Open Access Article
Open Access ArticleBacterial nanotubes mediate bacterial growth on periodic nano-pillars†
Yunyi
Cao
a,
Saikat
Jana
b,
Leon
Bowen
c,
Hongzhong
Liu
 d,
Nicholas S.
Jakubovics
d,
Nicholas S.
Jakubovics
 e and
Jinju
Chen
e and
Jinju
Chen
 *a
*a
aSchool of Engineering, Newcastle University, Newcastle Upon Tyne, NE1 7RU, UK. E-mail: Jinju.chen@ncl.ac.uk
bSchool of Biomedical Sciences, University of Leeds, LS2 9JT, UK
cDepartment of Physics, Durham University, Durham, DH1 3LE, UK
dSchool of Mechanical Engineering, Xi’an Jiaotong University, Xi’an 710054, China
eSchool of Dental Sciences, Newcastle University, Newcastle Upon Tyne, NE2 4BW, UK
First published on 21st July 2020
Abstract
Surface topography designed to achieve spatial segregation has shown promise in delaying bacterial attachment and biofilm growth. However, the underlying mechanisms linking surface topography to the inhibition of microbial attachment and growth still remain unclear. Here, we investigated bacterial attachment, cell alignment and biofilm formation of Pseudomonas aeruginosa on periodic nano-pillar surfaces with different pillar spacing. Using fluorescence and scanning electron microscopy, bacteria were shown to align between the nanopillars. Threadlike structures (“bacterial nanotubes”) protruded from the majority of bacterial cells and appeared to link cells directly with the nanopillars. Using ΔfliM and ΔpilA mutants lacking flagella or pili, respectively, we further demonstrated that cell alignment behavior within nano-pillars is independent of the flagella or pili. The presence of bacteria nanotubes was found in all cases, and is not linked to the expression of flagella or pili. We propose that bacterial nanotubes are produced to aid in cell–surface or cell–cell connections. Nano-pillars with smaller spacing appeared to enhance the extension and elongation of bacterial nanotube networks. Therefore, nano-pillars with narrow spacing can be easily overcome by nanotubes that connect isolated bacterial aggregates. Such nanotube networks may aid cell–cell communication, thereby promoting biofilm development.
1 Introduction
Bacterial cells can colonize surfaces and form biofilms that consist of microbial cells embedded in extracellular polymeric substances (EPS).1,2 The unique structure of biofilms protects bacteria from the surrounding environment, conferring a capacity for persistence against phagocytosis, oxidative stresses, nutrient/oxygen restriction, metabolic waste accumulation, interspecies competition, and conventional antimicrobial agents.3 Bacterial biofilms can cause persistent human infections and can foul the surface of medical devices.4,5 For example, Pseudomonas aeruginosa, an organism well-known for its capacity to form biofilms, is an opportunistic pathogen and is one of the top three causes of opportunistic human infections,6 causing nosocomial infections in catheter lines, or chronically infecting the lungs of cystic fibrosis patients.3,6 Biofilm formation by P. aeruginosa is responsible for antimicrobial tolerance and causes major problems for treatment of infections.7 Additionally, P. aeruginosa is intrinsically resistant to a variety of antibiotics and disinfectants and multi-drug resistant (MDR) strains have been identified.3,8 Therefore, it is important to develop biomaterials that can control biofilm growth thereby reduce infections. In particular, surface modifications that physically create rational surface topographies have attracted attention in recent years, and have shown to inhibit bacterial attachment and biofilm growth without the use of antimicrobials.2,9–11 A comprehensive understanding of the interactions between bacteria and materials with different surface topographies may pave the way for more effective strategies to control biofilm growth.Surface patterning is an important determinant of bacterial attachment. Bacterial attachment is favoured on recessed portions of patterned surfaces, and bacteria tend to attach preferentially to patterns in the micro or nanometre range rather than to smooth surfaces.11 Jeong et al.12 showed that Shewanella oneidensis recognized nanoscale structures and attached preferentially with alignment along the length direction of nanowires. Hochbaum et al.4 found that P. aeruginosa PA14 tended to maximum their contact area with the surface, forming a spontaneous cell alignment between periodic nano-pillars with a post pitch of 2.2, 0.9 and 0.7 μm. Subsequent work showed similar behaviour as P. aeruginosa aligned within subcellular-nanogratings,13Escherichia coli orientated towards surface line patterns,14 and Pseudomonas fluorescens were trapped preferentially in surface trenches.15 As such, topographical features with micrometre or submicrometer length scales (i.e., comparable with the length scale of the bacteria themselves) can influence the arrangement of adhered cells during the early stage of biofilm development.4,16,17 However, the underlying mechanism that results in the observed arrangements of cells is currently unclear,4,13,14 which hinders the development of an overarching understanding of bacterial cell–material interactions.
In this paper, the alignment, attachment of bacteria and biofilm growth were investigated on nano-pillars with systematic variations in dimensions. P. aeruginosa PAO1-mCherry were incubated with surfaces for 2 hours or 24 hours, and were visualized by fluorescence microscopy and scanning electron microscopy (SEM). By using bacterial mutants (PAO1 ΔfliM and ΔpilA), we show here that cell alignment of the initial attachment (∼2 h) is a general phenomenon within these bacterial strains. Additionally, bacterial nanotubes were observed via high-resolution SEM, and may play roles in cell–cell communication. After 24 hours, nanotube networks were apparent and may aid the connection of isolated cell clusters, by overcoming the pillars and masking surface topographies. We suggest that bacterial nanotubes may play critical structural functions in biofilm formation on patterned surfaces.
