Open Access Article
Open Access ArticleSpatiotemporal pattern formation in E. coli biofilms explained by a simple physical energy balance†
Philippe
Thomen‡
,
Jules D. P.
Valentin§
,
Anne-Florence
Bitbol
 and
Nelly
Henry
and
Nelly
Henry
 *
*
Sorbonne Université, CNRS, Laboratoire Jean Perrin (UMR 8237), 4 place Jussieu, F-75005 Paris, France. E-mail: nelly.henry@upmc.fr
First published on 5th December 2019
Abstract
While the biofilm growth mode conveys notable thriving advantages to bacterial populations, the mechanisms of biofilm formation are still strongly debated. Here, we investigate the remarkable spontaneous formation of regular spatial patterns during the growth of an Escherichia coli biofilm. These patterns reported here appear with non-motile bacteria, which excludes both chemotactic origins and other motility-based ones. We demonstrate that a minimal physical model based on phase separation describes them well. To confirm the predictive capacity of our model, we tune the cell–cell and cell–surface interactions using cells expressing different surface appendages. We further explain how F pilus-bearing cells enroll their wild type kindred, poorly piliated, into their typical pattern when mixed together. This work supports the hypothesis that purely physicochemical processes, such as the interplay of cell–cell and cell–surface interactions, can drive the emergence of a highly organized spatial structure that is potentially decisive for community fate and for biological functions.
Introduction
In nature, bacteria mostly live attached to surfaces, where they form organized communities exhibiting properties that significantly differ from those of their planktonic counterparts.1–4 These communities, called biofilms, convey notable thriving advantages to bacterial populations. They are widespread across diverse environments, including soils, river banks, animals, industrial setups, ship hulls.5,6 Biofilms directly influence the Earth's biochemical cycles,7 as well as human activities, with advantageous or detrimental impacts, thus calling for inventive strategies to control their development and functions.4,8Yet, their fundamentals, such as the mechanism underpinning the switch from planktonic to biofilm lifestyle, remain strongly debated. Biofilm formation is initiated by the binding of a few cells on a surface, where they quickly grow to form confined and locally concentrated communities embedded in extracellular matrix. The sophisticated lifestyle of these attached communities has inspired the idea that biofilms may constitute a developmental stage, or a response to a biological program.9,10 Yet, no biofilm-specific gene has been discovered so far, and many – if not all – functions exhibited by biofilm-dwelling cells have also been observed (albeit separately) in planktonic cultures under specific conditions.11–13 In addition, the confinement of the cells, intrinsic to the biofilm growth mode, induces multiple physicochemical gradients which can impact various biological functions.14–16 These non-trivial physicochemical conditions might suffice to explain the biofilm-specific properties but the complexity of their interplay with the biological functions obscures the understanding of fundamental causal relationships. To gain insight into these questions, we decided to focus on the remarkable spontaneous formation of regular spatial patterns by a growing E. coli biofilm.
Incited by the similarity of the observed features with the figures displayed in physics universal processes, we reasoned that analyzing these patterns should hint at general mechanisms and fundamental driving forces.
Ordered patterns emerge spontaneously at various scales and in a wide range of systems, including oscillatory chemical reactions and landscape-scale ecological organizations17–22 as well as purely physical systems, such as phase transitions and convection patterns in fluids.23 Abundant theoretical work has been conducted to describe the universal mechanisms involved in such pattern formation, including the study of reaction-diffusion systems with activation and inhibition (Turing patterns) or excitability (Belousov–Zhabotinsky reaction), and more generally the description of instabilities in nonlinear systems.23
By contrast, the emergence of collective patterns in bacterial systems has mainly been explained by chemotaxis-related mechanisms. Exquisitely regular spatial patterns organized as regular circular arrays of dense disk-shaped spots of cells have been observed upon central deposition of motile cells on a semi-solid agar plate containing nutrients.24–27 The phenomenon was comprehensively interpreted in terms of chemotaxis to a self-secreted attractant. These patterns develop on agar plates under specific conditions where E. coli secretes high levels of these attractants. In this context, Gosh and co-workers recently proposed a model including the mechanical interactions that can be mediated by the extracellular polymeric substances.28
Patterns in biofilms were also observed in Pseudomonas aeruginosa29 and explained as resulting from a tension between growth and competition for nutrients, in experimental conditions with strong gradients of oxygen and nutrient concentrations.
