One-pot synthesis of oligonucleotide-grafted polymeric nanoparticles†
Emi
Haladjova
 *a,
Iva
Ugrinova
b and
Stanislav
Rangelov
*a
*a,
Iva
Ugrinova
b and
Stanislav
Rangelov
*a
aInstitute of Polymers, Bulgarian Academy of Sciences, “Akad. G. Bonchev” St., Bl. 103-A, 1113 Sofia, Bulgaria. E-mail: ehaladjova@polymer.bas.bg; rangelov@polymer.bas.bg
bInstitute of Molecular Biology, Bulgarian Academy of Sciences, “Akad. G. Bonchev” St., Bl. 21, 1113 Sofia, Bulgaria
First published on 18th November 2019
Abstract
A feasible one-pot approach for constructing oligonucleotide-grafted polymeric nanoparticles is reported. The approach involves formation of mesoglobules from a thermoresponsive polymer, coating of the mesoglobules with a cross-linked polymeric shell, and grafting the latter with oligonucleotide strands. Dynamic and static light scattering are used to parameterize the novel constructs. They are relatively large structures with hydrodynamic radii and molar masses reaching 200 nm and 150.0 × 106 g mol−1, respectively. The oligonucleotide-grafted polymeric nanoparticles are of spherical morphology and moderately negative (−12.4 to −19.1 mV) ζ potential as revealed by AFM, TEM, and electrophoretic light scattering. In accordance with their large size, they are found to carry thousands of oligonucleotide strands per particle. The novel constructs are thermoresponsive. They undergo reversible collapse upon heating and swelling upon cooling, which is associated with changes in the grafting density and, hence, the conformation of the oligonucleotide strands from unextended at room temperature to a more extended one at elevated temperatures. The versatility of the approach is demonstrated by varying the type of the cross-linked shell and content of the oligonucleotide strands and, hence, the grafting density. Appropriate diversification and modifications are suggested as well.
Introduction
Spherical nucleic acids (SNAs) are nanostructures composed of a metallic or another inorganic core to the surface of which highly oriented oligonucleotide strands are covalently attached, thus forming a dense layer.1,2 The dense arrangement and high orientation as well as the three-dimensional architecture and small size (typically sub-100 nm) give rise to specific properties of SNAs that are different from those of their linear nucleic acid counterparts – enhanced resistance to enzymatic degradation, increased overall lifetime, higher binding affinity to complementary ligands, no need for transfection agents to traverse cell membranes, minimal cytotoxicity and non-specific immunogenic responses.2–5 Besides its role as a scaffold for assembling and orienting the oligonucleotides, the core, depending on its nature, provides additional physical and chemical properties – plasmonic, catalytic, scattering, and quenching. Thus, SNAs have emerged as highly modular and tailorable nanostructures that have been utilized in a number of biodiagnostic and therapeutic applications.1–12In response to concerns about the potential long-term toxicity and metabolic fate of the metallic and inorganic cores, strategies to design SNAs with organic cores have been developed.8–12 The organic cores further expand the versatility in the chemistry, structure, and properties of the resulting nanostructures. Typically, the approaches to synthesizing these constructs involve self-assembly of amphiphilic oligonucleotide strands,13 prepared by coupling of nucleic acids with organic molecules, proteins, drugs, and polymers8,11–22 or intercalation of amphiphilic conjugates into the hydrophobic cores of pre-formed copolymer micelles.12 In spite of the recent advancement, the design of SNAs with organic cores that are biocompatible and exhibit the hallmark properties of prototypical SNAs is still a great challenge.
Mesoglobules are typically well-defined, colloidally stable, and narrowly distributed spherical particles that are formed in dilute aqueous solution by polymers exhibiting LCST properties (LCST, lower critical solution temperature).23–26 These polymers are thermoresponsive and undergo a reversible soluble-to-insoluble state transition upon small changes in temperature:27,28 they are soluble at low temperatures but start to precipitate upon heating above a certain critical temperature. The size of the mesoglobules, which may vary from tens to hundreds of nanometers, can be easily controlled by the nature and molar mass of the thermoresponsive polymer, its initial concentration, the heating protocol, and the presence of additives. Upon cooling below the critical temperature, the mesoglobules fully disintegrate into individual macromolecules. The reversible formation and disintegration of mesoglobules is their most attractive feature for the fabrication of polymeric nanocapsules – nanostructures, composed of a hollow core and a polymeric shell, that have shown potential as drug and vaccine carriers, biomimetic nanoreactors, artificial organelles, and gene and protein delivery vectors.29–32 Furthermore, thermoresponsive polymers can be employed for loading and controlled release of active molecules33–35 and for creating a specific environment and protection of the payload,36 which, in the context of material and nanoparticle design, provides additional tailorability and versatility. The most widely studied thermoresponsive polymers are poly(N-isopropylacrylamide), poly(2-isopropyl-2-oxazoline), and polymers of oligo(ethylene glycol) (meth)acrylates.
