Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dendrimicelles with pH-controlled aggregation number of core-dendrimers and stability†
Junyou
Wang
 ab,
Liu
Lei
a,
Ilja K.
Voets
ab,
Liu
Lei
a,
Ilja K.
Voets
 c,
Martien A.
Cohen Stuart
c,
Martien A.
Cohen Stuart
 a and
Aldrik H.
Velders
a and
Aldrik H.
Velders
 *b
*b
aState-Key Laboratory of Chemical Engineering and Shanghai Key Laboratory of Multiphase Materials Chemical Engineering, East China University of Science and Technology, Shanghai 200237, People's Republic of China
bLaboratory of BioNanoTechnology, Wageningen University, Bornse Weilanden, Wageningen, The Netherlands. E-mail: aldrik.velders@wur.nl
cLaboratory of Macromolecular and Organic Chemistry, Department of Chemistry and Chemical Engineering, and Institute for Complex Molecular Systems, Eindhoven University of Technology, The Netherlands
First published on 17th August 2020
Abstract
We present a simple way to build up well-controlled coacervate-core dendrimicelles by assembly of anionic PAMAM dendrimers with a cationic-neutral diblock copolymer. Upon increasing pH, the formation of micellar structures shows constant size but the number of dendrimer molecules incorporated in one micelle decreases, following the charge stoichiometry formation rules; concomitantly, the salt stability increases. This study shows the straightforward tuning of macromolecular core-units and related micelle properties.
Complex coacervate core micelles (C3Ms) are nano-structures formed through electrostatic interaction between a polyion-neutral diblock copolymer and oppositely charged polyelectrolytes.1 This relatively new category of micelles is also known as: polyion complex micelles (PIC micelles), block ionomer complexes (BICs) or polyelectrolyte complexes (PECs).2–4 In contrast to classical micelles assembled from amphiphilic molecules, which are driven by hydrophobic attractions, and featuring a dense, solvent-free core filled completely by hydrophobic moieties, C3Ms are often dynamic structures. This is because the core of C3Ms consists of fluid-like coacervate complex containing a significant amount of water, and the final structure depends on many factors, such as pH, ionic strength, nature of charged groups, and polymer length.5–9 Obviously, these tunabilities make the C3Ms attractive candidates for designing responsive soft materials.10,11 The co-assembly of the oppositely charged components leads to an optimal micellization around charge stoichiometry, where equal number of positive and negative charges are incorporated in the micelle.12,13 This typical property offers interesting possibilities to control the aggregation number of the molecules in the micelle, e.g. one component can act as a template, and the requirement of charge stoichiometry then serves to adjust the aggregation number of the other component. The charge stoichiometry principle has been widely applied for guiding preparation of C3Ms, but not used as a control factor for tuning the properties, e.g., the aggregation number of the C3Ms.
Dendrimers represent a special class of polyelectrolyte macromolecules with a highly symmetric branched structure, explored in materials science and particularly biomedical applications.14–16 Unfortunately, the use of free dendrimer is strongly precluded by the constrained size and surface properties.17–20 Developing supramolecular structures involving dendrimers, in particular core–shell ‘dendrimicelles’ with delicate control of size, shape, structure and function eliminates this problem.21–24 More recently, we showed in a systematic study how 10 different generations of anionic PAMAM dendrimers interact with cationic-neutral block-copolymers. Interestingly, micelles were formed with the same size, containing a decreasing number of dendrimers upon increase of the generation number.25 Controlling and tuning dendrimer numbers in C3Ms is important for potential applications, e.g. in material or medicinal settings, when the dendrimers can be loaded with functional cargoes in their cavities.
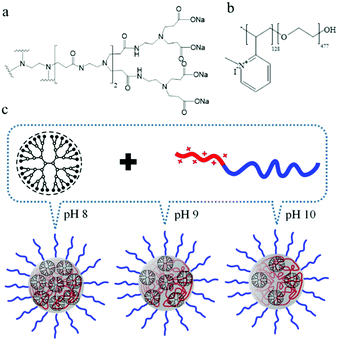
As the surface charge of dendrimers typically shows strong pH dependency, together with the charge stoichiometry rules for polyelectrolytes co-assembly, we now hypothesized the aggregation number of dendrimers in dendrimicelles, for a specific generation of PAMAM, could be tuned by changing the surface charge of the dendrimer. Here we demonstrate a simple strategy that allows to construct dendrimicelles with constant size, but pH-tunable dendrimer encapsulation and salt stability, Scheme 1. Present strategy holds potential for developing smart materials that allow multiple responses and controlled releases.
