang, W. Wu, P. Manghnani, S. Xu, Y. Wang, C. C. Goh, L. G. Ng and B. Liu, ACS Nano, 2019, 13, 4742–4751 CrossRef PubMed.
- W. B. Wu, D. Mao, F. Hu, S. D. Xu, C. Chen, C. J. Zhang, X. M. Cheng, Y. Y. Yuan, D. Ding, D. L. Kong and B. Liu, Adv. Mater., 2017, 29, 1700548 CrossRef PubMed.
- S. Xu, Y. Yuan, X. Cai, C.-J. Zhang, F. Hu, J. Liang, G. Zhang, D. Zhang and B. Liu, Chem. Sci., 2015, 6, 5824–5830 RSC.
- (a) T. Maisch, Mini-Rev. Med. Chem., 2009, 9, 974–983 CrossRef CAS PubMed; (b) H. Chen, W. Zhang, G. Zhu, J. Xie and X. Chen, Nat. Rev. Mater., 2017, 2, 1–8 Search PubMed.
- (a) C. Zhou, W. Xu, P. Zhang, M. Jiang, Y. Chen, R. T. K. Kwok, M. M. S. Lee, G. Shan, R. Qi, X. Zhou, J. W. Y. Lam, S. Wang and B. Z. Tang, Adv. Funct. Mater., 2019, 29, 18059864 Search PubMed; (b) R. Hu, F. Zhou, T. Zhou, J. Shen, Z. Wang, Z. Zhao, A. Qin and B. Z. Tang, Biomaterials, 2018, 187, 47–54 CrossRef CAS PubMed; (c) Y. Li, Z. Zhao, J. Zhang, R. T. K. Kwok, S. Xie, R. Tang, Y. Jia, J. Yang, L. Wang, J. W. Y. Lam, W. Zheng, X. Jiang and B. Z. Tang, Adv. Funct. Mater., 2018, 28, 180463242 Search PubMed; (d) M. Jiang, X. Gu, R. T. K. Kwok, Y. Li, H. H. Y. Sung, X. Zheng, Y. Zhang, J. W. Y. Lam, I. D. Williams, X. Huang, K. S. Wong and B. Z. Tang, Adv. Funct. Mater., 2018, 28, 1704589 CrossRef; (e) E. Zhao, Y. Chen, S. Chen, H. Deng, C. Gui, C. W. T. Leung, Y. Hong, J. W. Y. Lam and B. Z. Tang, Adv. Mater., 2015, 27, 4931–4937 CrossRef CAS PubMed; (f) M. M. Kang, C. C. Zhou, S. M. Wu, B. R. Yu, Z. J. Zhang, N. Song, M. M. S. Lee, W. H. Xu, F. J. Xu, D. Wang, L. Wang and B. Z. Tang, J. Am. Chem. Soc., 2019, 141, 16781–16789 CrossRef CAS PubMed; (g) M. M. Kang, T. K. Kwok. Ryan, J. G. Wang, H. Zhang, W. Y. Lam. Jacky, Y. Li, P. F. Zhang, H. Zou, X. G. Gu, F. Li and B. Z. Tang, J. Mater. Chem. B, 2018, 6, 3894–3903 RSC; (h) Q. Y. Li, Y. Li, T. L. Min, J. Y. Gong, L. L. Du, D. L. Phillips, J. K. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, R. T. K. Kwok, C. L. Ho, K. Li, J. G. Wang and B. Z. Tang, Angew. Chem., Int. Ed., 2020, 59, 10184–10188 Search PubMed.
- G. Valduga, G. Bertoloni, E. Reddi and G. Jori, J. Photochem. Photobiol., B, 1993, 21, 81–86 CrossRef CAS.
- J. Parsonnet, Environ. Health Perspect., 1995, 103, 263–268 Search PubMed.
- X. M. Zhen, L. Chudal, N. K. Pandey, J. Phan, X. Ran, E. Amandor, X. J. Huang, O. Johnson, Y. P. Ran, W. Chen, M. R. Hamblind and L. Y. Huang, Mater. Sci. Eng., C, 2020, 110659 CrossRef CAS PubMed.