More recently, a universal mechanism that does not invoke chemotaxis was proposed to explain the formation of regular patterns in bacterial systems.30 This mechanism relies on a decrease of bacterial motility with density, which promotes phase separation into a low-density and a high-density phase, and on logistic growth, which favours a single uniform density. The combination of these two antagonistic ingredients results into an arrested non-equilibrium phase separation, giving stable patterns.
The patterns we report here emerge robustly under conditions realistic to natural environments, namely immersed biofilms growing under continuous flow of nutrients. Such generic patterns could then possibly be present in spontaneous biofilm formation. Crucially, these patterns appear with non-motile bacteria, which excludes both chemotactic origins and other motility-based ones. The morphology formed by the F pilus-bearing cells as they build the biofilm is reminiscent of phase separation, e.g. in immiscible fluids, and indeed, we demonstrate that a minimal physical model based on phase separation describes them well. To confirm the predictive capacity of our model, we tune the cell–cell and cell–surface interactions using cells expressing surface appendages different from our initial F pilus-overexpressing strain. We further explain how F pilus-bearing cells enroll their wild type kindred into their typical pattern when mixed together. The good agreement between simulations and experimental observations convincingly comforts our model.
This work supports the hypothesis that purely physicochemical processes can determine sophisticated biofilm initiation. It shows how the interplay of cell–cell and cell–surface interactions can drive the emergence of a highly organized spatial structure that is potentially decisive for community fate. The universality of the process should shed light on the general principles and mechanisms grounding bacterial biofilm formation.
Material and methods
Bacterial strains and growth conditions
We used a wild type E. coli MG1655 strain (WT) CGSC 8003, a gift from the Ghigo lab (Pasteur Institute – France). This is a non-motile strain, due to the deletion of the gene coding for McaS, a small non-coding regulatory RNA involved in positive regulation of flagellar synthesis.31 We also used several variants of this strain: the variant genetically transformed to over-express the non-conjugative F pilus, the fluorescent variant, MG1655-gfp-F, carrying a gfp-mut3 gene inserted on the chromosome under the control of the lambda-promoter pR, the constitutive curli producers (MG1655-gfp-ompR234) and the constitutive Ag43 producers (MG1655-gfp-PcL-flu), which were obtained as in Beloin et al.32 All strains were grown at 37 °C overnight in lysogeny broth (LB) medium and diluted in M63B1 medium with 0.4% glucose up to exponential growth prior to biofilm initiation. When required, ampicillin (Amp, 100 μg mL−1), and tetracycline (Tet, 7.5 μg mL−1) were used in the overnight pre-cultures.Microfabrication and biofilm
Microscopy
We used an inverted NIKON TE300 microscope equipped with motorized x, y, z displacements and shutters. Timelapse bright field and fluorescence images were collected using a 20× S plan Fluor objective, NA 0.45 WD 8.2–6.9 using optical configurations detailed in ESI.†Image analysis and correlation function calculation
The images, initially acquired in 12-bits were converted into 8-bits and auto-scaled, then binarized using ImageJ automatic procedure. Next, to analyze emerging order in the images, we calculated the radial average of the autocorrelation of the image as follows: the auto-scaled and binarized images were treated using the Radially Averaged Autocorrelation ImageJ macro (see Fig. S1, ESI†) which calculates C(R) – the correlation function of an image – as C(R) = 〈fifj〉, where fi and fj are the values (0 or 1) for a pair of pixels separated by a distance R, and the average is over all such pairs. A value of 1 means perfect correlation, of −1 perfect anticorrelation and of 0 no correlation. R0, the first zero crossing value of the function, which gives the mean size of the pattern, was determined by taking the average of the R values of the last positive and first negative values of C(R).Flow cytometry (FCM)