Herein, we employ a novel synthetic approach for preparation of oligonucleotide-grafted polymeric nanoparticles. The approach relies on (i) generation of a core from a thermoresponsive polymer, (ii) coating the core with a cross-linked polymeric shell, and (iii) surface functionalization of the shell with oligonucleotide strands (Scheme 1). The resulting structures, which can be considered as intermediates for nanocapsule formation (that is, hollow SNAs) and platforms for further modification and loading with drugs, therapeutic proteins, and DNA, are reported for the first time. By quantifying the number of oligonucleotide strands on the surface, we assess the grafting density and estimate the conformation of the oligonucleotides. The latter is of particular importance since it can strongly influence the accessibility of the oligonucleotides for hybridization37,38 and the SNA-like properties of the particles.2 We evaluate the feasibility of the synthetic approach used and suggest appropriate diversification of the resulting nanoconstructs in terms of composition, structure, functions, and properties.
Experimental
Materials
N,N-Methylenbisacrylamide (BIS, 99%), 2,2′-azobis(2-methylpropionamidine)dihydrochloride (AAPH, 97%) and N-isopropylacrylamide (NIPAm, 97%) were purchased from Sigma-Aldrich and used as received. 2-Hydroxyethylmethacrylate (HEMA, 99%, Sigma-Aldrich) was purified by passing through a column filled with Al2O3. A methacrylamide modified oligonucleotide (5′-5TA ATA CGA CTC ACT ATA GGG-3′) with internal hexaethylene glycol (spacer 18) modification (21 bases, corresponding to Mw = 6707 g mol−1) was purchased from Biomers.net GmbH. The water used to obtain the polymer and oligonucleotide solutions was purified through deionization by a Millipore Direct-Q system. Poly[methoxydi(ethylene glycol)methacrylate] (PDEGMA) (Mn = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) was synthesized by group transfer polymerization under trisdimethylaminosulfonium bifluoride catalysis at room temperature. The polymerization procedure is described in detail elsewhere.39
000 g mol−1) was synthesized by group transfer polymerization under trisdimethylaminosulfonium bifluoride catalysis at room temperature. The polymerization procedure is described in detail elsewhere.39
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in order to remove the unreacted monomer, initiator, cross-linker, oligonucleotide, and non-entrapped and/or released PDEGMA.
000 in order to remove the unreacted monomer, initiator, cross-linker, oligonucleotide, and non-entrapped and/or released PDEGMA.
Methods
| Rh = kT/(6πηD0) | (1) |
| ζ = 4πηυ/ε | (2) |
The standard deviations in the static, dynamic, and electrophoretic light scattering data are up to 8%.
Results and discussion
Preparation of oligonucleotide-grafted polymeric nanoparticles
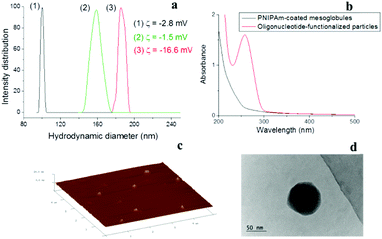
In the present study, mesoglobules formed at 65 °C by nanoprecipitation of a pre-cooled to 4–5 °C dilute aqueous solution of poly[methoxydi(ethylene glycol)methacrylate] (PDEGMA) served as the core for coating with a cross-linked polymeric shell. PDEGMA is a low-Tg (Tg = −40 °C42) thermoresponsive polymer, which exhibits LCST properties with a sharp phase transition in the 26–29 °C range.43–45 It belongs to the family of polymethacrylates with oligo(ethylene glycol) side chains, the biocompatibility of which allows applications as a biomaterial.46 In addition to the possibilities to control the size of the mesoglobules by controlling the initial concentration, applying various heating protocols, and adding and controlling the content of specific additives,43–45 other advantages of PDEGMA are the lack of hysteresis at the transition upon heating and cooling, as well as the negligible effects of additives such as surfactants and salts on the position of the phase transition temperature.47,48The construction of the polymeric shell was performed in two steps. The first step involved formation of a cross-linked polymer layer by pseudo-seeded radical polymerization49–51 of N-isopropylacrylamide (NIPAm) in the presence of N,N-methylenebis(acrylamide) (BIS) as a cross-linker, initiated by 2,2′-azobis(2-methylpropionamidine)dihydrochloride (AAPH). The shell thickness and cross-linking density were controlled by the initiator-to-monomer and cross-linker-to-monomer ratios, respectively. The second step involved surface functionalization of the resulting core–shell particles