We selected polyamidoamine (PAMAM) dendrimers as well defined pH-sensitive micelle core moiety in our study since they are water soluble, commercially available and have proven their efficacy in a variety of applications.26,27 Third generation (G3), carboxylate-terminated PAMAM dendrimers (also referred to as generation G2.5, as the synthesis lacks the final amidation step), carrying 32 –COONa groups,28 have a pH-dependent charge density due to protonation of the surface carboxylate ions (Scheme 1a). The diblock copolymer, P2MVP128-b-PEO477, contains quaternized pyridine groups on the charged block, giving a positive charge density independent of pH (Scheme 1b). Hence, via the degree of protonation of PAMAM the co-assembly of the charged blocks becomes dependent on pH. Typically, the association of cationic and anionic homopolymers results in a macroscopic phase separation, with the denser phase being the coacervate phase, as has been shown, e.g., in our previous studies.5,29 A micellar structure forms when a neutral block such as PEO is introduced into at least one of the charged blocks; since PEO does not participate in the phase separation, it can stop the continuous phase separation and restrict the coacervate complexes to sizes in the colloidal range. Indeed, addition of P2MVP homopolymer to a dendrimer solution induces a dramatic increase in light scattering intensity due to formation of droplets of varying size, indicating that macroscopic phase separation happens in solution. Addition of PEO homopolymer has no effect, while the addition of P2MVP128-b-PEO477 diblock copolymer to dendrimer solution leads to well-defined micelles, as confirmed by light and X-ray scattering data as discussed below. These observations suggest that micelle formation is indeed driven by the charged blocks and size-controlled by the neutral block (Scheme 1c). We call this micelle G3-C3Ms.
In order to verify the micellization is controlled by pH, we titrated the diblock copolymer into dendrimer solutions at different values of fixed pH, from 6 to 11. The dendrimer solution used was a 1 mL 0.025 mM one, corresponding to a surface group concentration of 0.8 mM. The variations of the light scattering intensity and hydrodynamic radius were recorded as a function of the amount of positive charge added into the dendrimer solution. No aggregation was found at pH 6, with hardly any change in light scattering intensity over the entire composition titration range (Fig. 1a). Previous studies have found that complexation between dendrimer and cationic polymers only happens when the charge density of dendrimer exceeds a critical value.30,31 The protonation degree of carboxylate groups on the dendrimer surface is modulated by pH,32 and apparently at pH 6 mainly (protonated) carboxylic acid groups are present and charge density is not high enough to form coacervate complexes.13,25 In addition, at this relatively low pH also the fraction of non-quaternized pyridines of the diblock copolymer can be protonated, interfering with the coacervation process of the dendrimers and diblock copolymer. At pH 7, the light scattering intensity has a peak at about 0.3 μmol diblock copolymer charge, suggesting formation of some sort of aggregates. Titrations at pH 8 to 11 give similar profiles; the titration curve at pH 10 is plotted in Fig. 1a as a representative example (for other pH values, see ESI,† Fig. S1). The intensity increases with increasing amount of positive charge and reaches a maximum around 0.81 μmol, which equals the amount of carboxylate groups in the dendrimer solution (0.8 μmol). This observation confirms that the carboxylate groups on the dendrimer surface are completely deprotonated at pH 10, which is consistent with previous reports.31 The intensity maximum corresponds to the preferred micelle composition, PMC,12 (the first peak in titration curve) where the charge stoichiometry is satisfied so that the negative charges of the dendrimer are compensated completely by the positive charges from the diblock copolymer. Micelles directly prepared by mixing dendrimer and diblock copolymer in the ratio corresponding to the optimum charge stoichiometry, provided light scattering data with similar intensity and particle size, corroborating thermodynamic control of the micelle formation.