Cell aggregation ability was evaluated by FCM using a Becton-Dickinson flow cytometer (FACScalibur) equipped with an Argon laser (488 nm). FCM acquisitions were performed on exponentially growing cell suspensions of the various strains used in this study. The signal of bacterial cells, including all aggregates and single cells, was sorted from background based on scattering intensities considering side (SSC) versus forward (FSC) scattering. The number of aggregates of the suspension, Nag, was determined from cell GFP fluorescence (channel FL1, band pass centered on 530 nm) plotted against FSC (details in Fig. S2, ESI†). The aggregation index was defined as iag = [Nag/(Nsc + Nag)]·Iag, where Nsc is the number of single cells and Iag, the mean GFP fluorescence intensity of the aggregates. Data were analyzed using FlowJo (Tree Star) multivariate analysis software.Numerical simulations
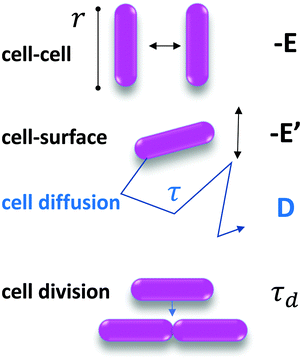
We constructed a minimal and generic model to explain the formation of patterns in biofilms. For simplicity, our agent-based simulations are performed in two dimensions on a triangular lattice with N2 vertices with periodic boundary conditions. On this lattice, we initially put cells at random vertices. These cells can diffuse with a diffusion coefficient D and interact with their nearest neighbors with an attractive interaction energy −E, measured in units of thermal energy kT (E > 0). Note that diffusion is passive in our model, reflecting the fact that our experiments employ a non-motile strain.Because experimentally, we observe that clusters of multiple bacteria hardly move, in our model, we neglect motion of clusters, and only simulate diffusion of individual cells, with a diffusion coefficient D that depends on the variant considered (see Fig. 6B), because it is affected by the cell-substrate interaction energy −E′.
In the absence of growth, our model is a simple lattice gas model, which features a liquid–gas phase transition. Equilibrium Monte Carlo simulations show that this phase transition occurs at E ∼ 1.25kT, in agreement with theoretical estimates based on nucleation theory34,35 and consistent with the mean-field approximation estimate of E = 1.2kT. This phase transition underlies pattern formation in our complete model with growth. In practice, during biofilm formation, we observe a phase separation between a high-density phase (the patches) and a low-density phase (the space with sparse bacteria around the patches).
To describe growth, we assume that cells that have no empty nearest neighbor site cannot divide, while all others divide with a division rate 1/τd. Upon division, the offspring cell fills a randomly chosen neighboring lattice site.
To simulate the evolution of this system, we perform agent-based simulations, in the framework of the Gillespie algorithm,36 which allows exact stochastic simulations without artificial discretization of time. We simulate the dynamics of the system assuming that detailed balance is satisfied, and considering a dynamics of traps, which sets the transition rates between all possible states of the system (additional details in ESI†).
Results
F pilus-expressing E. coli develop biofilms featuring regular patterns
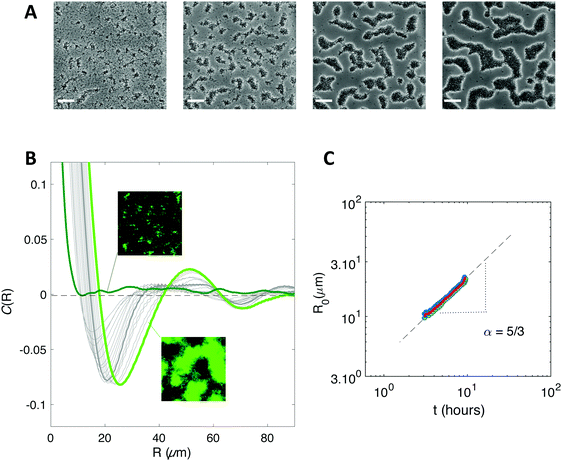