by oligonucleotide strands. For the latter, we choose an oligonucleotide with a non-specific base sequence, functionalized with a methacrylamide group (see the Experimental section), which allowed for their covalent attachment to the PNIPAm shell by a copolymerization reaction with an additional portion of NIPAm. The consecutive steps of core formation, shell construction, and functionalization of the latter are presented in Scheme 1, whereas the size variations, from which the shell thickness was estimated, are shown in Fig. 1a. The figure shows that the construction of the shell was associated with a step-wise increase of the apparent hydrodynamic diameter from 100 to 160 nm (associated with formation of a polymeric layer surrounding the core) and further to 186 nm (consistent with grafting of 21 bases long oligonucleotide strands). Furthermore, in situ analysis showed a shift of the ζ potential to a more negative value (ζ = −16.6 mV) also suggesting successful surface functionalization (Fig. 1a).
After the oligonucleotide grafting reaction was accomplished, the system was cooled down to room temperature and subjected to dialysis with a MWCO of 50 kDa membrane to remove non-entrapped and/or released PDEGMA and the unreacted monomer, initiator, and cross-linker as well as the unbound oligonucleotide. To demonstrate the effectiveness of purification by dialysis, gel electrophoresis (Fig. 2) and electrophoretic light scattering (Fig. S1, ESI†) characterization were performed. Fig. 2a shows gel images of the initial oligonucleotide (control sample, lanes 1 and 2) and of the oligonucleotide-functionalized particles (sample ACN1) after dialysis (lanes 3 and 4). The quantification of the unreacted oligonucleotide is presented in Fig. 2b. As seen, after dialysis for 5 h the unbound oligonucleotide was not detected (lanes 3 and 4 in Fig. 2a and b). With regard to the visualization of the oligonucleotide-functionalized particles, a wide area with an increased integrated density was observed instead of a single, sharp, shifted band. This finding is consistent with the heterogeneity of the particles in terms of both size distribution and oligonucleotide content. It is noteworthy that the total amount of the removed material consisted of less than 8% of the total weight of all reactants, indicating high effectiveness of the approach. Furthermore, the particles preserved their integrity upon cooling and during the purification, which demonstrated the stability of the cross-linked functionalized shell.
The successful incorporation of the oligonucleotides was further evidenced by UV-vis spectroscopy by measuring the absorbance of dispersions of the initial, that is, PNIPAm only coated, particles and that of the final oligonucleotide-functionalized particles at 260 nm (Fig. 1b). Furthermore, the oligonucleotide-functionalized particles were visualized by atomic force microscopy (AFM) and TEM (Fig. 1c and d). They were of spherical morphology and size (in dry form) consistent with dynamic light scattering data (vide infra). Given the covalent attachment of the oligonucleotide strands to the shell and the high cross-linking density of the latter, any further release of PDEGMA from the core and oligonucleotides from the shell has to be ruled out. It is noteworthy that the release of PDEGMA from the core can be controlled by controlling the thickness and cross-linking density of the shell.51 Full removal of PDEGMA would result in formation of coreless SNA structures.
To demonstrate the versatility of the approach, the PNIPAm shell was replaced with a shell composed of cross-linked poly(hydroxyethyl methacrylate) (PHEMA). It is noteworthy that the mechanisms of constructing/building the two shells as well as their structure and properties are different as shown elsewhere.52,53 The successful preparation of oligonucleotide-grafted nanoparticles was evidenced by independent techniques: DLS showed a step-wise increase of the particle size consistent with formation of the PHEMA shell and oligonucleotide grafting of the latter; meanwhile the ζ potential analysis gave a more negative value after grafting; spherical structures were readily apparent upon imaging by AFM; and UV absorbance of the purified sample appeared at 260 nm, which was consistent with oligonucleotide attachment to the PHEMA shell. Experimental findings demonstrating the successful formation of the oligonucleotide-grafted nanoparticles are presented in the ESI† (Fig. S3).