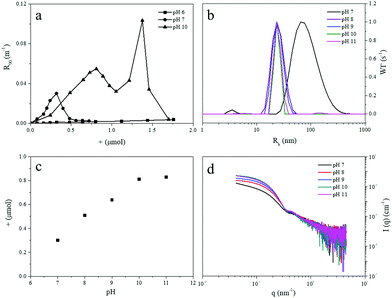 | ||
| Fig. 1 (a) Light scattering titration of dendrimer G3 with P2MVP128-b-PEO477 diblock copolymer at pH 6, 7, and 10, the intensity is plotted as a function of positive charge of diblock copolymer added to a 1 mL 0.8 mM dendrimer solution, corresponding to 0.8 μmol of surface groups. Titration curves at pH 8, 9, and 11 are similar with that of pH 10, see ESI.† (b) Size and size distribution of the formed micelles at different pH. (c) Added amount of positive charge of diblock copolymer at PMC point at different pH. (d) SAXS profiles of G3-C3Ms at different pH. | ||
Aggregates formed at pH 7 are about 75 nm in size, with a rather wide distribution around the mean (Fig. 1b). At pH 8–11 micelles are formed, with radii around 24 nm, and narrower size distributions. This is comparable to the size of C3Ms obtained with linear polyelectrolytes.33,34 The bigger size but low scattering intensity at pH 7 suggests that the aggregates may not be well-defined C3Ms, but rather floppy structures without clear core–shell structure, likely due to insufficient driving force. The PMC peak positions (Fig. 1c) demonstrate the required amount of positive charge to achieve the PMC. We see that these values increase upon going from pH 7 to pH 10, and stay constant upon further increase of pH to 11, confirming that the dendrimer indeed is already fully charged, i.e. deprotonated, at pH 10. Angular dependence and electrophoretic mobility results (Fig. S2, ESI†) indicate the formed micelles at different pH are spherical nanoparticles with neutral surface. Small-angle X-ray scattering profiles taken at the PMC (Fig. 1d) confirm that C3Ms are well-defined spherical core–shell structures for pH 8 to 10, but not for pH 7, consistent with the occurrence of irregular structures at this pH. By fitting the scattering curves for pH 8 to 10 with a polydisperse core–shell model, we find a core radius around 11 nm and a shell thickness of about 13 nm (Fig. S3 and Table S2, ESI†). Adding more diblock copolymers into the dendrimer solution, beyond the PMC, results in a second peak in both the intensity and radius (Fig. 1a), which we ascribe to a shape transition from spherical to elongated (worm-like) structures, as was established by analyzing the angle-dependent and depolarized light scattering data in a previous study.32 A similar shape transition was also found in titrations of different linear polyelectrolytes.35–37
The molar mass of micelles, Mmicelle, and the aggregation number Ndendrimer, and Npolymer of dendrimer and polymer, respectively, are obtained from partial Zimm analysis and plotted as a function of pH in Fig. 2 (see also Fig. S4, S5 and Table S3, ESI†). The floppy aggregates at pH 7 show a bigger molar mass, with more dendrimer and diblock copolymer incorporated in the structure. From pH 8 to pH 10, the aggregates (‘dendrimicelles’) show a constant number of diblock copolymers about 17 ± 1, but a decreasing number of dendrimers, from 82 to 60, which fits well with the decrease of the molar mass of G3-C3Ms. The micelles at pH 11 have similar molar mass and aggregation numbers as micelles at pH 10; the surface groups of dendrimer are obviously completely deprotonated already at pH 10.
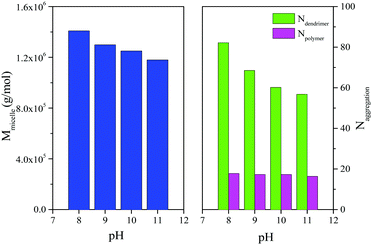 | ||
| Fig. 2 Micellar mass, Mmicelle and numbers of dendrimer and diblock copolymer incorporated in one micelle (aggregation number, N) as a function of pH. Note that samples are prepared separately under fixed pH. Calculation details and original numbers are shown in ESI.† | ||
Apparently, independently of pH at which the dendrimicelles were prepared, the G3-C3Ms maintain a constant number of diblock copolymer, but host a variable number of dendrimers in the core at different pH. This result can be explained in terms of forces that control micellization of surfactants and amphiphilic copolymers. The size of a stable micelle is controlled by a balance between two forces: the interfacial tension of the (complex) core (which depends on the chemical composition of the core, and the ionic strength), and the osmotic pressure between the corona chains (that essentially depends on length of the neutral (PEO) blocks). Hence, these two forces do not depend on dendrimer generation or pH, so micellar size, and the number of diblock copolymers are (roughly) constant. The diblock copolymers build up a kind of cage with fixed number of polymers and, as consequence, fixed overall cationic charges.