2 Experimental section
2.1 Fabrication of surface substrates with nano-pillars
Nano-pillars were fabricated via e-beam lithography and soft-lithography (double-moulding) methods (see Fig. S1, ESI†). All pillars were set to have a height of 2 μm. The top of pillars was set to have a diameter of 500 nm, and the space between adjacent top pillars was set to 5 μm, 2 μm and 1 μm. Initially 2D nano-patterns were designed by Klayout Editor software (https://www.klayout.de/), and e-beam lithography was used to fabricate nano-pillar arrays on silicon substrates, following the dry etch process, which has been described in our previous work9 and elsewhere.18–20 Notably, the shape of pillar was trapezoidal owing to the etching process and the diameter of pillar increases from 500 nm (top) to 1 μm (bottom) (see Fig. S2, ESI†). To clarify, we defined the pillar diameter and space between pillars only based on the top of pillars in this study, unless specifically noted.The silicon nano-pillar arrays were treated with an anti-sticking agent (tridecafluoro-1,1,2,2-tetrahydrooctyl)-trichlorosilane (Gelest Inc.) by exposure in a desiccator under vacuum for 30 minutes. To get negative replicas from the silicon substrates, a mixture of polydimethylsiloxane (PDMS) solution was prepared using SYLGARD 184 Elastomer Kit (Dow Corning Corporation, Midland, MI) with a base-to-curing agent ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (wt/wt). The pre-polymer solution was thoroughly mixed and degassed under vacuum for 30 minutes to eliminate air bubbles. The mixture was poured over the silicon substrates in a Petri dish and cured at 70 °C for 2 hours. After cooling at room temperature, the negative PDMS mould was gently peeled off from the substrate. To get the final replicas of the nano-pillars on silicon substrates, UV-curable epoxy (OG 142-87, Epoxy Technology, Inc.) was poured onto the negative PDMS mould fabricated above, and air bubbles were removed by a plastic disposable pipette. The poured UV-curable epoxy was covered with a pre-cleaned glass slide, and cured at ∼100 mW at 365 nm, for 20–25 minutes under a UV-lamp. After cooling to room temperature, the cured epoxy was demoulded by bending the PDMS mould. The epoxy replicas can be stored for about a month at room temperature without noticeable deformation.19
1 (wt/wt). The pre-polymer solution was thoroughly mixed and degassed under vacuum for 30 minutes to eliminate air bubbles. The mixture was poured over the silicon substrates in a Petri dish and cured at 70 °C for 2 hours. After cooling at room temperature, the negative PDMS mould was gently peeled off from the substrate. To get the final replicas of the nano-pillars on silicon substrates, UV-curable epoxy (OG 142-87, Epoxy Technology, Inc.) was poured onto the negative PDMS mould fabricated above, and air bubbles were removed by a plastic disposable pipette. The poured UV-curable epoxy was covered with a pre-cleaned glass slide, and cured at ∼100 mW at 365 nm, for 20–25 minutes under a UV-lamp. After cooling to room temperature, the cured epoxy was demoulded by bending the PDMS mould. The epoxy replicas can be stored for about a month at room temperature without noticeable deformation.19
2.2 Bacterial culture and biofilm formation of P. aeruginosa
P. aeruginosa PAO1-mCherry, PAO1 ΔfliM and ΔpilA were used throughout this study. PAO1-mcherry is the derivative of P. aeruginosa PAO1-N (Nottingham subline,21 kindly provided by Dr James Brown), which was engineered via chromosomal insertion (attTn7::ptac-mcherry) to constitutively express fluorescent proteins, and is a biofilm-forming bacterial strain that has been widely used.9,22,23 PAO1 mutant strains which lack flagella (∼ΔfliM) and lack the main Type IV pilus filament protein (∼ΔpilA) were kindly provided by Prof. Matthew Parsek (University of Washington).24–26 For bacterial adhesion and biofilm formation assays, cells were routinely cultured in Trypticase Soy Broth (TSB) (Melford Laboratories Ltd, UK), in a shaker at 180 rpm, 37 °C for 16 hours and then diluted to OD600 = 0.01 in 100× diluted TSB with a spectrophotometer (Biochrom Libra S11, Biochrom Ltd, Cambridge, UK). Prior to seeding, the epoxy nano-pillar substrates were added to wells of a 12-well culture plate. To assay bacterial adhesion to surfaces, 3 ml of the diluted bacterial culture was incubated with substrates for 2 hours at 37 °C and then removed for visualization. To examine the effect of nano-pillars on biofilm formation, 3 ml of diluted bacterial suspension was added to each sample, and incubated for 24 hours at 37 °C. In this study, at least three independent experiments were performed for each substrate type.2.3 Fluorescent microscope analysis
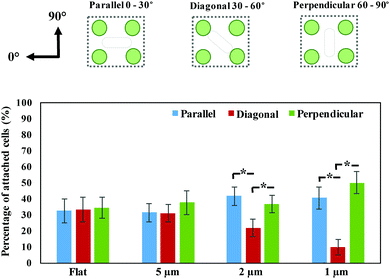
The substrates were removed from the wells with sterile forceps and gently rinsed three times with Phosphate Buffered Saline (PBS, pH = 7.4) to remove non-adherent or loosely adhered bacteria.2,9 The samples were then visualized by Olympus BX61 upright fluorescent microscope with a 20× lens. The area of periodic nano-pillars was initially identified using the bright-field channel. The attachment and alignment of bacterial cells on nano-pillars after 2 hours’ incubation was visualized by acquiring 2D fluorescent images under the focal plane. An area of 121.25 × 108.75 μm2 was randomly selected from the 2D fluorescent images. The fluorescent images were decomposed into the corresponding fast Fourier transforms (FFT) images via Matlab code. The Fourier Transform decomposes an image into its sine and cosine components, and each point represents a particular frequency contained in the spatial domain image. Therefore, the FFT images contain the peaks associated with the spatial frequencies of bacteria within the nano-pillars.4 The alignment of bacterial cells was further analyzed by an in-house made Matlab code, and categorized as Parallel (0–30°), Diagonal (30–60°) or Perpendicular (60–90°), according to the smallest angle difference between the cell and horizontal axis of nano-pillar pattern (defined as 0°). On the other hand, the surface coverage of bacteria on nano-pillars was determined by calculating the surface area of bacteria cells with ImageJ.9 For the biofilms formed after 24 hours, z-stacks were performed through the thickness of biofilm from six random locations on the surface. The biomass under each field of view (430.00 × 324.38 μm2) was determined by COMSTAT2 plugin (Lyngby, Denmark) in ImageJ.2.4 Scanning electron microscope analysis
In this study, bacterial attachment and biofilm formation on epoxy nano-pillar substrates were visualized with a Dual Beam Focussed Ion Beam Scanning Electron Microscope (FIB-SEM) system. The samples were washed with PBS and fixed in 2% glutaraldehyde in 3 M Sorenson's phosphate buffer overnight at 4 °C. The samples were transferred to a new plate and dehydrated through a series of ethanol solutions of 25% (v/v), 50%, 75%, and 100%, followed by critical point drying. Then the samples were sputter-coated with 16 nm platinum coating using a Cressington 328 ultra-high quality coater to improve imaging quality in the Dual Beam system, following the visualization of SEM. The beam voltage and current were set to 5 kV and 0.34 nA, respectively.2.5 Statistical analysis
Data are represented by mean values with standard errors throughout. Statistical differences between samples were determined by One-way ANOVA with Tukey's test for multiple comparisons. P < 0.05 was considered statistically significant in this study, as indicated by the symbols in the representative figures. The Pearson correlation analysis was used to determine the linear correlation between statistics as noted in main texts.3 Results and discussion
3.1 Bacterial alignment within nano-pillars after 2 hours
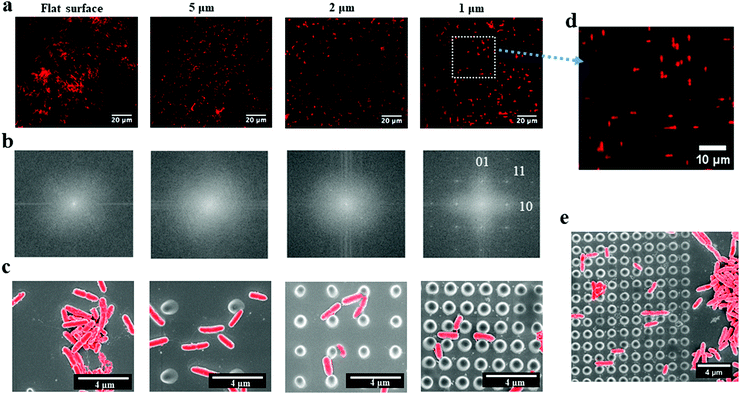