The key factors that govern the SNA-like properties are the surface density and orientation of the oligonucleotide strands. In an attempt to increase the oligonucleotide grafting density, we performed the above experiments with 1.8-fold larger amounts of oligonucleotide. The contents and concentrations of the PDEGMA, initiator, monomer, and cross-linker as well as the molar ratios between them were the same for all preparations. Experimental evidence is presented in Fig. S4 (ESI†). To facilitate the presentation and discussion, the following abbreviation codes were introduced: ACN1, ACN2, ACH1, and ACH2, where 1 and 2 correspond to the smaller and larger oligonucleotide contents, respectively, whereas ACN and ACH refer to shells of PNIPAm and PHEMA, respectively.
Determination of the total oligonucleotide amount by measuring the absorbance at 260 nm (Fig. 1b, see also the ESI†) confirmed that the incorporation was effective and the purification did not cause a loss of oligonucleotide larger than just a few percent, corresponding to more than 90% effectiveness of oligonucleotide grafting. It is noteworthy that at an excess larger than 1.8, unproportionally large amounts of unreacted oligonucleotides were removed, implying that saturation of the surface density was reached. Concerns about the vulnerability of DNA to attack of radicals raised the question of the stability of the oligonucleotides during the grafting reaction. We incubated a non-functionalized oligonucleotide with the free radical-generating initiator at 65 °C for different time intervals, thus simulating the reaction conditions of oligonucleotide attachment. Gel electrophoresis showed a single major band for each sample at the same position as that of the control, indicating no damage (Fig. S2, ESI†).
Characterization and parameterization of the oligonucleotide-grafted nanoparticles and determination of the oligonucleotide grafting density
For determination of the oligonucleotide grafting density, we employed an approach involving precise determination of the molar masses (Mw) and hydrodynamic radii (Rh) of the particles by static and dynamic light scattering. DLS measurements were performed at 9 angles in the 50–130° range and for 4–6 dispersions with increasing concentrations for each of the samples. Typically the samples exhibited monomodal relaxation time distributions (Fig. 3a), indicating the existence of only one population of particles, linear dependence of the relaxation rate on sin2(θ/2) (Fig. 3b), reflecting translational diffusion of these particles, and very slight concentration dependence of the diffusion coefficient (Fig. 3c), from which D0 (the diffusion coefficient at infinite dilution, which is used to calculate Rh) was determined. The SLS data were treated using the Berry plot method (Fig. 4). Besides Mw, the analysis provides also information about the second virial coefficient (A2) and the radius of gyration (Rg). The latter combined with Rh gives the quantity Rg/Rh, which is indicative of the particle geometry and morphology.54–56 The measurements were performed at 25, 37, and 65 °C. More examples from the DLS and SLS studies are given in the ESI† (Fig. S5). The static and dynamic light scattering parameters are summarized in Table 1.| Sample | Shell-forming polymer | Temperature (°C) | Oligonucleotide amount (μmol) | M w × 10−6 (g mol−1) | A 2 × 105 (mL mol g−2) | R g (nm) | D 0 × 1012 (m2 s−1) | R h (nm) | R g/Rh | ζ (mV) |
|---|---|---|---|---|---|---|---|---|---|---|
| ACN1 | PNIPAM | 25 | 0.045 | 136.0 | −0.061 | 87.5 | 2.96 | 82.9 | 1.06 | −16.6 |
| ACN2 | PNIPAM | 25 | 0.081 | 146.0 | −0.063 | 205.0 | 1.40 | 175.1 | 1.17 | −17.4 |
| ACN2 | PNIPAM | 37 | 0.081 | 149.0 | −0.34 | 121.0 | 2.64 | 124.1 | 0.98 | — |
| ACH1 | PHEMA | 37 | 0.030 | 65.2 | −1.8 | 128.0 | 2.46 | 133.5 | 0.96 | −12.4 |
| ACH2 | PHEMA | 37 | 0.054 | 71.0 | −2.3 | 203.0 | 1.64 | 199.6 | 1.02 | −19.1 |
As seen from Table 1, relatively large particles with hydrodynamic radii up to 200 nm and molar masses reaching 150 × 106 g mol−1 are formed. The molar masses of the particles based on PNIPAm (ACN) are ca. two times larger than those based on PHEMA (ACH), which most probably reflects the different mechanisms of shell formation.52,53 It is noteworthy that the molar masses practically did not change upon increasing temperature as demonstrated with ACN2; only the size of the particles changed (vide infra). The values of A2 are invariably negative, but very small in magnitude, implying slightly unfavorable particle–solvent interactions. The radii of gyration are consistent with the hydrodynamic radii, giving rise to Rg/Rh values slightly deviating from unity, which is compatible with a morphology of relatively compact spherical particles with thin shells.54–56 Although the data in Table 1 may reflect some reproducibility issues, the effect of the larger quantities of oligonucleotide strands grafted on the shells of the particles is visible from the increase of Mw, Rh, and Rg as well as from the shift of the ζ potential to more negative values.
