Open Access Article
Open Access ArticleProgrammable patterns in a DNA-based reaction–diffusion system†
Sifang
Chen
 ab and
Georg
Seelig
*bcd
ab and
Georg
Seelig
*bcd
aDepartment of Physics, University of Washington, USA
bMolecular Engineering & Sciences Institute, University of Washington, USA. E-mail: gseelig@uw.edu
cDepartment of Electrical & Computer Engineering, University of Washington, USA
dPaul G. Allen School for Computer Science & Engineering, University of Washington, USA
First published on 27th March 2020
Abstract
Biology offers compelling proof that macroscopic “living materials” can emerge from reactions between diffusing biomolecules. Here, we show that molecular self-organization could be a similarly powerful approach for engineering functional synthetic materials. We introduce a programmable DNA embedded hydrogel that produces tunable patterns at the centimeter length scale. We generate these patterns by implementing chemical reaction networks through synthetic DNA complexes, embedding the complexes in the hydrogel, and triggering with locally applied input DNA strands. We first demonstrate ring pattern formation around a circular input cavity and show that the ring width and intensity can be predictably tuned. Then, we create patterns of increasing complexity, including concentric rings and non-isotropic patterns. Finally, we show “destructive” and “constructive” interference patterns, by combining several ring-forming modules in the gel and triggering them from multiple sources. We further show that computer simulations based on the reaction–diffusion model can predict and inform the programming of target patterns.
Introduction
Programmable matter research aims to engineer functional materials that can autonomously transform their appearances or physical properties in response to environmental stimuli and user-defined inputs. Top down methods like 3D-printing have enabled the development of shapeshifting biomimetic constructs that are sensitive to heat, light, or water.1,2 Advances in micro-robotics have led to modular robotic swarms that can self-organize into two- and three-dimensional structures.3,4 But we are still far from creating truly programmable macroscopic matter. Currently, synthetic materials and systems either rely on components too large to be integrated into material fabrics, as with modular robot systems, or have limited functions, as with 3D printed materials.Chemical computing offers a tantalizing alternative. Biological patterning processes like camouflaging and morphogenesis suggest that complex and environmentally responsive systems could arise from the self-organization of information-bearing agents like molecules or cells.5,6 Engineering molecular systems to predictably form complex patterns like those seen in biology would clearly have significant implications for programmable materials research.
The mathematical model of reaction–diffusion provides a framework for designing and engineering programmable structures through chemical computing.6–9 In this model, spatial patterns can emerge from local interactions between diffusing agents.10 Simulations developed within this framework have successfully replicated complex biological patterns,11,12 suggesting a path toward model-guided engineering of autonomous self-organizing systems. However, experimental de novo realizations of pattern formation have been sparse.
Early examples of synthetic pattern formation include the Belousov–Zhabotinsky (BZ) chemical oscillator,13,14 which generates macroscopic spatiotemporal patterns via a series of redox reactions. While the mechanics of BZ reactions are well understood, we cannot control reaction kinetics or program the resulting patterns to display target behaviors. Synthetic biologists have genetically engineered quorum-sensing bacteria to create stripes and traveling waves.15–17 Such results showcase the potential of a biochemical approach to programming self-organized pattern formation, but the precision of patterning is still limited because the engineered reaction networks operate in a background of evolved and not fully understood cellular machinery. Cell-free biochemical reaction networks are another promising alternative but systems engineered to date, although capable of generating a wide range of patterns in aqueous reactors,18–21 still largely rely on catalysis of evolved enzymes and have limited programmability.
DNA is unique, even among biopolymers, because DNA interactions are programmable by sequence.22,23 DNA origami and related self-assembly technologies take advantage of this predictability for the construction of 2- and 3-dimensional objects of varying sizes and complexity.24,25 This work has culminated in macroscopic materials with nanometer-scale addressability. But DNA origamis, periodic crystals26 and DNA-crosslinked hydrogels27–34 are either expensive because DNA acts as the primary structural component or lack long-range order because interactions are localized. Thus, to recapitulate the diversity and scale of biological patterns and materials with DNA alone we still need to develop approaches that extend to the centimeter scale and beyond.