To fill the cage and neutralize all the charges, we either need many dendrimers with relatively few surface charges (lower pH), or few dendrimers with many surface charges (high pH). This is equivalent to the condition, n+ = n−, or NpZp = NdZd, where Np and Nd are the numbers of cationic block and dendrimer per micelle, and Zp and Zd are the charge numbers per molecule, respectively. Similar results were found in our previous study on PAMAM dendrimer-based C3Ms, in which the dendrimer aggregation number was manipulated by dendrimer generation used, at constant number of diblock copolymers.32 For the micelles prepared at the different pH values 11, 10, 9 and 8, the net dendrimer negative charge, Zd, is 32, 32, 28 and 25, respectively. Clearly, the pH dependent deprotonation degree is not following the same trend as of single carboxylic acids; the close proximity of many carboxylate groups increase the pKa of neighbouring carboxylic acid groups. Although the overall change in Zd of the dendrimers is significant, it is not enough in this system to allow for a subtle pH-responsive micelle rupture; something we could achieve more readily with supramolecular and redox-responsive micelle-core systems.38,39
Since G3-C3Ms at lower pH incorporate more dendrimers in the core than micelles at higher pH, one might initially expect a bigger micellar size at low pH, which however seems not to be the case judging the light scattering data (Fig. 1b). Estimating the number of dendrimers, and hence total dendrimer volume, increases by about 30%, from 60 at pH 10 to 82 at pH 8, the core size of the micelles can vary only by a factor of (82/60)1/3 = 1.1, so by about 10% in radius. Such a slight change is possibly not well detected in our experiments. Another possibility is that the core size does not change while the core density increases with increasing dendrimer number, implying less water in the core. Again, this change cannot be large either, as the coacervate complexes in the micellar core contain more than 60% of water (polymer density is relatively low) so a small decrease in water content may not be picked up.5 Evidently, the noticeable change in dendrimer number does not have important consequences for either core size or core density.
It is well known that micelles from polyelectrolytes are sensitive to ionic strength and fall apart completely above a critical salt concentration, Cs,cr.40–42 In Fig. 3 the Cs,cr of dendrimicelles is plotted, obtained from salt titrations (ESI,† Fig. S6 and Table S5). Clearly, the stability increases from 75 mM to 200 mM upon increasing pH from 7 to 10 or 11. The high salt stability at high pH relates to the stronger driving force due to the fact that more negative charges per dendrimer are involved in the micellization. The CSC of 200 mM at high pH is similar to the CSC found for the G3 in the previous study.25 Interestingly, at pH 7 here, the G3 dendrimer has about a negative charge of 25, which is still higher than the charge of a G2 PAMAM (16), but the CSC of the G2 was found to be 160 mM.25 This might hint towards the importance of charge density, rather than just charge alone.
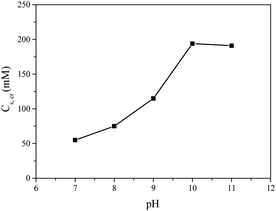 | ||
| Fig. 3 Critical salt concentration Cs,cr of G3-C3Ms formed at different pH. Micelles were prepared at the PMC ratio. | ||
In conclusion, we have shown interesting aspects of micelles formed by the electrostatic interaction between the anionic PAMAM dendrimer and a cationic-neutral diblock copolymer. The spherical micelles are core–shell structures with a core size around 11 nm and shell thickness of about 13 nm. The micellar size and number of diblock copolymer incorporated in one micelle is constant, but the dendrimer number and the stability against salt can be tuned by changing pH. These fascinating properties are related to the tunable electrostatic driving force and the co-assembly of two polyelectrolytes, which cannot be achieved in the assembly of single amphiphilic macromolecules. Our findings increase options for polyelectrolyte self-assembly, particularly with non-linear and nano-scaled structures such as dendrimers. The control and tunability of dendrimicelles also allows for building up multi-functional supra-micelles, e.g., by controlled packing of dendrimers with different cargo on, or inside dendrimers,43–45 for delivery and release applications, or for formation of 2d superstructures.46–48 Variations in dendrimer type, generation and block-copolymer will provide a plethora of permutations to explore and exploit. To create better controlled micelle structures, and to fine tune the responsiveness of micelles to triggers such as temperature, pH, salt, light.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank PCC at WUR for use of their DLS equipment & support.Notes and references
- I. K. Voets, A. de Keizer and M. A. Cohen Stuart, Adv. Colloid Interface Sci., 2009, 147–148, 300 CrossRef CAS PubMed.