The average number of oligonucleotide strands per particle was determined from the molar mass data, total particle concentration, and oligonucleotide content. A numerical example of the calculation is given in the ESI.† Then, the oligonucleotide grafting density was calculated based on the size of the structures from DLS, assuming a spherical morphology. The number of oligonucleotide strands per particle ranged from 1144 to 5500 (Fig. 5a). This is significantly higher than reference micellar SNAs, liposomal SNAs, and prototypical gold nanoparticle SNAs,6,11,16,57 which are also given in Fig. 5a for comparison, and is fully consistent with the larger size of the present oligonucleotide-grafted polymeric nanoparticles. As the maximum possible surface coverage is dependent on the particle size and morphology with the more curved particles being able to support more strands/chains than the less curved ones, these results were converted into grafting density, σ, expressed as the number of strands per nm2 (Fig. 5b). At 25 and 37 °C the grafting density values ranged between 0.0044 and 0.034 nm−2, which are comparable but slightly lower than those of the reference samples, ranging in the 0.011–0.106 nm−2 interval.
 | ||
| Fig. 5 (a) Number of oligonucleotide strands per particle and (b) grafting density, σ, of the strands for the oligonucleotide-grafted polymeric nanoparticles (ACN1, ACN2, ACH1, and ACH2) at 25, 37, and 65 °C and reference SNAs (A–E) at room temperature. Closed symbols – oligonucleotide-grafted polymeric nanoparticles. Open symbols – reference SNAs: A and B – ref. 11; C – ref. 57; D – ref. 16; E – ref. 6. The black, green, and blue closed symbols in (b) refer to 25, 37, and 65 °C, respectively. σtr is the grafting density at the transition from the unextended to the brush conformation. | ||
Thermoresponsive behavior of oligonucleotide-grafted polymeric nanoparticles
The present oligonucleotide-grafted polymeric nanoparticles exhibited a dramatic thermoresponsive behavior as most of the materials from which they are built are LCST polymers. Fig. 6 shows typical profiles of the size variations during abrupt heating–cooling cycles. The profiles are very deep, corresponding to considerable shrinkage of the particles upon heating and swelling upon cooling. The heating to 65 °C resulted in an increase in σ, since the shrinkage is associated with a decrease of the size and, hence, the total surface area of the particles. Fig. 5b clearly shows that the values of σ of the present oligonucleotide-grafted polymeric nanoparticles at 65 °C are slightly higher than those of the reference SNAs with organic cores and comparable to the prototypical gold nanoparticle SNAs. A striking result was that in no case were the critical values of the grafting density at the transition from the unextended to the brush conformation (σtr) reached. Indeed, if the oligonucleotide strands are regarded as surface-anchored random coils,58–61 then the transition from the unextended to the brush configuration can be determined by σtr = 1/RF2, where RF is the Flory radius (see the ESI†); the calculated σtr values were within the 0.155–0.182 nm−2 range, depending on the oligonucleotide composition, as indicated by a line in Fig. 5b. It is obvious from Fig. 5b that the values of σ for the present ACN and ACH samples as well as those of the reference samples fall into the region below the σtr line, implying that in neither of those nanoconstructs were the oligonucleotide strands in the brush regime. Nevertheless, structures with such a surface coverage have been repeatedly shown to manifest their unique SNA-like properties.1–13,18,57Though not crossing the threshold σtr, the oligonucleotide strands undergo a reversible transition from an unextended conformation at room temperature into a more extended one at 65 °C (Scheme 2). A very interesting observation was that the particles grew in size at each subsequent cooling (Fig. 6). Here, we may speculate on some delayed relaxation of the oligonucleotide strands to the unextended conformation, which, combined with the fast swelling of the core, might be associated with larger structural rearrangement leading to an increase in size. This phenomenon was not further explored but it is noteworthy that when leaving the samples to anneal by slow cooling to room temperature, their initial dimensions were recovered (Fig. 6).