To address this need, recent work has begun to explore the feasibility of DNA-only reaction–diffusion patterns.29,34–37 Toehold-mediated DNA strand displacement has proved to be a convenient framework for implementing complex reaction sequences using synthetic DNA in well-mixed test tubes.38,39 Using the principles of strand displacement, researchers have created sophisticated reaction networks that perform computation like neural networks,40,41 diagnostic classifiers,42 dynamic 3D nanostructures43,44 and even approximate the dynamics of formal, mathematically specified chemical reaction networks (CRNs).45–48 Building on these results, theoretical work has argued that a wide range of patterns is achievable if DNA-based CRNs are embedded in a spatial reactor.49,50 Chirieleison et al. took an important step toward experimentally demonstrating pattern formation with DNA strand displacement-based CRNs and engineered an edge detection system.51 However, despite the advances made in these projects, the state of art for programming macroscopic features still lags that of their microscopic counterparts.
Here, we report the design and synthesis of DNA embedded hydrogels for programmable spatial patterning at the centimeter length scale. Patterns are generated via the reaction–diffusion of DNA complexes separately embedded in porous hydrogel and predefined cavities in the gel. Using this system, patterns of varying geometries can be generated and quantitatively tuned by controlling the reaction rates of species. To further demonstrate programmability, we show that the dynamic behavior of these spatial patterns can be predicted by computer simulations.
Constructing a pulse-generator
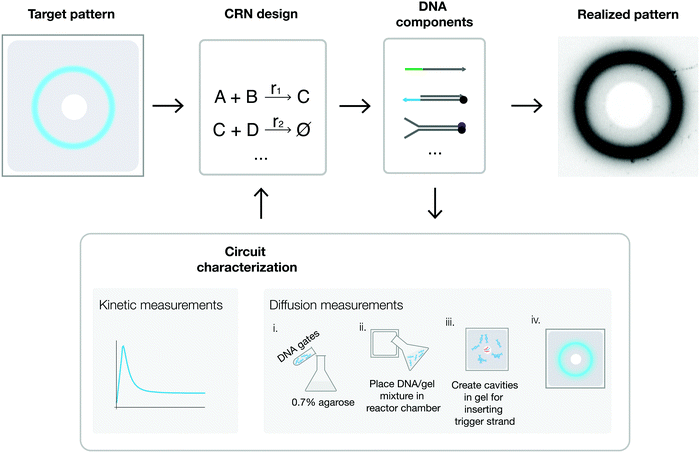
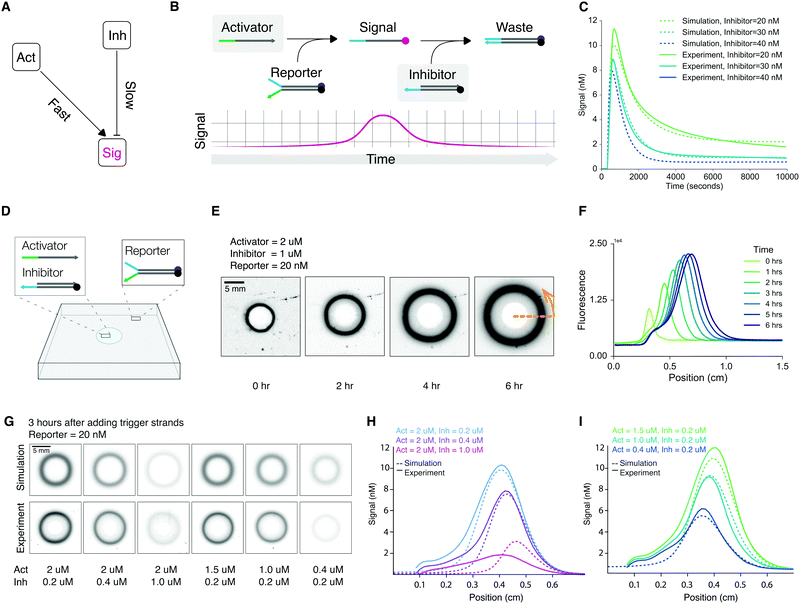
Fig. 1 shows the workflow of our DNA-based programmable patterning system. A simple ring-forming module forms the basic building block for all other patterns realized in this work (Fig. 1A). Rings are an archetype for studying synthetic pattern formation14,16 and, as we will show, form an ideal starting point for generating more complex patterns. To implement this module, we begin by formulating a pulse-generating CRN (Fig. 1B). We then realize this CRN using DNA strand displacement-based complexes (Fig. 1C). Next, we embedded DNA in hydrogel by suspending DNA complexes in an agarose solution and molding the mixture into thin sheets. Finally, we load initiator strands into cavities in the hydrogel to trigger programmed pattern formation (Fig. 1D). A predictive spatial model built in Visual DSD52 informs the concentrations of initiator strands required for generating target patterns. Input parameters for the model include reaction constants and diffusion coefficients inferred from spectrometry and gel experiments (Fig. 1D). Depending on the initial conditions (concentrations of initiating strands) and boundary conditions (shape and placement of cavities), gels embedded with identical DNA gates can be programmed to display a variety of spatial dynamics (Fig. 1E).The core CRN for pulse formation consists of three reactants: activator, reporter, and inhibitor (Fig. 2A). An activator is a single-stranded DNA molecule used to initiate a reaction cascade. A reporter is a partially double-stranded DNA complex with a fluorophore-labeled signal (top) strand and a quencher-labeled bottom strand that is fully complementary to the activator. An inhibitor is a partially double-stranded DNA complex with an unmodified protector strand and a longer quencher strand that is fully complementary to the signal strand (Fig. 2B). We entered the desired domain structures into NUPACK Design53 to generate compatible sequences for building the pulse module (Section S1, ESI†). These sequences are listed in Table S1 (ESI†). In a well-mixed setting, this three-component reaction module produces a single pulse via a two-step reaction (Fig. 2B): first, activators trigger fluorescence by releasing signal strands from reporters through toehold-meditated strand displacement; then, the signal is absorbed and repressed by the inhibitor.