P. aeruginosa PAO1-mCherry, a rod-shaped bacterium, was grown for 2 hours on the periodic nano-pillars with varying spaces (∼5 μm, 2 μm and 1 μm). All the nano-pillars had a diameter of about 500 nm, a height of 2 μm, and were periodically arranged in an array with square symmetry. We found that the initial attachment of P. aeruginosa exhibited preferences in cell alignment that were dependent on the spaces between pillars, as shown in the fluorescent microscopy images (Fig. 1a). These fluorescent images only contained the red signals meaning the aligned rod-shaped cells, thereby neglecting the background of pillars. Therefore, their corresponding fast Fourier transform (FFT) images (Fig. 1b) can indicate the spatial orientations of attached cells, as shown in white positional peaks within FFT images. For nano-pillars with spacing of 5 μm that is much larger than the cell dimensions, bacterial attachment to the surface was apparently random and no specific orientation was observed. In addition, the FFT images of cells within 5 μm-spacing nano-pillars showed no orientational order, akin to attachment on the flat surface, which had only faint positional ordering peaks showing as a white spot (Fig. 1b). When the space of nano-pillars decreased to 2 μm, the white peaks in the FFT images seemed to show a tendency towards the perpendicular [01] and parallel [10] ordering directions (Fig. 1b). Surprisingly, when the top space between nano-pillars decreased further to 1 μm that approaches the dimensions of P. aeruginosa, bacteria exhibited specific cell alignment (or orientation) and predominantly attached parallel or perpendicular to nano-pillars (Fig. 1d). The corresponding FFT image (Fig. 1b) also showed a clear transition that the faint central spot of FFT extended towards the [10] and [01] ordering peaks when the pillar spaces decreased, indicating preferential cell orientation and alignment on the surfaces. The SEM images (Fig. 1c) also confirmed the different bacterial alignment with the decreasing of nano-pillar spaces. An SEM image at the edge of the nano-pillar array with the space of 1 μm showed clearly the preferential alignment of cells on the nano-pillar area compared with the flat area (Fig. 1e). The abrupt change at the interface suggested that the change in cell orientation were specifically related to the surface topography features. Therefore, we hypothesized that the preferential orientation/alignment behaviour of cells when attaching onto nano-pillar were attributed to the different pillar spaces, and nano-pillars with smaller spaces that comparable to bacterial size would have a more significant effect.An in-house made MATLAB code was used to quantify the cell orientation/alignment of P. aeruginosa on the periodic nano-pillars. These data confirmed that the spacing of nano-pillars has profound effects on the different cell orientation/alignment. For the nano-pillar with 5 μm spacing, the cell orientation exhibited a near-uniform distribution of attachment angles (Fig. 2) (p > 0.05), which is similar to the angle distribution as found on flat surface. If the space of nano-pillars decreased to 2 μm, more cells orientated as “Parallel (0–30°)” (41.58 ± 5.75% of the total attached cells) and “Perpendicular (60–90°)” (36.56 ± 5.30%), than “Diagonal (30–60°)” (21.86 ± 5.36%) (p < 0.05). When the space of nano-pillars decreased further to 1 μm, it became even clearer that most cells orientated as “Parallel (0–30°)” (40.42 ± 8.36%) or “Perpendicular (60–90°)” (44.37 ± 8.76%) (p < 0.05), which is consistent with the fluorescent microscopy images shown in Fig. 1d. The quantification above confirmed that the periodic nano-pillars with smaller spaces have profound effects on the cell orientation/alignment when attaching onto the nano-pillar surfaces, which is consistent with previous investigations.4
3.2 Bacterial attachment was delayed on nano-pillars of smaller spaces
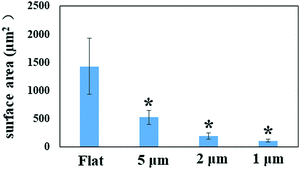
By quantifying the fluorescent signal in the microscope images shown in Fig. 1a, the nano-pillar space was found to be positively correlated with the extent of initial bacterial attachment (Fig. 3, r > 0.98 for all surfaces, Pearson correlation analysis). The total attachment of P. aeruginosa cells on flat and nano-pillar surfaces was ranked in the order: flat surface > 5 μm-spacing > 2 μm-spacing > 1 μm-spacing. Additionally, all the nano-pillar surfaces harboured less surface area covered by bacteria, as compared with the flat surface (p < 0.05). On the other hand, both 2 μm-spacing and 1 μm-spacing nano-pillar surfaces had fewer attached bacterial cells as compared with 5 μm-spacing structures (p < 0.05). The difference between attachment to 2 μm-spacing and 1 μm-spacing nano-pillar surfaces was not significant (p = 0.13). Therefore, in addition to the effects on cell orientation/alignment of P. aeruginosa, the periodic nano-pillars were also found to be inhibitory to initial bacterial attachment. Regarding the preference of bacteria attachment to specific dimensions of surface topographies and how it may be trapped between pillars or grooves, the same principle of maximizing the contact area between bacteria and matrix applies to cocci.2,9,27Next, we sought to look more closely at the impact of nano-pillars with different spaces on initial bacterial attachment. We used SEM at a high magnification to visualize the interaction of P. aeruginosa with surfaces (Fig. 4a–f). In addition, we analyzed multiple SEM images (n = 20) of P. aeruginosa cells to measure cell dimensions and found the diameter was 0.54 ± 0.10 μm and the length was 1.37 ± 0.81 μm in this study. It was noted that most bacterial cells preferentially colonized the areas between nano-pillars (Fig. 1c and 4), which was attributed to additional colonization sites in these areas compared with the top of nano-pillars.2,28 For 5 μm-spacing nano-pillar surface, since the spacing between pillars is much larger than the bacterial cell size, up to 10 bacterial cells can deposit between parallel/perpendicular pillars while arranged flat against the surface (Fig. 1c and 4a, b). Also, 23.14 ± 10.18% of the attached cells (based on SEM images, n = 10; cell number = 314) were found to contact the sidewalls of nano-pillars. When the nano-pillar space decreased to 2 μm, which is near to the length of P. aeruginosa, up one to two bacteria can lie within the parallel/perpendicular pillars (Fig. 1c and 4c, d). Longer cells were able to contact two pillars; however, two bacterial cells can also squeeze between the pillars as shown in Fig. 4c and d. On the other hand, 88.66 ± 11.34% of attached cells (based on SEM images, n = 10; cell number = 98) were in contact with the sidewalls of nano-pillars on the 2 μm-spacing nano-pillar surfaces. When the pillar spaces further decreased to 1 μm, and the bottom space between adjacent trapezoidal pillars can further deceased to 500 nm (Fig. S2, ESI†), either top or bottom space is closer to the diameter of P. aeruginosa. Therefore, 98.82 ± 1.18% cells squeezed between the pillars (based on SEM images, n = 10; cell number = 76), as the space only allowed up to one bacterial cell to lie in between nano-pillars, thereby resulting in cell alignment (Fig. 4e and f). Surfaces with a smaller space between pillars provide fewer colonization sites for bacterial cells, and this can inhibit initial bacterial attachment as previously reported.2,28 Bacteria tend to maximize their contact area with surface textures, and nano-pillars act as topographical extensions of the substrate. Therefore, cells preferentially make contacts with nano-pillars, and consequently align within the periodic nano-pillars. Within the same nano-patterned area, the density of nano-pillars increases with the decreasing of the spacing between neighbouring nano-pillars, providing extra colonization sites for bacterial cells. This possibly explained why the attached cells on 2 μm-spacing and 1 μm-spacing nano-pillar surfaces were not significantly different. On a denser patterned surface bearing nano-pillars with spaces that comparable to bacterial size, bacteria cells can have more chances to contact nano-pillars, thereby attach preferentially and irreversibly re-position between nano-pillars.