- (a) D. E. Dolmans, D. Fukumura and R. K. Jain, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef CAS PubMed; (b) J. Moan and Q. Peng, Anticancer Res., 2002, 23, 3591–3600 Search PubMed; (c) N. Zhao, B. Wu, X. Hu and D. Xing, Biomaterials, 2017, 141, 40–49 CrossRef CAS PubMed; (d) S. Xu, Y. Yuan, X. Cai, C.-J. Zhang, F. Hu, J. Liang, G. Zhang, D. Zhang and B. Liu, Chem. Sci., 2015, 6, 5824–5830 RSC.
- O. T. Fackler and R. Grosse, J. Cell Biol., 2008, 181, 879–884 CrossRef CAS PubMed.
- (a) E. Zhao, Y. Chen, H. Wang, S. Chen, J. W. Lam, C. W. Leung, Y. Hong and B. Z. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 7180–7188 CrossRef CAS PubMed; (b) W. Zhang, Y. Huang, Y. Chen, E. Zhao, Y. Hong, S. Chen, J. W. Y. Lam, J. Hou and B. Z. Tang, ACS Appl. Mater. Interfaces, 2019, 11, 10567–10577 CrossRef CAS PubMed; (c) G. Feng, Y. Yuan, H. Fang, R. Zhang, B. Xing, G. Zhang, D. Zhang and B. Liu, Chem. Commun., 2015, 51, 12490–12493 RSC; (d) M. Gao, Q. Hu, G. Feng, N. Tomczak, R. Liu, B. Xing, B. Z. Tang and B. Liu, Adv. Healthcare Mater., 2015, 4, 659–663 CrossRef CAS PubMed.
- (a) C. Schweitzer and R. Schmidt, Chem. Rev., 2003, 103, 1685–1757 CrossRef CAS PubMed; (b) L. P. Silva, A. Núnez-Montenegro, C. M. Magalhães, P. J. O. Ferreira, D. Duarte, P. Gonzalez-Berdullas, J. E. Rodríguez-Borges, N. Vale and J. C. G. Esteves da Silva, Eur. J. Med. Chem., 2019, 183, 111683 CrossRef PubMed.
- (a) N. Li, Y. Y. Liu, Y. Li, J. B. Zhuang, R. R. Cui, Q. Gong, N. Zhao and B. Z. Tang, ACS Appl. Mater. Interfaces, 2018, 10, 24249–24257 CrossRef CAS PubMed; (b) N. Zhao, S. Chen, Y. Hong and B. Z. Tang, Chem. Commun., 2015, 51, 13599–13602 RSC; (c) D. Wang, M. M. S. Lee, G. Shan, R. T. Kwok, J. W. Y. Lam, H. Su, Y. Cai and B. Z. Tang, Adv. Mater., 2018, 30, 1802105 CrossRef PubMed.
- D. Wang, H. Su, R. T. K. Kwok, X. Hu, H. Zou, Q. Luo, M. M. S. Lee, W. Xu, J. W. Y. Lam and B. Z. Tang, Chem. Sci., 2018, 9, 3685–3693 RSC.
- C. Zhu, Q. Yang, L. Liu, F. Lv, S. Li, G. Yang and S. Wang, Adv. Mater., 2011, 23, 4805–4810 CrossRef CAS PubMed.
- (a) X. Li, H. Bai, Y. Yang, J. Yoon, S. Wang and X. Zhang, Adv. Mater., 2019, 31, 1805092 Search PubMed; (b) F. Hu, S. Xu and B. Liu, Adv. Mater., 2018, 30, 1801350 CrossRef PubMed; (c) H. Chen, S. L. Li, M. Wu, K. Kenry, Z. M. Huang, C. S. Lee and B. Liu, Angew. Chem., Int. Ed., 2019, 58, 1–6 CrossRef.
- (a) H. Bai, H. Chen, R. Hu, M. Li, F. Lv, L. Liu and S. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 31550–31557 CrossRef CAS PubMed; (b) Y. Wang, T. S. Corbitt, S. D. Jett, Y. Tang, K. S. Schanze, E. Y. Chi and D. G. Whitten, Langmuir, 2012, 28, 65–70 CrossRef CAS PubMed.