To test the pulse module, we added reporter and inhibitor to a solution, triggered the reaction by adding the activator, and measured fluorescence changes using a spectrofluorometer. As designed, we can program the pulse shape by changing reactant concentrations. Specifically, pulse amplitude and duration depend on the rates of signal activation and inhibition: when we lowered the activator-to-inhibitor ratio in the solution, we observed a corresponding decrease in pulse amplitude and duration (Fig. 2C).
We developed a computational model in Visual DSD to simulate the pulse module (Sections S2 and S3, ESI†). The model consists of two reversible bimolecular reactions: signal activation (activator and reporter react to produce signal) and signal inhibition (signal is absorbed by inhibitor). Model parameters include reaction rate constants of signal activation and inhibition, as measured from separate spectrometry experiments (Fig. S1–S4, ESI†). Our simulation confirms the module as a pulse generator (Fig. 2C) and further refines the rate constants to improve prediction accuracy (Section S3, and Table S2, ESI†).
Programming single-ring patterns
Next, we set out to test spatial pattern formation. We synthesized DNA embedded hydrogel sheets by suspending reporters in 0.7% agarose solution. We chose a 0.7% agarose gel because it provides a good combination of structural support and low melting point compatible with embedding double-stranded DNA complexes. We cast the gels into thin sheets by pouring the mixed solutions into acrylic reactors. Fig. S5 and S6 (ESI†) illustrate this process in detail.To initiate pattern formation, we loaded activators and inhibitors into a circular cavity at the center of the DNA embedded hydrogel sheet (Fig. 2D). As the diffusion fronts of activators and inhibitors advance, they react with the embedded reporters to trigger an outwardly propagating pulse, leading to the formation of a ring pattern. Thus, the ring pattern is a direct result of the interplay between diffusion and reaction. Fig. 2E shows the formation of a ring pattern over the course of 6 hours. Additionally, radially averaged intensity profiles provide quantitative information about pattern geometry not readily discernable from gel images alone. We found that the width and peak intensity of the ring grow over time as reporters are being triggered (Fig. 2F). These results are expected. The rings grow wider in accordance with the reaction–diffusion of reporters. The rings grow more intense because as more activators diffuse into the hydrogel, more reporters will be triggered, which leads to increasing peak intensity until a maximum as determined by the concentration of reporter embedded in the gel.
Like the amplitude and duration of a signal pulse measured in a well-mixed solution, the intensity and width of a ring also depend on the initial conditions of the DNA embedded hydrogel sheet. Reducing the initial concentration of activators resulted in rings with decreased widths and peak intensities (Fig. 2G). Radially averaged intensity profiles show that the inhibitor concentration controls the position of the trailing edge (Fig. 2H), while the activator concentration controls the position of the leading edge (Fig. 2I). To make meaningful comparisons, we established standard curves to convert fluorescence values to concentration units for spectrometry and gel image data (Fig. S7 and S8, ESI†).
Using Visual DSD, we built a predictive reaction–diffusion model to simulate pattern formation (Section S4, ESI†). The model uses rate constants inferred from spectrometry data (Table S2, ESI†) and assumes a common diffusion coefficient for all DNA complexes in our gel matrix (derivation of the diffusion coefficient is described in detail in Section S2 and Fig. S9, ESI†). The simulation results are displayed alongside corresponding gel images and show good quantitative agreement with the experimentally observed patterns (Fig. 2G).