3.3 Bacterial nanotubes aid in the cell–cell connections on nano-pillars after 2 hours
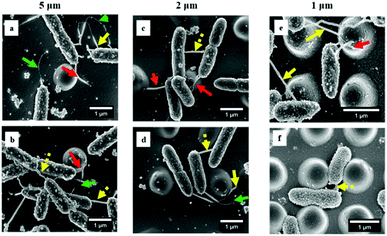
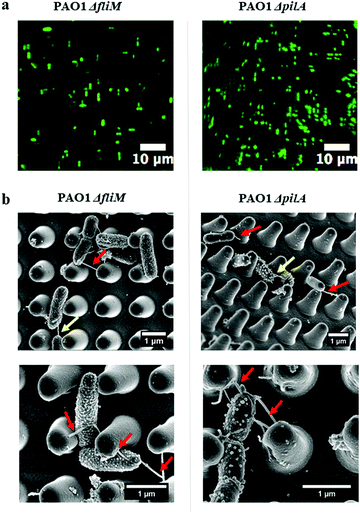
Interestingly, the high-resolution SEM images (Fig. 4) showed tubular structures (hereafter referred to as ‘nanotubes’) projecting from cell surfaces at different positions. These bacterial nanotubes were measured to be several micrometers in length and about 20–100 nm in diameter, consistent with dimensions previously reported for these structures.29 Strikingly, we observed that “root-like” extending nanotubes projected from a single cell surface and elongated to a distance of a few microns, sufficient to bridge the sidewalls of nano-pillars (Fig. 4, red arrows). We observed that extending nanotubes sometimes interconnected distal cells (Fig. 4, yellow arrows). Even these ‘‘long-distance’’ intercellular nanotubes occasionally made contacts with the nano-pillars. Also, ‘‘short-distance’’ intercellular nanotubes (∼1 μm in length) were visible between cells lying in proximity (Fig. 4, dashed yellow arrows), and connected the neighbouring cells. Notably, long extending or intercellular nanotubes frequently exhibited both bright and dark regions, which might be attributed to the different focal positions under the SEM. The nanotubes originated from cell surfaces at a higher focal position, the emergence sites were usually brighter (Fig. 4, red and yellow arrows), akin to the thickness of short intercellular nanotubes (Fig. 4, dashed yellow arrows). The dark regions of longer nanotubes (Fig. 4, green arrows) appeared thinner than the short intercellular ones. To improve the visualizations, we used indium tin oxide (ITO) glass substrates and identical culture conditions to further characterize the nanotube networks under the SEM. The excellent conductivity of ITO-glasses enabled nanotubes to be viewed without coating samples (Fig. 5). The complex nanotube networks were still visible, and exhibited uniform thickness of around 20 nm for all nanotube types (Fig. 5). This confirmed that the coating thickness and the different focal planes contributed to the apparently dissimilar nanotube morphology within nano-pillars observed when samples had been coated with platinum. On ITO-glasses, a single cell lacking nearby neighbours still produced extending nanotubes that formed a web-like network (Fig. 5a). By contrast, when other cells were close, intercellular nanotubes emerged between neighbouring cells, appearing as either long or short structures similar to the cells within nano-pillars (Fig. 5b and c). The observations above indicated that the development of nanotube networks is prevalent when bacteria are grown on a solid surface, and may mediate cell attachment.The transition from reversible to irreversible adhesion of P. aeruginosa involves cell repositioning to a longitudinal position via cell appendages such as flagella or pili, as cells that are bound by their pole are capable of spinning on their axis or crawling to maximise the contact area between the cells and the surface.11 To investigate whether these nanotubes were either flagella or pili and whether they are involved in cell alignment within the nano-pillars, mutants lacking genes essential for synthesis of either flagella or pili (PAO1 ΔfliM and ΔpilA) were used. ΔfliM bacteria do not possess flagella and exhibit impaired swimming and swarming motilities thereby lacking cell spinning; and ΔpilA mutant exhibit a major deficit in twitching motility thereby cannot ‘walk’ or ‘crawl’ over surfaces.24,30 We grew these bacterial mutants on the nano-pillars with the space of 1 μm for 2 hours, under culture conditions identical to those used for the wild type P. aeruginosa PAO1-mCherry. As shown in Fig. 6a, cell alignment of either PAO1 ΔfliM or ΔpilA was similar to the wild type, and cell alignment was predominantly parallel or perpendicular to the arrangement of pillars. This indicates that there were no effects of appendage knockouts (i.e. flagella and pili) on the cell alignment behaviour within nano-pillars. Notably, the nanotubes were also readily visible on bacterial mutants (Fig. 6b), ruling out the possibility that these nanotubes are flagella or pili. Our investigations above indicated that cell alignment may be a general phenomenon, occurring in examples of wild-type bacteria and in the absence of flagella or pili, which was also consistent with the findings in other studies.4,13
Overall, the investigation above showed that the surface topography at the micro- and nanoscale that is comparable to the bacterial size, can affect bacterial alignment and attachment. It is likely that cells try to maximize contact area with the surface topography, presumably to achieve a stronger and more stable attachment, which results in a specific alignment behaviour of the attached cells. “Bacterial nanotubes” may mediate bacterial growth on periodic nano-pillars. Similar observations have also been reported recently showing different morphologies compared with cell appendages such as flagella or pili.29,31–34 By using bacterial mutants (ΔfliM and ΔpilA), we ruled out effects of the appendage knockouts on nanotube formation. However, further characterization of the composition of P. aeruginosa bacterial nanotubes may need sophisticated techniques such as Cryo-EM and/or total internal reflection fluorescence (TIRF) with super-resolution structured illumination microscopy (SIM).35 Nevertheless, the occurrence of nanotubes in both flagella and pilus mutants suggested that cell alignment is related to interactions with the cell surfaces or biofilm components closely associated with the cell wall rather than to these appendages. Here, we did not show direct evidence that nanotubes mediate cell alignment within the nano-pillars. It is plausible that either extending or intercellular nanotube networks can greatly increase the cell surface area and enhance its ability to sense surrounding environment.31 Additionally, our high-resolution SEM images provide evidence that nanotubes can aid cell–cell connections after bacterial growth on surfaces even over a short time (2 hours).