We initiated biofilm growth by introducing E. coli cells constitutively expressing the F pilus, which promotes biofilm formation, inside a millifluidic channel continuously supplied with growth medium at a flow rate of 1 mL h−1. This flow rate and this geometry were previously shown to set shear values below the threshold that prevents direct bacteria settlement.33 Biofilm development was monitored by collecting time-lapse images over the first 6 to 8 hours after cell injection in the channel. We noticed dynamical patterns similar to those observed in phase separation process, e.g. the spinodal decomposition of binary immiscible fluids,37 suggesting that some physical order underpinned the formation of these figures (Fig. 1A). To quantitatively assess the observed patterns, we computed the spatial autocorrelation function C(R) of the images of a growing biofilm during the first hours of its development. About 1 to 2 hours after flow start, the correlation curves displayed the damped oscillations typical of regular patterns (Fig. 1B). The typical domain size of the pattern, R0, the value of R when the correlation function C(R) first reaches zero, increased as the biofilm grew (Fig. 1C). Note that our imaging protocol provided accurate results only above a characteristic size of 10 μm.Inspired by studies of phase separation, we plotted R0 as a function of time over the pattern coarsening period and adjusted the data to the power law R0(t) = β + tα following Huse's generalization of the classical spinodal decomposition growth law.38 Here, α is the growth law exponent and β is a phenomenological constant taking into account diffusion through the bulk and transport at the interface of the domains.39,40
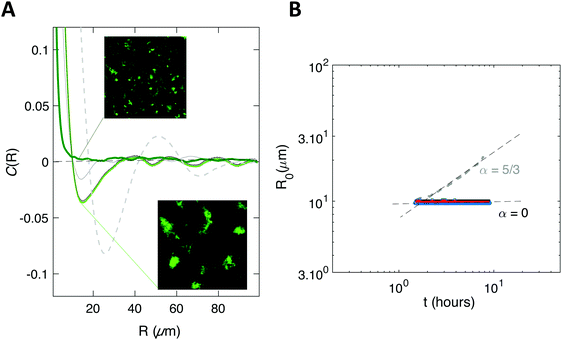
We found exponents comprised between 5/3 and 7/2 – much higher than the one (1/3) characterizing the scaling of de-mixing liquid phases known as Ostwald ripening.38,41 This faster growth can be explained by the fact that cells are dividing while the pattern forms. To evaluate the impact of cell division on pattern formation, we considered non-dividing cells. We initiated biofilm formation as before, except that the injected cells were treated with a sodium azide solution at non-lethal concentration that blocked cell division, and that we included no carbon source (no glucose) in the supplied medium. Visible patterns formed with autocorrelation functions typical of ordered patterns (Fig. 2A). However, these patterns then remained steady (Fig. 2B), in contrast with the ones formed by the dividing cells.
Model description
Thus motivated, we developed a model based on the minimal ingredients of phase separation and growth to test their ability to reproduce the formation and time evolution of the experimentally observed biofilm patterns (see Fig. 1A).We consider that bacteria interact together upon contact with an interaction energy −E (with E > 0), and diffuse in two dimensions on the substrate, with a diffusion coefficient D that depends on the cell–surface interaction energy −E′ (with E′ > 0), as summarized in Fig. 3.
To model growth, we assume that cells divide with a division rate 1/τd (related to td, the doubling time, by 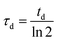 ), provided that they are not completely surrounded by other cells yet.
), provided that they are not completely surrounded by other cells yet.
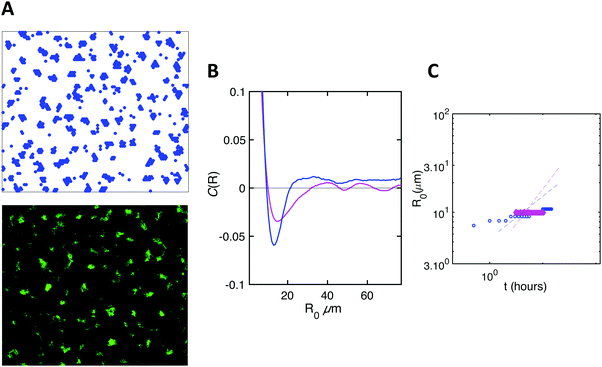
The key thermodynamic parameter, which sets the equilibrium state, is E. However, from a kinetic point of view, the diffusion coefficient D of bacteria on the surface is also crucial. Combined with the size of a bacterium, r, it sets a timescale 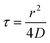 . The energy −E′ of interaction of bacteria with the substrate impacts D, and thus effectively rescales time. We performed agent-based simulations of biofilm formation within this minimal and generic model (see Methods and ESI†).
. The energy −E′ of interaction of bacteria with the substrate impacts D, and thus effectively rescales time. We performed agent-based simulations of biofilm formation within this minimal and generic model (see Methods and ESI†).