Building tunable concentric rings
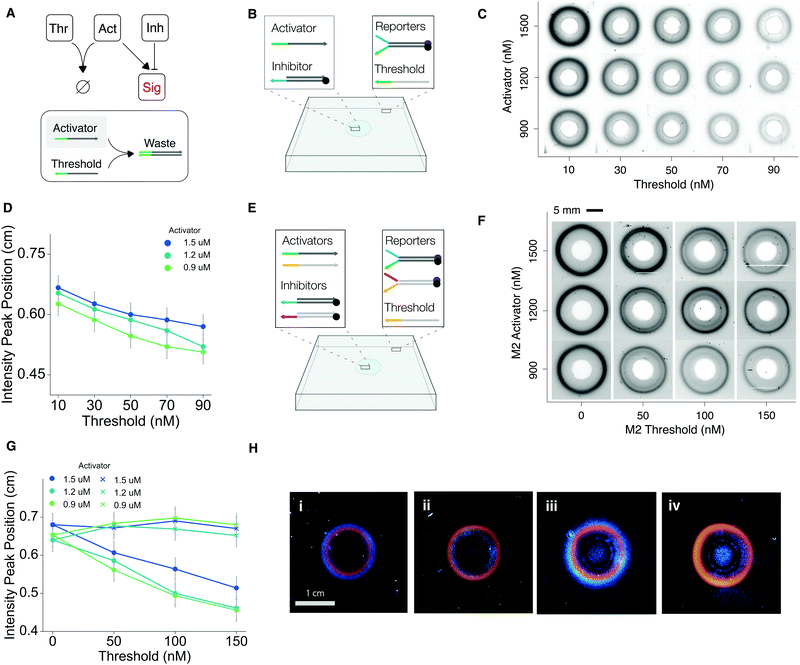
Next, we asked whether we could control the radius of the ring pattern by adding a threshold component to our core CRN. The threshold is a single-stranded DNA that is fully complementary to the activator (Fig. 3A). Because hybridization between the activator and the threshold is faster than the reaction between the activator and the reporter, the threshold effectively acts as a sink to the activator.To study the effect of threshold in a spatial setting, we embedded threshold along with reporters in the hydrogel (Fig. 3B). Activators diffusing into the gel are annihilated upon encountering the threshold. Thus, signal activation only occurs once a region has been depleted of unreacted threshold. We prepared gels embedded with different threshold concentrations (Fig. 3C and Fig. S10, ESI†). We found that for gels triggered with identical activator and inhibitor concentrations, higher threshold concentrations reduced the radius proportionally; for gels embedded with a nonzero amount of threshold, decreasing the activator concentration also reduced ring radius, but less effectively than increasing the threshold (Fig. 3D). Here, we define the radius as the distance between the position of peak intensity and the center of the gel, as measured from radially averaged intensity profiles (Fig. S10, ESI†). We updated our predictive computational model to include the threshold, using empirically derived rate constants and diffusion coefficient (Fig. S11, ESI†). Comparisons between the radially averaged intensity profiles of gel images and spatial simulations show that the simulation performs well for predicting ring patterns under different initial conditions (Fig. S12, ESI†).
Leveraging our ability to program the ring radius, we proceeded to build programmable patterns of two concentric rings. We designed a second ring-forming module with the same components as the first module but orthogonal sequences (Section S1 and Table S1, ESI†). Next, we combined the reporters for these two modules, labeled M1 and M2, in the same hydrogel and added threshold for M2 only (Fig. 3E). We prepared twelve DNA embedded hydrogels, corresponding to four M2 threshold and three M2 activator concentration levels (Fig. 3F), while maintaining the same concentrations of M1 components across all twelve gels. To trigger pattern formation, we loaded both M1 and M2 activators and inhibitors in the cavity. The outer ring's radius remained largely unchanged across experiments. Meanwhile, the inner ring's radius was proportional to the levels of activator and threshold, with high activator and low threshold values corresponding to larger radii (Fig. 3G). For gels without the threshold, we found that changing M2 activator concentrations alone had no effect on the radius of the inner ring (Fig. 3G), further validating the essential role of threshold for changing ring radius.