- (a) C. Zhou, M. Jiang, J. Du, H. Bai, G. Shan, R. T. K. Kwok, J. H. C. Chau, J. Zhang, J. W. Y. Lam, P. Huang and B. Z. Tang, Chem. Sci., 2020, 11, 4730–4740 RSC; (b) R. Hu, F. Zhou, T. Zhou, J. Shen, Z. Wang, Z. Zhao, A. Qina and B. Z. Tang, Biomaterials, 2018, 187, 47–54 CrossRef CAS PubMed; (c) M. Kang, R. T. K. Kwok, J. Wang, H. Zhang, J. W. Y. Lam, Y. Li, P. Zhang, H. Zou, X. Gu, F. Li and B. Z. Tang, J. Mater. Chem. B, 2018, 6, 3894–3903 RSC.
- M. Wainwright and K. B. Crossley, J. Chemother., 2002, 14, 431–443 CrossRef CAS PubMed.
- (a) X. Li, H. Bai, Y. Yang, J. Yoon, S. Wang and X. Zhang, Adv. Mater., 2019, 31, 1805092 Search PubMed; (b) F. Hu, S. Xu and B. Liu, Adv. Mater., 2018, 30, 1801350 CrossRef PubMed.
- (a) M. A. Pereira, M. A. F. Faustino, J. P. C. Tome, M. G. P. M. S. Neves, A. C. Tome, J. A. S. Cavaleiro, A. Cunha and A. Almeida, Photochem. Photobiol. Sci., 2014, 13, 680–690 RSC; (b) T. Wu, A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy and D. Kahne, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11754–11759 CrossRef CAS PubMed; (c) H. Ma, Y. Ma, L. Lei, W. Yin, Y. Yang, T. Wang, P. Yin, Z. Lei, M. Yang, Y. Qin and S. Zhang, ACS Sustainable Chem. Eng., 2018, 6, 15064–15071 CrossRef CAS.
- A. Minnock, D. I. Vernon, J. Schofield, J. Griffiths, J. H. Parish and S. B. Brown, Antimicrob. Agents Chemother., 2000, 44, 522–527 CrossRef CAS PubMed.
- (a) N. Zhao, P. Li, J. Zhuang, Y. Liu, Y. Xiao, R. Qin and N. Li, ACS Appl. Mater. Interfaces, 2019, 11, 11227–11237 CrossRef CAS PubMed; (b) Y. Zheng, H. Lu, Z. Jiang, Y. Guan, J. Zou, X. Wang, R. Cheng and H. Gao, J. Mater. Chem. B, 2017, 5, 6277–6281 RSC.
- (a) X. Zhen, L. Chudal, N. K. Pandey, J. Phan, X. Ran, E. Amandor, X. Huang, O. Johnson, Y. Ran, W. Chen, M. R. Hamblind and L. Huang, Mater. Sci. Eng., C, 2020, 110, 110659 CrossRef CAS PubMed; (b) L. Huang, G. Szewczyk, T. Sarna and M. R. Hamblin, ACS Infect. Dis., 2017, 3, 320–328 CrossRef CAS PubMed.
- (a) J. Mosinger and B. Mosinger, Experientia, 1995, 51, 106–109 CrossRef CAS PubMed; (b) L. Y. Huang, L. Ma, W. J. Xuan, X. M. Zhen, H. Zheng, W. Chen and M. R. Hamblin, J. Biomed. Nanotechnol., 2019, 15, 2142–2148 CrossRef CAS PubMed.
- (a) B. Purushothaman, J. Choi, S. Park, J. Lee, A. A. S. Samson, S. Hong and J. M. Song, J. Mater. Chem. B, 2019, 7, 65–79 RSC; (b) T. Qi, B. Chen, Z. Wang, H. Du, D. Liu, Q. Yin, B. Liu, Q. Zhang and Y. Wang, Biomaterials, 2019, 213, 119219 CrossRef CAS PubMed.
- (a) J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed; (b) Z. Xu and L. Xu, Chem. Commun., 2016, 52, 1094–1119 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0tb00888e |
| This journal is © The Royal Society of Chemistry 2020 |