- A. Harada and K. Kataoka, Macromolecules, 1995, 28, 5294 CrossRef CAS.
- A. V. Kabanov, T. K. Bronich, V. A. Kabanov, K. Yu and A. Eisenberg, Macromolecules, 1996, 29, 6797 CrossRef CAS.
- J. F. Gohy, S. K. Varshney, S. Antoun and R. Jerome, Macromolecules, 2000, 33, 9298 CrossRef CAS.
- E. Spruijt, A. H. Westphal, J. W. Borst, M. A. Cohen Stuart and J. van der Gucht, Macromolecules, 2010, 43, 6476 CrossRef CAS.
- C. E. Sing and S. L. Perry, Soft Matter, 2020, 16, 2885 RSC.
- S. Srivastava and M. V. Tirrell, Adv. Chem. Phys., 2016, 161, 499 CrossRef CAS.
- J. van der Gucht, E. Spruijt, M. Lemmers and M. A. Cohen Stuart, J. Colloid Interface Sci., 2011, 361, 407 CrossRef CAS PubMed.
- W. M. Aumiller and C. D. Keating, Adv. Colloid Interface Sci., 2017, 239, 75 CrossRef CAS PubMed.
- C. Facciotti, V. Saggiomo, S. van Hurne, A. Bunschoten, R. Kaup and A. H. Velders, Supramol. Chem., 2020, 32, 30 CrossRef CAS.
- J. Y. Wang, A. H. Velders, E. Gianolio, S. Aime, F. J. Vergeldt, H. Van As, Y. Yan, M. Drechsler, A. de Keizer, M. A. Cohen Stuart and J. van der Gucht, Chem. Commun., 2013, 49, 3736 RSC.
- S. van der Burgh, A. de Keizer and M. A. Cohen Stuart, Langmuir, 2004, 20, 1073 CrossRef CAS PubMed.
- Y. Yan, N. A. M. Besseling, A. de Keizer, A. T. M. Marcelis, M. Drechsler and M. A. Cohen Stuart, Angew. Chem., Int. Ed., 2007, 46, 1807 CrossRef CAS PubMed.
- S. H. Medina and M. E. H. El-Sayed, Chem. Rev., 2009, 109, 3141 CrossRef CAS PubMed.
- M. A. Mintzer and M. W. Grinstaff, Chem. Soc. Rev., 2011, 40, 173 RSC.
- F. Grohn, Soft Matter, 2010, 6, 4296 RSC.
- A. Sousa-Herves, E. Fernandez-Megia and R. Riguera, Chem. Commun., 2008, 3136 RSC.
- J. Khandare, M. Calderon, N. M. Dagia and R. Haag, Chem. Soc. Rev., 2012, 41, 2824 RSC.
- W. C. She, K. Luo, C. Y. Zhang, G. Wang, Y. Y. Geng, L. Li, B. He and Z. W. Gu, Biomaterials, 2013, 34, 1613 CrossRef CAS PubMed.
- D. A. Tomalia, Soft Matter, 2010, 6, 456 RSC.
- H. R. Stapert, N. Nishiyama, D. L. Jiang, T. Aida and K. Kataoka, Langmuir, 2000, 16, 8182 CrossRef CAS.
- R. Haag, Angew. Chem., Int. Ed., 2004, 43, 278 CrossRef CAS PubMed.
- S. Svenson and D. A. Tomalia, Adv. Drug Delivery Rev., 2005, 57, 2106 CrossRef CAS PubMed.
- F. Reinhold, U. Kolb, I. Lieberwirth and F. Grohn, Langmuir, 2009, 25, 1345 CrossRef CAS PubMed.
- J. Y. Wang, I. K. Voets, R. Fokkink, J. van der Gucht and A. H. Velders, Soft Matter, 2014, 10, 7337 RSC.
- D. Astruc, E. Boisselier and C. Ornelas, Chem. Rev., 2010, 110, 1857 CrossRef CAS PubMed.