To better visualize the two-ring patterns, we replaced the FAM fluorophore in M2 with Cy5, such that M1 and M2 signals have distinct colors. We used this improved visualization to show that it is possible to program the order in which the rings appear. Using the same setup as the previous experiment, we prepared two gels containing either M1 threshold (Fig. 3H(i)) or M2 threshold (Fig. 3H(ii)), but not both. The gels were subjected to identical conditions otherwise. In Gel I, M1 signal activation lags M2 signal activation, resulting in an orange ring (FAM, M1) encircled by a blue ring (Cy5, M2). In Gel II, the order of the rings is reversed. Alternatively, we embedded two gels with both M1 and M2 threshold and triggered them with either higher M1 activator (Fig. 3H(iii)) or higher M2 activator (Fig. 3H(iv)). This experiment shows that we can program the order of rings by either varying the concentration of initiator strands or the composition of the DNA hydrogel.
Beyond isotropic patterns
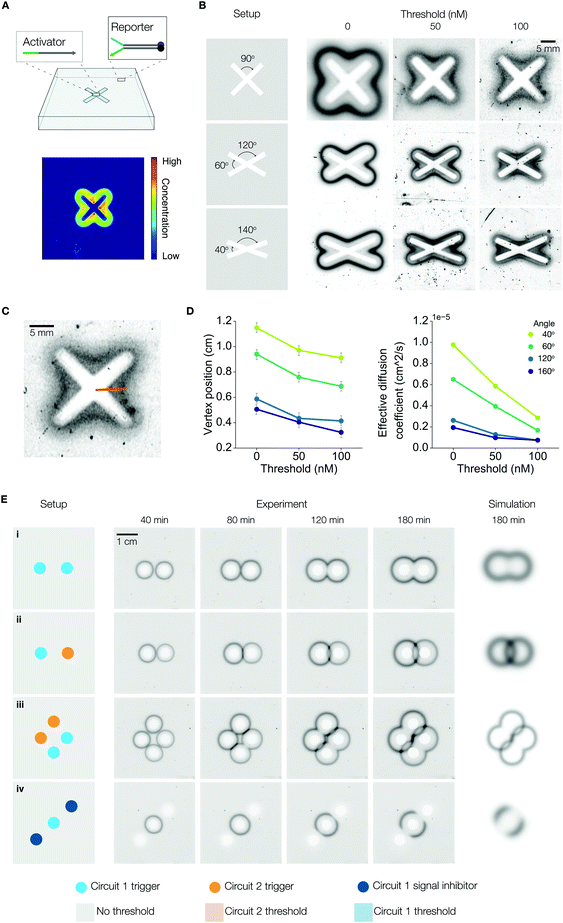
So far, we have only considered programmable pattern formation with isotropic boundary conditions. Next, we went beyond this simple geometry and generated anisotropic patterns by changing the shapes and placements of cavities. First, we loaded activators into an “X” shaped cavity in a gel embedded with only reporters (Fig. 4A). Interestingly, the X-shaped cavity led to an asymmetric concentration gradient of reactants. Positions near the center of the X received reactants from two separate directions (i.e. the two legs of the X adjacent to the center), whereas positions near the tips of the X received reactants from only one direction. Because the rate of reporter activation is proportional to the concentration of reactants, this gel configuration resulted in high signal at the center and low signal at the tips (Fig. 4A). We then amplified the asymmetry by embedding threshold in the hydrogels and, separately, by changing the angles between the legs of the X-shaped cavity. Adding threshold amplifies the time difference between signal activation at the tips and at the center, while decreasing the angle increases activator concentration in the interior of the angle, which leads to faster signal activation.We prepared nine DNA embedded hydrogels, which were divided into three groups based on their embedded threshold concentrations. Each group was further divided into three gels based on the angles in the “X” cavity on the gel (Fig. 4B). We measured the distance from the vertex to the center of the gel for different time points, cavity angles, and threshold concentrations (Fig. 4C). We also plotted the vertex distances for different time points and fitted the data to Fick's equation to find the “effective diffusion coefficient”, a reaction–diffusion dependent parameter we use to quantify the speed of signal propagation (Fig. S15, ESI†). For the same angle, both vertex distance (at the last time point) and the effective diffusion coefficient show an approximately linear dependence in threshold concentration (Fig. 4D).