Comparison of experimental results and model's predictions
In our simulations, we observed the spontaneous formation of spatial patterns with gradually increasing order. Starting from randomly placed cells, clusters formed and joined into labyrinth-like patterns that thickened before merging as growth continued. This is qualitatively in good agreement with the patterns observed experimentally (Fig. 4A). Moreover, the spatial autocorrelation functions of our simulation data closely matched the experimental curves at intermediate times, as shown in Fig. 4B. The coarsening of the simulated patterns also follows a power law, similarly to the experimental case (Fig. 4C). However, the power law exponent obtained from simulations – about 4/3 – is smaller than the one found experimentally – comprised between 5/3 and 5/2 depending on the experiment (Fig. S3, ESI†). Note that the exact value of the exponent may depend on the experimental details, e.g. after injection into the channel, cells do not immediately start dividing but experience a lag phase, whose duration depends on the details of the initial cell culture and injection. Moreover, cell division does not really start before the flow is turned on and nutrients are supplied. Nevertheless, for simplicity, experimental data analysis is performed taking the origin of time when cells are injected into the channel, which tends to increase the coarsening exponent.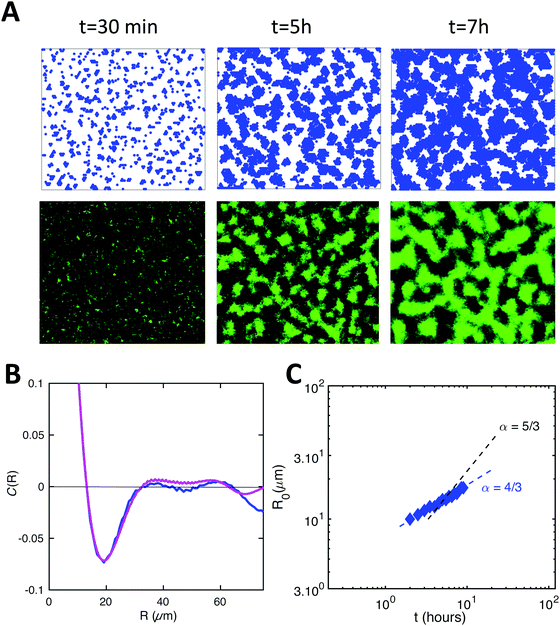 | ||
| Fig. 4 Numerical results for 2D simulations satisfyingly reproduce biofilm patterns. (A) Simulated (upper row) and experimental images (lower row) of pattern formation at different times. (B) Spatial autocorrelation curves at time 5 h from simulation (blue line) and experiment (magenta line). (C) Coarsening of the simulated patterns, assessed as in the experiments (blue diamonds); the dashed line shows the experimental result from Fig. 2B. Simulation parameters: E = 2kT; D = 0.013 μm2 s−1; τd = 156 min; r = 3 μm. | ||
We also simulated the situation where no division occurs, and obtained a very good agreement with the experimental data obtained in the absence of carbon source (Fig. 5).
 | ||
| Fig. 5 Simulations reproduce the emergence of an order in the absence of growth. Same as in Fig. 4. | ||
Overall, the behaviour predicted by the model is in good agreement with the patterning observed experimentally.