3.4 The growth of P. aeruginosa biofilm is aided via bacterial nanotubes on periodic nano-pillars after 24 hours
By using fluorescence microscopy and SEM (Fig. 7a and b), we investigated P. aeruginosa growth on periodic nano-pillars after 24 hours. Firstly, we evaluated whether nano-pillars can delay biofilm growth as a consequence of impaired attachment within nano-pillars. The total biomass on the flat surface was found to be almost 1.5 times, twice and 1.8 times more than that on nano-pillar surfaces (5 μm-spacing, 2 μm-spacing and 1 μm-spacing, respectively) (see Fig. 7c). The flat surface harbored more P. aeruginosa biofilm clusters shown as a 3D structure with well-connected nanotube filament networks as shown in the SEM images (Fig. 7b1-2). Smaller biofilm clusters with nanotube networks were also found between the nano-pillars on the 5 μm-spacing structure (Fig. 7b3). In addition, small aggregates comprising approximately 7 cells were found near the pillar, and were connected to each other via the nanotube filament network. Similar observations were also found on the 2 μm-spacing structure (Fig. 7b5-6 and Fig. S3c, ESI†) and the biomass was significantly lower than that on 5 μm-spacing structure (Fig. 7c, p < 0.05). Surprisingly, we observed that bacterial cells filled into the 1 μm-spacing structure and started forming biofilm clusters at the top layer of nano-pillars (Fig. 7b7-8). The biomass on this surface (18.68 ± 2.70 μm3 μm−2) appeared greater than that on 2 μm-spacing structure (14.99 ± 2.66 μm3 μm−2) although differences were not significant (Fig. 7c, p = 0.61). Notably, the presence of a dense and much more complex web of nanotube filaments surrounding the cells was observed on this surface (Fig. 7b8 and Fig. S3d, ESI†). Similar to the adherence of P. aeruginosa within nano-pillars after 2 hours, extending nanotubes after 24 hours bridged the sidewalls of nano-pillars (Fig. 7b, red arrows) and some intercellular nanotubes bridged the neighboring cells (Fig. 7b, yellow arrows). Surprisingly, some nanotubes reached up to 10 μm (or even longer) via migrating across the nano-pillars (Fig. 7b and Fig. S3, ESI†), even though there were no bacterial cells settling ont the pillars. These long nanotubes were able to connect distantly isolated cells. It was likely that the observed nanotube filament networks became more complex over time and were helping cells to connect each other and form either bacterial aggregates or clusters.We next set out to characterize biofilm growth on the periodic nano-pillar surfaces over time. The first interesting observation was that, despite the different spaces of nano-pillars, P. aeruginosa still progressed through the typical early stage of biofilm development (Fig. S3, ESI†), although the biomass on nano-pillar surfaces was lower than that on the flat surface (Fig. 7c). At this stage, cells did not show any preferential orientation behavior as cells start to form aggregates or clusters, involving a much more complicated dynamic process. We observed that a preliminary biofilm cluster with a 3D structure formed between the nano-pillars with 5 μm space (Fig. 7b3 and Fig. S3b, ESI†), with a morphology which was akin to that on the flat surface (Fig. S3a, ESI†). By contrast, on 2 μm-spacing or 1 μm-spacing nano-pillars, P. aeruginosa developed smaller and more heterogeneous bacterial clusters on recessed portions of patterned surfaces, possibly because nano-pillars isolated the bacterial cells or aggregates (Fig. S3c and d, ESI†). One way to inhibit early-stage biofilm is employing a specific surface topography to hinder cell body contacts.1 Here, periodic nano-pillars with varying spaces engineered the initial bacterial attachment as cells only attached within the confined pillar spaces, and nano-pillars with a smaller space had lower attachment. Therefore, bacterial cells were apparently isolated and separated by the nano-pillars. However, the height of pillars is around 2 μm, which is similar to the dimension of P. aeruginosa cells. Therefore, multiple bacterial cells can easily deposit between nano-pillars (Fig. 7b5–8). This is apparent on nano-pillars with 1 μm and 2 μm spaces, which showed that cells covered the pillar gaps and formed multi-layered bacterial clusters either around or on top of the nano-pillars (Fig. S3c and d, ESI†). Additionally, the accumulated cells can either mask the surface chemistry or smooth the surface topography, and serve as a conditioning film to provide nutrients and adhesion receptors for subsequent bacterial attachment.2 Collectively, nano-pillars can delay biofilm growth owing to the isolation of cells within the structure, while they may be less effective over time with forming small biofilm clusters.