Finally, we generated ring interference patterns by placing cavities at multiple locations in the gel. Table S3 (ESI†) lists the setup of each experiment in detail. Gels I–IV in Fig. 4E were synthesized from the same DNA embedded hydrogel solution containing M1 and M2 reporters. We applied different initial and boundary conditions to each gel to generate distinct patterns. For gel I, we loaded M1 activators and inhibitors in both cavities. We found that signals interfered destructively in regions where the rings intersected. This happens because the areas enclosed by the rings are devoid of reporters but are replete with inhibitors. Hence, as the two rings emanating from different cavities collide, their signal strands get consumed by the inhibitors from the opposite cavity. In contrast, when we loaded one of the cavities with M2 activators and inhibitors, we observed constructive interference of signals because the two diffusion fronts carry orthogonal reactants. Gel III was configured to display a combination of constructive and destructive signal interference. We created 3 cavities for gel IV. The center cavity was loaded with M1 activators and inhibitors; the two peripheral cavities were loaded with M1 inhibitors only. Since the signal and inhibitor diffusion fronts will annihilate each other wherever they collide, the resulting pattern is an incomplete ring with openings facing the directions of the peripheral cavities. Fig. S15 (ESI†) shows gels V and VI, where we used color coding and thresholding to induce two orthogonal rings of different radii that intersect.
Conclusion
We demonstrated a major advance in engineering programmable materials at the macroscale, using the reaction and diffusion of synthetic DNA strands to achieve quantitative and modular control over spatial patterns in hydrogels. To show proof of concept, we focused on a relatively simple pattern generated by non-catalytic CRNs. Incorporating more complex reaction networks, such as introducing feedback and cascading mechanisms, could produce dynamics like traveling waves and Turing patterns.49 We could also control diffusion by embedding appropriate complementary strands in the hydrogel to selectively slow down the diffusion of target DNA complexes, a possibility that has been explored in previous work.54 Similar mechanisms could be used to convert transient patterns into permanent patterns by immobilizing signal strands with complementary capture sequences. As with most hydrogel systems, gel desiccation is a concern. However, we only observed appreciable evaporation after more than 10 hours of imaging. Further, evaporation could be significantly reduced by covering the hydrogels with a film. Our system still relies on external spatial input and is therefore not fully autonomous and self-organizing. However, it is conceivable that a similar approach could be used to realize Turing patterns by combining nonlinear dynamics with control over diffusion rates.Scalise et al. proposed making complex DNA-based programmable patterns by sequentially applying modular filters to an initially simple input pattern.50 Our work has taken steps toward experimentally realizing such systems by predictably transforming simple input patterns into more complex output patterns. Thus, the workflow presented here can serve as an experimental basis for future projects exploring more complicated patterning systems.
In the longer term, we foresee applications where integrating chemical computing with additive manufacturing could expand the functionalities of existing 3D printed biomimetic materials. One could also imagine substituting fluorophores with other functional molecules, such as nanoparticles55 and quantum dots,56 to synthesize novel materials with useful properties. Our work expands on previous research efforts in synthetic chemistry, synthetic biology, and DNA nanotechnology. We believe this research presents a convincing case for using chemical computing in developing programmable matter.
Author contributions
S. C. and G. S. conceived and developed the project. S. C. performed experiments and computer simulations. S. C. and G. S. analyzed the data and wrote the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We would like to thank Neil Dalchau and Andrew Phillips from Microsoft Research for their valuable inputs on computational modeling. We also thank Yuan-Jyue Chen, Randolph Lopez, and Gourab Chatterjee for providing helpful advice. This work was support by ONR Award N000141612139 to G. S.References
- D. Raviv, et al., Active printed materials for complex self-evolving deformations, Sci. Rep., 2015, 4, 7422 CrossRef PubMed.
- A. Sydney Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Biomimetic 4D printing, Nat. Mater., 2016, 15, 413–418 CrossRef CAS PubMed.
- M. Yim, et al., Modular self-reconfigurable robot systems [Grand challenges of robotics], IEEE Robot. Autom. Mag., 2007, 14(1), 43–52 Search PubMed.
- M. Rubenstein, A. Cornejo and R. Nagpal, Programmable self-assembly in a thousand-robot swarm, Science, 2014, 345(6198), 795–799 CrossRef CAS PubMed.
- R. Hanlon, Cephalopod dynamic camouflage, Curr. Biol., 2007, 17(11), 400–404 CrossRef PubMed.
- A. J. Koch and H. Meinhardt, Biological pattern formation: From basic mechanisms to complex structures, Rev. Mod. Phys., 1994, 66, 1481–1507 CrossRef.
- A. M. Turing, The Chemical Basis of Morphogenesis, Philos. Trans. R. Soc., B, 1952, 237, 37–72 Search PubMed.