 | ||
| Scheme 2 Schematic presentation showing possible re-arrangements and transitions in oligonucleotide-grafted polymeric nanoparticles upon abrupt heating and slow cooling. | ||
Conclusions
In summary, we report on a synthetic approach for preparation of oligonucleotide-grafted polymeric nanoparticles, which involves formation of mesoglobules from a thermoresponsive polymer, coating of the mesoglobules with a cross-linked polymeric shell, and functionalization of the shell by grafting with oligonucleotide strands. The versatility of the approach is demonstrated by varying the type of the shell-forming polymer (PNIPAm and PHEMA) and the oligonucleotide content. Further diversification can be achieved by judicious selection of the core-forming material (thermoresponsive biocompatible polymers, polymer–drug and polymer–peptide conjugates, complexes with DNA, etc.) and the type and amount of the cross-linking agent (endowing biodegradability, and controlling the cross-linking density) as well as the type, length, composition, and base sequence of the nucleic acid (DNA, RNA, single- and double-stranded, specific sequences). We anticipate that this synthetic approach can be extended to a variety of SNA nanostructures exhibiting strong potential for therapeutic developments such as hollow SNAs, carriers of biologically active substances, platforms for controllable, continuous release of nucleic acids, gene regulation constructs, and diagnostic and imaging tools. The new class of oligonucleotide-grafted polymeric nanoparticles were found to carry thousands of oligonucleotide strands per particle, corresponding to a grafting density at room temperature slightly lower than that of the reference SNAs. Upon heating, the grafting density increased due to the collapse of the thermoresponsive moieties, resulting in shrinkage of the particles. These drastic structural changes and particularly the increase in grafting density were associated with stretching of the oligonucleotide strands. Although adopting a more extended conformation, the latter did not enter the brush regime; neither did the oligonucleotides of the reference samples. Such a conformation, however, may be beneficial for enhancement of the accessibility of the oligonucleotides for hybridization37,38 and explains very well the repeatedly demonstrated cooperative hybridization properties of various SNA structures.2,8,12,13,18 A distinctive feature of the present oligonucleotide-grafted polymeric nanoparticles is their considerably larger but still of biological relevance dimensions compared to the prototypical SNAs. Hopefully not losing many of the biological properties associated with the latter such as rapid cellular uptake, enhanced enzyme stability, minimal immune response, capability of transfecting cells without transfecting agents, etc., a set of extra properties related to the thermal sensitivity, polymeric nature, larger dimensions, and core–shell structure of the novel oligonucleotide-grafted constructs will be added.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Fund (Bulgaria) Project No. DN19/8-2017. The authors are grateful to Prof. P. Simon (Rhine-Waal University of Applied Sciences) for providing PDEGMA polymers and Dr M. Kyulavska (Institute of Polymers, BAS) for performing AFM.Notes and references
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607 CrossRef CAS.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, J. Am. Chem. Soc., 2012, 134, 1376 CrossRef CAS.
- C. H. Chung, J. H. Kim, J. Jung and B. H. Chung, Biosens. Bioelectron., 2013, 41, 827 CrossRef CAS.
- C. H. Choi, L. Hao, S. P. Narayan, E. Auyeung and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7625 CrossRef CAS.
- M. D. Massich, D. A. Giljohann, A. L. Schmucker, P. C. Patel and C. A. Mirkin, ACS Nano, 2010, 4, 5641 CrossRef CAS.
- T. A. Taton, C. A. Mirkin and R. L. Letsinger, Science, 2000, 289, 1757 CrossRef CAS.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027 CrossRef CAS.
- R. J. Banga, N. Chernyak, S. P. Narayan, S. T. Nguyen and C. A. Mirkin, J. Am. Chem. Soc., 2014, 136, 9866 CrossRef CAS.
- J. D. Brodin, E. Auyeung and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4564 CrossRef CAS.
- K. J. Watson, S. J. Park, J. H. Im, S. T. Nguyen and C. A. Mirkin, J. Am. Chem. Soc., 2001, 123, 5592 CrossRef CAS.
- Z. Li, Y. Zhang, P. Fullhart and C. A. Mirkin, Nano Lett., 2004, 4, 1055 CrossRef CAS.
- R. J. Banga, B. Meckes, S. P. Narayan, A. J. Sprangers, S. T. Nguyen and C. A. Mirkin, J. Am. Chem. Soc., 2017, 139, 4278 CrossRef CAS.
- C. Zhang, L. Hao, C. M. Calabrese, Y. Zhou, C. H. J. Choi, H. Xing and C. A. Mirkin, Small, 2015, 11, 5360 CrossRef CAS.