The second interesting observation is that the 1 μm-spacing nano-pillars were not effective in delaying biofilm growth since more biofilm biomass (Fig. 7c), was observed compared with the 2 μm-spacing nano-pillars after 24 hours. Our preceding results have shown that the smallest space (∼1 μm) has the lowest attachment (Fig. 3b). Therefore, there seems to be another separate effect that mediates bacterial growth on nano-pillars. We noticed that biofilm clusters developed within the confined spaces, while some separated bacterial cells or aggregates were connected via the nanotube networks. Notably, the nanotube networks were still visible for the bacterial mutants (PAO1 ΔfliM and ΔpilA) after 24 hours, and showed similar morphology to biofilms of wild-type bacteria (Fig. S4, ESI†). Unlike the nanotubes which only contact the sidewalls of nano-pillars after 2 hours, the nanotubes of wild-type or mutants after 24 hours elongated into web-like networks via migrating over nano-pillars (Fig. S4, red arrows, ESI†). It is likely that these nanotubes can explore the local geometry by binding onto the nano-pillars, and increase the cell surface area resulting in improved connections with neighbouring or distal cells. To better characterize the nanotube networks without the shielding of cell clusters, we allowed bacteria to attach within nano-pillars after 2 hours; after washing with PBS to remove loosely attached cells, we supplied fresh TSB and cultured for a further 24 hours (Fig. S5, ESI†). Strikingly, we observed the elongation of nanotube networks that connected the nano-pillars one by one. Within the nano-pillars of 2 μm space, nanotubes were observed that continuously connected around 4–10 pillars (Fig. S5a, ESI†). By contrast, the nanotubes continuously connected around 20–30 pillars within the nano-pillars of 1 μm space (Fig. S5b, ESI†). This indicated that nano-pillars with smaller spaces could enhance the connections between nanotubes. Here, we speculated that the nano-pillars acted as nodes within the nanotube networks to promote their extension and elongation. Nano-pillars of 1 μm-space have more pillars within the same projected area and smaller spaces; thereby can provide additional surface area for continuously spreading nanotubes along the nano-pillars. Therefore, the nano-pillars with the space of 1 μm cannot effectively isolate the cell clusters, as the nano-pillars can be easily overcome by the nanotubes that connected the bacterial aggregates far away. Here, the separated bacterial cells or aggregates can possibly communicate via the connected nanotube networks instead of direct cell body contacts, thereby promoting further biofilm development. This hypothesis is consistent with the observations showing increased biofilm growth and more complex nanotube networks on 1 μm-spacing nano-pillars.
Materials introduced into the body will rapidly become coated with host cells and/or body fluids such as saliva or serum, which can influence microbial adhesion.36 However, these coatings vary between individuals and even from one individual at different times of day.36 Therefore, to maintain consistency in our measurements, we did not attempt to model the interaction with host components. Nevertheless, further development of these surfaces will require a rigorous assessment of the impact of host components on bacterial attachment and nanotube formation. Recent studies have shown that it is possible to develop materials that promote host cell adhesion while still retaining the capacity to inhibit the early stages of attachment of bacteria such as P. aeruginosa and Gram-positive cocci.37,38 Flow can also affect bacteria attachment. Flow not only provides a higher supply rate of bacteria to the surface but also increases the shear stress imposed on adherent bacteria.38
It has been reported that nanopillars with different geometries (e.g. cone-shape, cylindrical) could achieve similar antibiofilm performance when the feature sizes were well modulated.39,40 It is unclear whether the conical geometry may be superior over a cylindrical shape. There is a lack of information about the pure “geometry effect” in the literature. Indeed, such a comparison would be difficult because the size effect is always associated with geometry effect.
In this study, we have focussed on the early stages of attachment and colonisation of surfaces (up to 24 hours) by bacteria. Whilst in some applications, the inhibition of adhesion is very important, this does not necessarily translate to long-term anti-bacterial efficacy of the material. For example, lotus-leaf inspired superhydrophobic surfaces have trapped air within the micro or nanostructures that restricts the direct contact between the solid surfaces and microorganisms, thereby inhibiting initial bacterial attachment.9,41–43 However, recent studies have shown that the prolonged immersion (1–4 hours) of superhydrophobic surfaces can result in the loss of the air-layer and subsequent bacterial growth may exceed that of the corresponding control materials.44–46 Therefore, it is not expected that the nanopillars in the present study can have long-term antibiofilm performance. However, it is very interesting to understand how the nanotubes produced by bacteria will mediate bacteria cluster formation, which is the major focus in current study.
Bacterial nanotubes or nanotube networks have been found within various bacterial species, suggesting that their existences is widespread in nature. For example, nanotubes of B. subtilis cells were formed within several minutes after bacteria grew on a solid surface, and appeared as both intercellular tubes and extending tubes.35 In addition, cryo-EM analysis showed that nanotubes directly emanate from the cytoplasmic cell membrane, consisting of chains of consecutive constricted segments harboring a continuous lumen.31,35 Also, these nanotube networks can serve as a route for exchange of cellular molecules within and between species.29 Extracellular nanotube-like networks have also been implicated in long-range extracellular electron transport in Geobacter sulfurreducens, Shewanella oneidensis MR-1, Pelotomaculum thermopropionicum and Methanothermobacter thermoautotrophicus.47–51 Additionally, nanotube-like networks of S. oneidensis MR-1 have been found to be extensions of the outer membrane which are associated with outer membrane vesicles, structures ubiquitous in Gram-negative bacteria, rather than pilin-based structures as previously thought.52 Similarly, there is evidence that nanotube networks produced by Myxococcus xanthus consist of outer membrane vesicle chains, which connect cells spatially and transfer outer membrane proteins in a contact-dependent manner, thereby promote biofilm growth.53 Even though observations of nanotubes within various bacteria have been reported, little is known about the mechanism of nanotube formation. A gene implicated in nanotube formation of B. subtilis is ymdB, encoding a calcineurin-like phosphodiesterase; ymdB mutants exhibited a marked deficiency in nanotube production.31,35 YmdB can repress the expression of motility genes and induce the expression of genes associated with biofilm formation, hence controlling the switch from a motile to a multicellular sessile lifestyle.31 Additionally, recent studies revealed that the export apparatus of B. subtilis or E. coli flagella, designated CORE, can communally serve for the generation of both flagella and nanotubes.32,33 Mutants lacking CORE genes produce very limited nanotube networks and are deficient in the associated intercellular molecular trafficking.32 Whilst the mechanism of nanotube formation remains unclear, it is likely that the development of nanotube networks may be an early stage in biofilm formation. Bacterial nanotubes provide the foundation for unhampered intercellular molecular flow via bridging the cells.31 Various SEM images of bacterial biofilms have indicated the potential existence of bacterial nanotubes as prominent bridges between cells.31,54 Our preliminary SEM images of Staphylococcus epidermidis which was incubated under different timeframes, also showed the prevalent occurrence of nanotubes on different surfaces (Fig. S6–S9, ESI†), which appeared to bridge and connect cells. Gram-positive S. epidermidis does not produce flagella or pili, yet tube-like structures were still visible (Fig. S6–S9, ESI†). Although similar nanotubes have sometimes been described as bacteria fibrils,54 this is not entirely consistent with our observations of extending nanotubes within biofilm growth (Fig. S9, ESI†). However, mature bacterial biofilms are complex and heterogeneous structures, and the substantial mass of EPS may shield the nanotubes, thereby making it difficult to decipher the nature of these connections.