- M. Lovrak, et al., Free-standing supramolecular hydrogel objects by reaction–diffusion, Nat. Commun., 2017, 8, 1–10 CrossRef PubMed.
- S. Kondo and T. Miura, Reaction–diffusion model as a framework for understanding biological pattern formation, Science, 2010, 329(5999), 1616–1620 CrossRef CAS PubMed.
- A. Gierer and H. Meinhardt, A theory of biological pattern formation, Kybernetik, 1972, 12, 30–39 CAS.
- J. E. Pearson, Complex patterns in a simple system, Science, 1993, 261(5118), 189–192 CrossRef CAS PubMed.
- L. Yang and I. R. Epstein, Oscillatory Turing Patterns in Reaction–Diffusion Systems with Two Coupled Layers, Phys. Rev. Lett., 2003, 90(17), 178303 CrossRef PubMed.
- A. N. Zaikin and A. M. Zhabotinsky, Concentration wave propagation in two-dimensional liquid-phase self-oscillating system, Nature, 1970, 225, 535–537 CrossRef CAS PubMed.
- V. V. Yashin and A. C. Balazs, Pattern formation and shape changes in self-oscillating polymer gels, Science, 2006, 314(5800), 798–801 CrossRef CAS PubMed.
- S. Basu, Y. Gerchman, C. H. Collins, F. H. Arnold and R. Weiss, A synthetic multicellular system for programmed pattern formation, Nature, 2005, 434, 1130–1134 CrossRef CAS PubMed.
- J. J. Tabor, et al., A Synthetic Genetic Edge Detection Program, Cell, 2009, 137(7), 1272–1281 CrossRef PubMed.
- C. Liu, et al., Sequential Establishment of Stripe Patterns in an Expanding Cell Population, Science, 2011, 334, 238–241 CrossRef CAS PubMed.
- A. Padirac, T. Fujii, A. Estévez-Torres and Y. Rondelez, Spatial waves in synthetic biochemical networks, J. Am. Chem. Soc., 2013, 135(39), 14586–14592 CrossRef CAS PubMed.
- A. S. Zadorin, et al., Synthesis and materialization of a reaction–diffusion French flag pattern, Nat. Chem., 2017, 9, 990–996 CrossRef CAS PubMed.
- G. Gines, et al., Microscopic agents programmed by DNA circuits, Nat. Nanotechnol., 2017, 12, 351–359 CrossRef CAS PubMed.
- A. Dupin and F. C. Simmel, Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits, Nat. Chem., 2019, 11, 32–39 CrossRef CAS PubMed.
- V. A. Bloomfield, D. M. Crothers and I. J. Tinoco, Nucleic Acids: Structures, Properties, and Functions, University Science Books, 2000 Search PubMed.
- J. SantaLucia and D. Hicks, The Thermodynamics of DNA Structural Motifs, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 415–440 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- N. C. Seeman, Nanomaterials Based on DNA, Annu. Rev. Biochem., 2010, 79, 65–87 CrossRef CAS PubMed.
- J. Zheng, et al., From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal, Nature, 2009, 461, 74–77 CrossRef CAS PubMed.
- S. H. Um, et al., Enzyme-catalysed assembly of DNA hydrogel, Nat. Mater., 2006, 5, 797–801 CrossRef CAS PubMed.
- J. B. Lee, et al., Multifunctional nanoarchitectures from DNA-based ABC monomers, Nat. Nanotechnol., 2009, 4, 430–436 CrossRef CAS PubMed.
- F. C. Simmel and R. Schulman, Self-organizing materials built with DNA, MRS Bull., 2017, 42, 913–919 CrossRef CAS.
- D. C. Lin, B. Yurke and N. A. Langrana, Mechanical properties of a reversible, DNA-crosslinked polyacrylamide hydrogel, J. Biomech. Eng., 2004, 126, 104–110 CrossRef PubMed.
- D. C. Lin, B. Yurke and N. A. Langrana, Inducing reversible stiffness changes in DNA-crosslinked gels, J. Mater. Res., 2005, 20, 1456–1464 CrossRef CAS.
- M. Hagiya et al., On DNA-based gellular automata, Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Springer Verlag, 2014, vol. 8553 LNCS, pp. 177–189 Search PubMed.
- A. Cangialosi, et al., DNA sequence–directed shape change of photopatterned hydrogels via high-degree swelling, Science, 2017, 357(6356), 1126–1130 CrossRef CAS PubMed.
