Holographic immunoassays: direct detection of antibodies binding to colloidal spheres†
Abstract
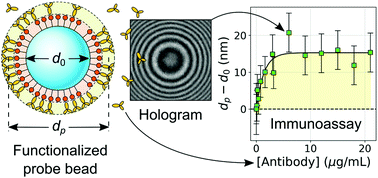
The size of a probe bead reported by holographic particle characterization depends on the proportion of the surface area covered by bound target molecules and so can be used as an assay for molecular binding. We validate this technique by measuring the kinetics of irreversible binding for the antibodies immunoglobulin G (IgG) and immunoglobulin M (IgM) as they attach to micrometer-diameter colloidal beads coated with protein A. These measurements yield the antibodies’ binding rates and can be inverted to obtain the concentration of antibodies in solution. Holographic molecular binding assays therefore can be used to perform fast quantitative immunoassays that are complementary to conventional serological tests.
- This article is part of the themed collection: Soft Matter Most Popular 2020


 Please wait while we load your content...
Please wait while we load your content...