Tuning the interaction energies suppresses the patterns, confirming model predictions
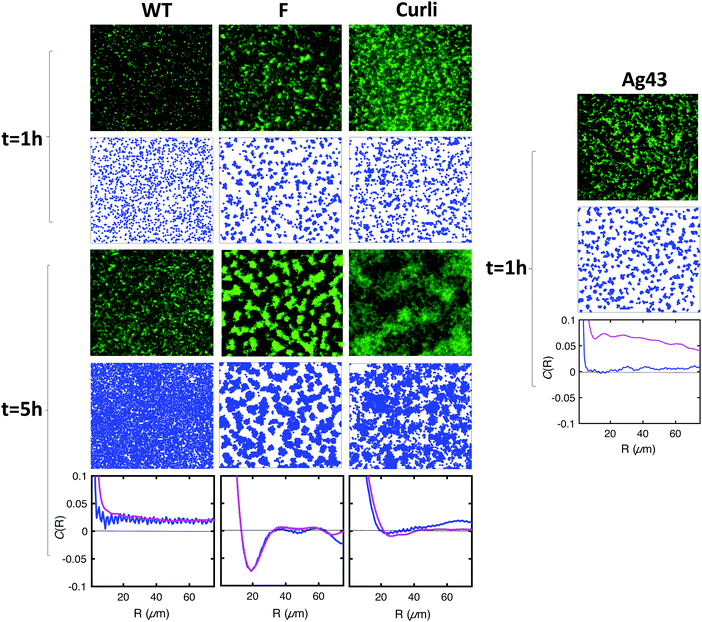
To further test the predictions of our model, we experimentally tuned the cell–cell and cell–substrate interactions. For this, we employed E. coli variants with the same genetic background as our initial F pilus-expressing strain, but displaying different surface appendages. Specifically, we used the wild type strain (WT) which does not overexpress any surface appendage, the Ag43 variant, which overexpresses the autotransporter antigen 43, and the Curli variant, which overexpresses the aggregative amyloid proteins Curli. These changes are expected to significantly alter both cell–cell and cell–surface interactions.32,42,43Using flow cytometry measurements as previously described,32,44 we made a quantitative evaluation of the ability of the cells to self-associate. We defined an aggregation index, iagg, which takes into account both the aggregate mean size and the number of aggregates found in a planktonic culture of exponentially growing cells at 37 °C – corresponding to the conditions used to initiate the biofilm – (see ESI† and Fig. S2). We found that the 4 strains rank in order of increasing ability to self-associate as follows: WT < Curli < F < Ag43 (Fig. 6A). We also observed that the index of aggregation of the Curli variant significantly increased when the culture was transferred to 30 °C, in good agreement with previous findings of optimal Curli expression at 30 °C (35).
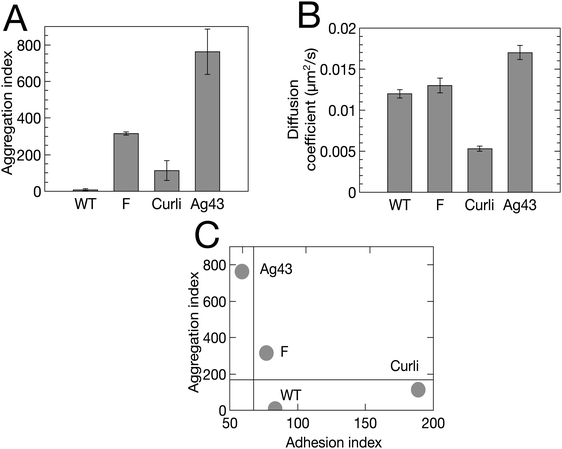 | ||
| Fig. 6 Four E. coli variants exhibit distinct adhesive and aggregative properties. (A) Aggregation index, iagg, derived from flow cytometry measurement (Fig. S2, ESI†). (B) Diffusion coefficient, D, derived from single tracking (Table S1, ESI†). Bivariate plot of aggregation index (iagg) versus adhesion index (iadh = 1/D). | ||
In addition, we ranked the four strains in increasing order of their cell–surface interaction strength using an adhesion index, iadh, defined as the inverse of the surface diffusion coefficient derived from single cell surface displacement trajectories recorded within the first 20 min after cell injection in the channel (see ESI†). We obtained Ag43 < F < WT < Curli (Fig. 6B).
Considering these indexes, we took E(WT) = 0, E(Curli) = kT and E(Ag43) = 4kT, where kT denotes the thermal energy scale, for our simulations of these various strains (recall that E(F) was taken equal to 2kT, see Fig. 4). Our simulations also took into account the measured values of the diffusion coefficients D for each strain (Tables S1 and S2, ESI†).
Typical biofilms formed by the 4 different strains are shown in Fig. 7 together with simulations obtained using cell–cell interaction energy and diffusion coefficient values matching the properties of the four strains. Note that the matching of interaction energies is not fully quantitative given the complexity of the relationship of the aggregation index to this parameter. However, our results are robust to reasonable variations of these interaction energies, the most important point being whether E is above or below the phase transition value of 1.25kT (see Materials and methods). Spatial autocorrelation functions were calculated (Fig. 7) for both experimental and numerical cell distributions. Noticeable differences between the four strains appeared after a few hours of growth and the simulated patterns exhibited an excellent agreement with the experimental figures, except in the case of Ag43, which cannot be described by our model as it detaches from the surface. Indeed, its energy of interaction with the surface is low and pushes the system in a regime where the shear stress imposed by the flow cannot be neglected anymore. We found that only the F-expressing strain gives rise to the oscillating autocorrelation function characteristic of regular pattern formation. No order emerged from WT and Curli samples, whether in experiments or in simulations.