- T. Hosoya, I. Kawamata, S. Nomura, M. Ichiro and S. Murata, Pattern Formation on Discrete Gel Matrix Based on DNA Computing, New Gener. Comput., 2019, 37, 97–111 CrossRef.
- J. Zenk, et al., Stable DNA-based reaction–diffusion patterns, RSC Adv., 2017, 7, 18032–18040 RSC.
- S. S. Wang and A. D. Ellington, Pattern Generation with Nucleic Acid Chemical Reaction Networks, Chem. Rev., 2019, 119(10), 6370–6383 CAS.
- K. Abe, I. Kawamata, S. I. M. Nomura and S. Murata, Programmable reactions and diffusion using DNA for pattern formation in hydrogel medium, Mol. Syst. Des. Eng., 2019, 4, 639–643 RSC.
- D. Y. Zhang and E. Winfree, Control of DNA strand displacement kinetics using toehold exchange, J. Am. Chem. Soc., 2009, 131(47), 17303–17314 CrossRef CAS PubMed.
- D. Y. Zhang and G. Seelig, Dynamic DNA nanotechnology using strand-displacement reactions, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- L. Qian, E. Winfree and J. Bruck, Neural network computation with DNA strand displacement cascades, Nature, 2011, 475, 368–372 CrossRef CAS PubMed.
- K. M. Cherry and L. Qian, Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks, Nature, 2018, 559, 370–376 CrossRef CAS PubMed.
- R. Lopez, R. Wang and G. Seelig, A molecular multi-gene classifier for disease diagnostics, Nat. Chem., 2018, 10, 746–754 CrossRef CAS PubMed.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Three-dimensional structures self-assembled from DNA bricks, Science, 2012, 338(6111), 1177–1183 CrossRef CAS PubMed.
- W. B. Rogers and V. N. Manoharan, Programming colloidal phase transitions with DNA strand displacement, Science, 2015, 347(6222), 639–642 CrossRef CAS PubMed.
- D. Soloveichik, G. Seelig and E. Winfree, DNA as a universal substrate for chemical kinetics, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(12), 5393–5398 CrossRef CAS PubMed.
- L. Cardelli, Two-domain DNA strand displacement, Math. Struct. Comput. Sci., 2013, 23, 247–271 CrossRef.
- Y. J. Chen, et al., Programmable chemical controllers made from DNA, Nat. Nanotechnol., 2013, 8, 755–762 CrossRef CAS PubMed.
- N. Srinivas, J. Parkin, G. Seelig, E. Winfree and D. Soloveichik, Enzyme-free nucleic acid dynamical systems, Science, 2017, 358(6369), eaal2052 CrossRef PubMed.
- N. Dalchau, G. Seelig and A. Phillips, Computational Design of Reaction-Diffusion Patterns Using DNA-Based Chemical Reaction Networks, DNA Computing and Molecular Programming, Springer, Cham, 2014, pp. 84–99 Search PubMed.
- D. Scalise and R. Schulman, Designing modular reaction–diffusion programs for complex pattern formation, Technology, 2014, 02, 55–66 CrossRef.
- S. M. Chirieleison, P. B. Allen, Z. B. Simpson, A. D. Ellington and X. Chen, Pattern transformation with DNA circuits, Nat. Chem., 2013, 5, 1000–1005 CrossRef CAS PubMed.
- M. R. Lakin, S. Youssef, F. Polo, S. Emmott and A. Phillips, Visual DSD: A design and analysis tool for DNA strand displacement systems, Bioinformatics, 2011, 27, 3211–3213 CrossRef CAS PubMed.
- J. N. Zadeh, et al., NUPACK: Analysis and design of nucleic acid systems, J. Comput. Chem., 2011, 32, 170–173 CrossRef CAS PubMed.
- P. B. Allen, X. Chen, Z. B. Simpson and A. D. Ellington, Modeling scalable pattern generation in DNA reaction networks, Nat. Comput., 2014, 13, 583–595 CrossRef CAS PubMed.
- W. B. Rogers, W. M. Shih and V. N. Manoharan, Using DNA to program the self-assembly of colloidal nanoparticles and microparticles, Nat. Rev. Mater., 2016, 1, 16008 CrossRef CAS.
- I. L. Medintz, H. T. Uyeda, E. R. Goldman and H. Mattoussi, Quantum dot bioconjugates for imaging, labelling and sensing, Nat. Mater., 2005, 4, 435–446 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sm02413a |
| This journal is © The Royal Society of Chemistry 2020 |