4 Conclusions
In summary, the cell alignment, bacterial attachment and biofilm formation of clinically relevant strains of P. aeruginosa were investigated on periodic nano-pillar surfaces. Over a short time (∼2 hours), bacterial cells showed lower attachment on the nano-pillar surfaces owing to cells preferentially attaching within the confined spaces of nano-pillars. In particular, the 1 μm-spacing nanopillars had a strong influence on the orientation of cells, which predominantly attached in parallel or perpendicular directions to the nano-pillars.The most interesting findings is that bacterial nanotubes (membranous intercellular bridges) contact either the nanopillars or other cells. By using the bacterial mutants (ΔfliM and ΔpilA) lacking flagella or pili, we further demonstrated that such cell alignment behavior within nano-pillars is independent of flagella or pili, and is possibly due to the cell's preference to maximize their contact area with the surface, where the pillars act as topographical extensions of the substrate. Additionally, nanotubes occurred in the wild-type and bacteria mutants, indicating that the formation of bacterial nanotubes is not dependent on flagella or pili.
Smaller bacterial clusters were formed in between nano-pillars after 24 hours, and were likely to be isolated by the nano-pillars. Therefore, the bacterial growth of P. aeruginosa after 24 hours was delayed on periodic nano-pillars, with reduced biofilm biomass compared with the flat surfaces. However, the 1 μm-spacing nano-pillars, which showed the lowest bacterial attachment after 2 hours, were not effective in delaying biofilm growth after 24 hours. Nano-pillars with smaller spaces appeared to help the extension and elongation of bacterial nanotube networks. Therefore, nano-pillars can be easily overcome by the nanotubes that connected the isolated bacterial aggregates. Such nanotube networks can possibly aid cell–cell communication, thereby promoting further biofilm development. The findings above will be helpful for designing more effective biomaterial surfaces for controlling biofilm growth. In particular, determining a threshold for controlling bacterial nanotube development will be important for isolating bacterial clusters which will be our future work. In addition, the nano-pillar surfaces described here provide a useful tool for investigating the elongation of nanotubes, and may pave a way for future characterization of bacterial nanotubes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Y. Cao acknowledges the PhD studentship (Research Excellence Academy funding scheme) from Newcastle University. J. Chen acknowledges funding from the Engineering and Physical Sciences Research Council (EP/K039083/1) and EPSRC Partnering for GCRF (EP/R512692/1). J. Chen and H. Liu also acknowledge funding from Royal Society-Newton Mobility Grant (IEC\NSFC\191070). We acknowledge Prof. Matthew Parsek (University of Washington) and Dr James Brown (University of Nottingham) kindly shared the bacterial mutant strains used in this study. We also acknowledge the technical assistance and useful discussions with Dr Nadia Rostami, Dr Rebecca Jones and Ekaterina Kozhevnikova.Notes and references
- M. Kargar, Y.-R. Chang, H. Khalili Hosseinabad, A. Pruden and W. A. Ducker, ACS Biomater. Sci. Eng., 2016, 2, 1039–1048 CrossRef CAS.
- Y. Cao, B. Su, S. Chinnaraj, S. Jana, L. Bowen, S. Charlton, P. Duan, N. S. Jakubovics and J. Chen, Sci. Rep., 2018, 8, 1071 CrossRef PubMed.
- M. F. Moradali, S. Ghods and B. H. A. Rehm, Front. Cell. Infect. Microbiol., 2017, 7, 39 CrossRef PubMed.
- A. I. Hochbaum and J. Aizenberg, Nano Lett., 2010, 10, 3717–3721 CrossRef CAS PubMed.
- F. Song and D. Ren, Langmuir, 2014, 30, 10354–10362 CrossRef CAS PubMed.
- C. K. Stover, X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik and M. Lagrou, Nature, 2000, 406, 959 CrossRef CAS PubMed.
- T. Rasamiravaka, Q. Labtani, P. Duez and M. El Jaziri, BioMed. Res. Int., 2015, 2015, 759348 Search PubMed.
- R. Smith and J. Coast, BMJ [Br. Med. J.], 2013, 346, 1493 Search PubMed.
- Y. Cao, S. Jana, L. Bowen, X. Tan, H. Liu, N. Rostami, J. Brown, N. S. Jakubovics and J. Chen, Langmuir, 2019, 35, 14670–14680 CrossRef CAS PubMed.
- F. Song, H. Koo and D. Ren, J. Dent. Res., 2015, 94, 1027–1034 CrossRef CAS PubMed.
- C. Berne, C. K. Ellison, A. Ducret and Y. V. Brun, Nat. Rev. Microbiol., 2018, 16, 616–627 CrossRef CAS PubMed.
- H. E. Jeong, I. Kim, P. Karam, H.-J. Choi and P. Yang, Nano Lett., 2013, 13, 2864–2869 CrossRef CAS PubMed.
- C. Q. Lai, Langmuir, 2018, 34, 4059–4070 CrossRef CAS PubMed.
- H. Gu, A. Chen, X. Song, M. E. Brasch, J. H. Henderson and D. Ren, Sci. Rep., 2016, 6, 29516 CrossRef CAS PubMed.
- C. Díaz, P. L. Schilardi, R. C. Salvarezza and M. F. L. De Mele, Colloids Surf., B, 2011, 82, 536–542 CrossRef PubMed.
- C. Díaz, P. Schilardi, R. Salvarezza and M. F. L. de Mele, Colloids Surf., B, 2011, 82, 536–542 CrossRef PubMed.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Appl. Environ. Microbiol., 2013, 79, 2703–2712 CrossRef CAS PubMed.
- B. Pokroy, A. K. Epstein, M. Persson-Gulda and J. Aizenberg, Adv. Mater., 2009, 21, 463–469 CrossRef CAS.
- P. Kim, W. E. Adorno-Martinez, M. Khan and J. Aizenberg, Nat. Protoc., 2012, 7, 311 CrossRef CAS PubMed.
- P. Kim, A. K. Epstein, M. Khan, L. D. Zarzar, D. J. Lipomi, G. M. Whitesides and J. Aizenberg, Nano Lett., 2011, 12, 527–533 CrossRef PubMed.
- J. Sidorenko, T. Jatsenko and M. Kivisaar, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2017, 797, 26–37 CrossRef PubMed.
- K. A. McFarland, E. L. Dolben, M. LeRoux, T. K. Kambara, K. M. Ramsey, R. L. Kirkpatrick, J. D. Mougous, D. A. Hogan and S. L. Dove, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8433–8438 CrossRef CAS PubMed.
- M. Weigert, A. Ross-Gillespie, A. Leinweber, G. Pessi, S. P. Brown and R. Kümmerli, Evol. Appl., 2017, 10, 91–101 CrossRef CAS PubMed.
- J. C. Conrad, M. L. Gibiansky, F. Jin, V. D. Gordon, D. A. Motto, M. A. Mathewson, W. G. Stopka, D. C. Zelasko, J. D. Shrout and G. C. Wong, Biophys. J., 2011, 100, 1608–1616 CrossRef CAS PubMed.