- B. J. Hong, I. Eryazici, R. Bleher, R. V. Thaner, C. A. Mirkin and S. T. Nguyen, J. Am. Chem. Soc., 2015, 137, 8184 CrossRef CAS.
- X. Y. Tan, X. G. Lu, F. Jia, X. F. Liu, Y. H. Sun, J. K. Logan and K. Zhang, J. Am. Chem. Soc., 2016, 138, 10834 CrossRef CAS PubMed.
- C. M. Calabrese, T. J. Merkel, W. E. Briley, P. S. Randeria, S. P. Narayan, J. L. Rouge, D. A. Walker, A. W. Scott and C. A. Mirkin, Angew. Chem., Int. Ed., 2015, 54, 476 CAS.
- J. D. Brodin, A. J. Sprangers, J. R. McMillan and C. A. Mirkin, J. Am. Chem. Soc., 2015, 137, 14838 CrossRef CAS.
- R. J. Banga, S. A. Krovi, S. P. Narayan, A. J. Sprangers, G. Liu, C. A. Mirkin and S. T. Nguyen, Biomacromolecules, 2017, 18, 483 CrossRef CAS.
- T. Schnitzler and A. Herrmann, Acc. Chem. Res., 2012, 45, 1419 CrossRef CAS.
- A. M. Rush, M. P. Thompson, E. T. Tatro and N. C. Gianneschi, ACS Nano, 2013, 7, 1379 CrossRef CAS.
- L. Peng, C. S. Wu, M. X. You, D. Han, Y. Chen, T. Fu, M. Ye and W. H. Tan, Chem. Sci., 2013, 4, 1928 RSC.
- K. Isoda, N. Kanayama, D. Miyamoto, T. Takarada and M. Maeda, React. Funct. Polym., 2011, 71, 367 CrossRef CAS.
- A. Gorelov, A. Du Chesne and K. Dawson, Phys. A, 1997, 240, 443 CrossRef CAS.
- K. Dawson, A. Gorelov, E. Timoshenko, Y. Kuznetsov and A. Du Chesne, Phys. A, 1997, 244, 68 CrossRef CAS.
- V. Aseyev, H. Tenhu and F. M. Winnik, Adv. Polym. Sci., 2006, 196, 1 CrossRef CAS.
- G. Zhang and C. Wu, Adv. Polym. Sci., 2006, 195, 101 CrossRef CAS.
- G. Chen and A. Hoffman, Nature, 1995, 373, 49 CrossRef CAS.
- G. Chen and A. Hoffman, Macromol. Chem. Phys., 1995, 196, 1251 CrossRef CAS.
- P. Tanner, P. Baumann, R. Enea, O. Onaca, C. Palivan and W. Meier, Acc. Chem. Res., 2011, 44, 1039 CrossRef CAS.
- D. Lensen, D. M. Vriezema and J. C. M. van Hest, Macromol. Biosci., 2008, 8, 991 CrossRef CAS.
- S. F. M. van Dongen, H.-P. M. de Hoog, R. J. R. W. Peters, M. Nalani, R. J. M. Nolte and J. C. M. van Hest, Chem. Rev., 2009, 109, 6212 CrossRef CAS.
- J. Cui, M. P. van Koeverden, M. Müllner, K. Kempe and F. Caruso, Adv. Colloid Interface Sci., 2014, 207, 14 CrossRef CAS.
- D. Schmaljohann, Adv. Drug Delivery Rev., 2006, 58, 1655 CrossRef CAS.
- S. Ganta, H. Devalapally, A. Shahiwala and M. Amiji, J. Controlled Release, 2008, 126, 187 CrossRef CAS PubMed.
- M. Kurisawa, M. Yokoyama and T. Okano, J. Controlled Release, 2000, 69, 127 CrossRef CAS.
- H. W. Spiess, in Hierarchical macromolecular structures: 60 years after the Staudinger Nobel prize I, ed. V. Percec, Springer International Publishing, 2013, Probing macromolecular and supramolecular structure, dynamics, and function by magnetic resonance, p. 295 Search PubMed.
- S. R. N. Pena, S. Raina, G. P. Goodrich, N. V. Fedoroff and C. D. Keating, J. Am. Chem. Soc., 2002, 124, 7314 CrossRef CAS.
- W. J. Parak, T. Pellegrino, C. M. Micheel, D. Gerion, S. C. Williams and A. P. Alivisatos, Nano Lett., 2003, 3, 33 CrossRef CAS.