- D. A. Tomalia, J. B. Christensen and U. Boas, Dendrimers, dendrons, and dendritic polymers: discovery, applications, and the future, Cambridge University Press, NY, 2012 Search PubMed.
- Carboxyl-terminated PAMAM dendrimers have same number of repeat units and surface groups with amine-terminated PAMAM for the same generation. Hence, instead of using half-generation number, we use integer number to represent it.
- J. Y. Wang, M. A. Cohen Stuart and J. van der Gucht, Macromolecules, 2012, 45, 8903 CrossRef CAS.
- H. Zhang, P. L. Dubin, J. Ray, G. S. Manning, C. N. Moorefield and G. R. Newkome, J. Phys. Chem. B, 1999, 103, 2347 CrossRef CAS.
- Y. Li and P. L. Dubin, Macromolecules, 1995, 28, 8426 CrossRef CAS.
- N. Miura, P. L. Dubin, C. N. Moorefield and G. R. Newkome, Langmuir, 1999, 15, 4245 CrossRef CAS.
- Y. Lee and K. Kataoka, Soft Matter, 2009, 5, 3810 RSC.
- J. Y. Wang, A. de Keizer, H. P. van Leeuwen, Y. Yan, F. Vergeldt, H. van As, P. H. H. Bomans, N. Sommerdijk, M. A. Cohen Stuart and J. van der Gucht, Langmuir, 2011, 27, 14776 CrossRef CAS PubMed.
- Y. Yan, N. A. M. Besseling, A. de Keizer, M. Drechsler, R. Fokkink and M. A. Cohen Stuart, J. Phys. Chem. B, 2007, 111, 11662 CrossRef CAS PubMed.
- T. T. H. Pham, J. Y. Wang, M. W. T. Werten, F. Snijkers, F. A. de Wolf, M. A. Cohen Stuart and J. van der Gucht, Soft Matter, 2013, 9, 8923 RSC.
- J. Y. Wang, A. de Keizer, R. Fokkink, Y. Yan, M. A. Cohen Stuart and J. van der Gucht, J. Phys. Chem. B, 2010, 114, 8313 CrossRef CAS PubMed.
- C. Facciotti, V. Saggiomo, A. Bunschoten, R. Fokkink, J. B. ten Hove, J. Y. Wang and A. H. Velders, Soft Matter, 2018, 14, 9542 RSC.
- C. Facciotti, V. Saggiomo, A. Bunschoten, J. B. ten Hove, M. T. M. Rood, F. W. B. van Leeuwen and A. H. Velders, ChemSystemsChem, 2020, e1900032 Search PubMed.
- X. F. Yuan, A. Harada, Y. Yamasaki and K. Kataoka, Langmuir, 2005, 21, 2668 CrossRef CAS PubMed.
- Y. Yan, A. de Keizer, M. A. Cohen Stuart, M. Drechsler and N. A. M. Besseling, J. Phys. Chem. B, 2008, 112, 10908 CrossRef CAS PubMed.
- J. Y. Wang, M. A. Cohen Stuart, A. T. M. Marcelis, M. Colomb-Delsuc, S. Otto and J. van der Gucht, Macromolecules, 2012, 45, 7179 CrossRef CAS.
- M. V. Gomez, J. Guerra, A. H. Velders and R. M. Crooks, J. Am. Chem. Soc., 2009, 131, 341 CrossRef CAS PubMed.
- A. Ruggi, C. Beekman, D. Wasserberg, V. Subramaniam, D. N. Reinhoudt, F. W. B. van Leeuwen and A. H. Velders, Chem. – Eur. J., 2011, 17, 464 CrossRef CAS PubMed.
- J. During, A. Holzer, U. Kolb, R. Branscheid and F. Grohn, Angew. Chem., Int. Ed., 2013, 52, 8742 CrossRef PubMed.
- J. B. ten Hove, J. Wang, F. W. B. van Leeuwen and A. H. Velders, Nanoscale, 2017, 9, 18619 RSC.
- J. B. ten Hove, J. Y. Wang, M. N. van Oosterom, F. W. B. van Leeuwen and A. H. Velders, ACS Nano, 2017, 11, 11225 CrossRef CAS PubMed.
- J. B. ten Hove, F. W. B. van Leeuwen and A. H. Velders, Nat. Commun., 2018, 9, 5207 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sm00458h |
| This journal is © The Royal Society of Chemistry 2020 |