- J. Garbe, B. Bunk, M. Rohde and M. Schobert, BMC Microbiol., 2011, 11, 102 CrossRef CAS PubMed.
- C. R. Armbruster, C. K. Lee, J. Parker-Gilham, J. de Anda, A. Xia, K. Zhao, K. Murakami, B. S. Tseng, L. R. Hoffman and F. Jin, eLife, 2019, 8, e45084 CrossRef CAS PubMed.
- L. Jin, W. Guo, P. H. Xue, H. N. Gao, M. Zhao, C. Zheng, Y. L. Zhang and D. Han, Nanotechnology, 2015, 26, 055702 CrossRef PubMed.
- M. Lorenzetti, I. Dogša, T. A. Stošicki, D. Stopar, M. Kalin, S. Kobe and S. A. Novak, ACS Appl. Mater. Interfaces, 2015, 7, 1644–1651 CrossRef CAS PubMed.
- G. P. Dubey and S. Ben-Yehuda, Cell, 2011, 144, 590–600 CrossRef CAS PubMed.
- J. Bruzaud, J. Tarrade, A. Coudreuse, A. Canette, J.-M. Herry, E. T. de Givenchy, T. Darmanin, F. Guittard, M. Guilbaud and M.-N. Bellon-Fontaine, Colloids Surf., B, 2015, 131, 59–66 CrossRef CAS PubMed.
- A. K. Baidya, S. Bhattacharya, G. P. Dubey, G. Mamou and S. Ben-Yehuda, Curr. Opin. Microbiol., 2018, 42, 1–6 CrossRef CAS PubMed.
- S. Bhattacharya, A. K. Baidya, R. R. Pal, G. Mamou, Y. E. Gatt, H. Margalit, I. Rosenshine and S. Ben-Yehuda, Cell Rep., 2019, 27(334–342), e310 Search PubMed.
- R. R. Pal, A. K. Baidya, G. Mamou, S. Bhattacharya, Y. Socol, S. Kobi, N. Katsowich, S. Ben-Yehuda and I. Rosenshine, Cell, 2019, 177(683–696), e618 Search PubMed.
- S. Pirbadian, S. E. Barchinger, K. M. Leung, H. S. Byun, Y. Jangir, R. A. Bouhenni, S. B. Reed, M. F. Romine, D. A. Saffarini and L. Shi, Biophys. J., 2015, 108, 368a CrossRef.
- G. P. Dubey, G. B. M. Mohan, A. Dubrovsky, T. Amen, S. Tsipshtein, A. Rouvinski, A. Rosenberg, D. Kaganovich, E. Sherman and O. Medalia, Dev. Cell, 2016, 36, 453–461 CrossRef CAS PubMed.
- M. Dorkhan, L. E. C. de Paz, M. Skepo, G. Svensater and J. R. Davies, Microbiology-Sgm, 2012, 158, 390–397 CrossRef CAS PubMed.
- R. Fraioli, P. M. Tsimbouri, L. E. Fisher, A. H. Nobbs, B. Su, S. Neubauer, F. Rechenmacher, H. Kessler, M. P. Ginebra, M. J. Dalby, J. M. Manero and C. Mas-Moruno, Sci. Rep., 2017, 7, 16363 CrossRef PubMed.
- M. R. Nejadnik, H. C. van der Mei, H. J. Busscher and W. Norde, Appl. Environ. Microbiol., 2008, 74, 916–919 CrossRef CAS PubMed.
- M. N. Dickson, E. I. Liang, L. A. Rodriguez, N. Vollereaux and A. F. Yee, Biointerphases, 2015, 10, 021010 CrossRef PubMed.
- K. Modaresifar, S. Azizian, M. Ganjian, L. E. Fratila-Apachitei and A. A. Zadpoor, Acta Biomater., 2019, 83, 29–36 CrossRef CAS PubMed.
- V. K. Truong, H. K. Webb, E. Fadeeva, B. N. Chichkov, A. H. F. Wu, R. Lamb, J. Y. Wang, R. J. Crawford and E. P. Ivanova, Biofouling, 2012, 28, 539–550 CrossRef CAS PubMed.
- J. W. Ma, Y. K. Sun, K. Gleichauf, J. Lou and Q. L. Li, Langmuir, 2011, 27, 10035–10040 CrossRef CAS PubMed.
- P. F. Tang, W. Zhang, Y. Wang, B. X. Zhang, H. Wang, C. J. Lin and L. H. Zhang, J. Nanomater., 2011, 2011, 1004 Search PubMed.
- G. B. Hwang, K. Page, A. Patir, S. P. Nair, E. Allan and I. P. Parkin, ACS Nano, 2018, 12, 6050–6058 CrossRef CAS PubMed.
- K. Ellinas, D. Kefallinou, K. Stamatakis, E. Gogolides and A. Tserepi, ACS Appl. Mater. Interfaces, 2017, 9, 39781–39789 CrossRef CAS PubMed.
- R. S. Friedlander, H. Vlamakis, P. Kim, M. Khan, R. Kolter and J. Aizenberg, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5624–5629 CrossRef CAS PubMed.
- S. Sure, M. L. Ackland, A. A. J. Torriero, A. Adholeya and M. Kochar, Microbiology, 2016, 162, 2017–2028 CrossRef CAS PubMed.
- N. S. Malvankar and D. R. Lovley, ChemSusChem, 2012, 5, 1039–1046 CrossRef CAS PubMed.
- G. Reguera, K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen and D. R. Lovley, Nature, 2005, 435, 1098 CrossRef CAS PubMed.
- R. J. Steidl, S. Lampa-Pastirk and G. Reguera, Nat. Commun., 2016, 7, 12217 CrossRef CAS PubMed.
- M. Maruthupandy, M. Anand, G. Maduraiveeran, A. S. H. Beevi and R. J. Priya, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6, 045007 Search PubMed.
- S. Pirbadian, S. E. Barchinger, K. M. Leung, H. S. Byun, Y. Jangir, R. A. Bouhenni, S. B. Reed, M. F. Romine, D. A. Saffarini and L. Shi, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12883–12888 CrossRef CAS PubMed.
- J. P. Remis, D. Wei, A. Gorur, M. Zemla, J. Haraga, S. Allen, H. E. Witkowska, J. W. Costerton, J. E. Berleman and M. Auer, Environ. Microbiol., 2014, 16, 598–610 CrossRef CAS PubMed.
- C. Takahashi, G. Kalita, N. Ogawa, K. Moriguchi, M. Tanemura, Y. Kawashima and H. Yamamoto, Anal. Bioanal. Chem., 2015, 407, 1607–1613 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sm00602e |
| This journal is © The Royal Society of Chemistry 2020 |