- P. Simon, W. Radke and A. Mueller, Macromol. Symp., 2006, 240, 83 CrossRef CAS.
- S. Provencher, Macromol. Chem., 1979, 180, 201 CrossRef CAS.
- Y. Niebel, M. Buschmann, M. Lavertu and G. De Crescenzo, Biomacromolecules, 2014, 15, 940 CrossRef CAS.
- S. Han, M. Hagiwara and T. Ishizone, Macromolecules, 2003, 36, 8312 CrossRef CAS.
- S. Rangelov, P. Simon, N. Toncheva-Moncheva, Ph. Dimitrov, B. Gajewska and Ch. Tsvetanov, Polym. Bull., 2012, 68, 2175 CrossRef CAS.
- R. Trzcinska, D. Szweda, S. Rangelov, P. Suder, J. Silberring, A. Dworak and B. Trzebicka, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 3104 CrossRef CAS.
- B. Trzebicka, D. Szweda, S. Rangelov, A. Kowalczuk, B. Mendrek, A. Utrata-Wesolek and A. Dworak, J. Polym. Sci., Part A: Polym. Chem., 2013, 51, 614 CrossRef CAS.
- J.-F. Lutz, J. Polym. Sci., Part A: Polym. Chem., 2008, 46, 3459 CrossRef CAS.
- O. Akdemir, N. Badi, S. Pfeifer, Z. Zarafshani, A. Laschewsky, E. Wischerhoff and J.-F. Lutz, in Controlled/living radical polymerization: progress in ATRP, ed. K. Matyjaszewski, ACS Symposium Series 1023, American Chemical Society, Washington, DC, 2009, Design of thermoresponsive materials by ATRP of oligo(ethylene glycol)-based (macro)monomers, p. 189.
- J.-F. Lutz, in New smart materials via metal mediated macromolecular engineering, ed. E. Khosravi, Y. Yagci and Y. Savelyev, NATO Science for Peace and Security Series A: Chemistry and Biology, Springer, Dordrecht, 2009, Synthesis of smart materials by ATRP of oligo(ethylene glycol) methacrylates, p. 37 Search PubMed.
- E. B. Mock, H. De Bruyn, B. S. Hawkett, R. G. Gilbert and Ch. F. Zukoski, Langmuir, 2006, 22, 4037 CrossRef CAS.
- J. W. Kim and K. D. Suh, J. Ind. Eng. Chem., 2008, 14, 1 CrossRef CAS.
- E. Haladjova, N. Toncheva-Moncheva, M. D. Apostolova, B. Trzebicka, A. Dworak, P. Petrov, I. Dimitrov, S. Rangelov and Ch. B. Tsvetanov, Biomacromolecules, 2014, 15, 4377 CrossRef CAS.
- N. Toncheva, C. B. Tsvetanov, S. Rangelov, B. Trzebicka and A. Dworak, Polymer, 2013, 54, 5166 CrossRef CAS.
- E. Haladjova, S. Rangelov, C. B. Tsvetanov and P. Simon, Polymer, 2014, 55, 1621 CrossRef CAS.
- W. Burchard, Adv. Polym. Sci., 1983, 48, 1–124 CrossRef CAS.
- W. Burchard, Light Scattering: Principles and Development, ed. W. Brown, Clarendon Press, Oxford, 1996, p. 439 Search PubMed.
- A. Thurn, W. Burchard and R. Niki, Colloid Polym. Sci., 1987, 265, 653 CrossRef CAS.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Anal. Chem., 2006, 78, 8313 CrossRef CAS.
- A. Plumridge, S. P. Meisburger and L. Pollack, Nucleic Acids Res., 2017, 45, e66 CrossRef CAS.
- M. C. Murphy, I. Rasnik, W. Cheng, T. M. Lohman and T. Ha, Biophys. J., 2004, 86, 2530 CrossRef CAS.
- D. B. McIntosh, G. Duggan, Q. Gouil and O. A. Saleh, Biophys. J., 2014, 106, 659 CrossRef CAS.
- S. P. Meisburger, J. L. Sutton, H. Chen, S. A. Pabit, S. Kirmizialtin, R. Elber and L. Pollack, Biopolymers, 2013, 99, 1032 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Gel electrophoresis and characterization data from UV-vis, AFM, TEM, DLS, SLS, and electrophoretic light scattering. See DOI: 10.1039/c9sm01796h |
| This journal is © The Royal Society of Chemistry 2020 |