These results indicate that the pattern morphology depends on the basic physical parameters of our model in the way predicted. This represents a strong validation of our model.
Recruitment of WT cells in the pattern formed by F-expressing cells
We observed experimentally that WT cells which by themselves did not form ordered spatial distributions (Fig. 7), were recruited to patterns in the presence of F-expressing cells. Specifically, we initiated biofilm formation by mixing GFP-expressing WT cells with F-expressing cells devoid of fluorescent label at different F to WT ratios, namely 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 1
1; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. The results displayed in Fig. 8A show that in the three mixtures WT type cells (the fluorescent ones) appear to self-organize with a degree of order similar to the pure F system (see Fig. 1 and 4), albeit decreasing in the case of the 1
3. The results displayed in Fig. 8A show that in the three mixtures WT type cells (the fluorescent ones) appear to self-organize with a degree of order similar to the pure F system (see Fig. 1 and 4), albeit decreasing in the case of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 F to WT cell mixture (Fig. 8A). Note that the observed fluorescent cell clusters comprise both non-fluorescent (F) and fluorescent cells (WT) and display a fluorescent intensity which reflects the cell type ratio.
3 F to WT cell mixture (Fig. 8A). Note that the observed fluorescent cell clusters comprise both non-fluorescent (F) and fluorescent cells (WT) and display a fluorescent intensity which reflects the cell type ratio.
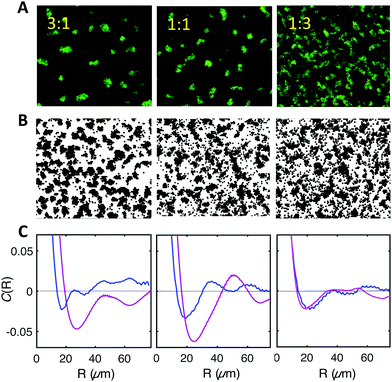 | ||
Fig. 8 F-pilus expressing cells recruit WT cells to their characteristic pattern. (A) Experimental images of biofilms formed by mixing F and WT cells at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (left); 1 1 (left); 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (middle), 1 1 (middle), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (right) ratios. (B) Simulation results in the same conditions. For comparison with the experimental images, WT and F cells have been represented in black without any color distinction (see Fig. S4 for the colored representation, ESI†). (C) Comparison of the spatial correlation functions obtained from experimental images (magenta) and simulations (blue) for the time t = 3 h 30 min for the three mixtures shown in (A) and (B). 3 (right) ratios. (B) Simulation results in the same conditions. For comparison with the experimental images, WT and F cells have been represented in black without any color distinction (see Fig. S4 for the colored representation, ESI†). (C) Comparison of the spatial correlation functions obtained from experimental images (magenta) and simulations (blue) for the time t = 3 h 30 min for the three mixtures shown in (A) and (B). | ||
In order to test whether our model could account for this interesting behavior, we simulated the experimental mix, considering a F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) WT interaction energy E′′ = 1.5kT, slightly smaller than that between F cells but larger than that between WT cells. The experimentally measured diffusion coefficients (Tables S1 and S2, ESI†) were taken into account for each cell type. Remarkably, the simulations also reported some degree of patterning, in qualitative agreement with experiments (Fig. 8B and C). However, quantitative agreement was less good than in the homogeneous systems. Indeed, the characteristic lengths and times as well as the amplitudes of the correlation provided by the model were smaller than the ones found in the experiments, thus suggesting that the interplay of the interactions might be slightly different in the real system.
WT interaction energy E′′ = 1.5kT, slightly smaller than that between F cells but larger than that between WT cells. The experimentally measured diffusion coefficients (Tables S1 and S2, ESI†) were taken into account for each cell type. Remarkably, the simulations also reported some degree of patterning, in qualitative agreement with experiments (Fig. 8B and C). However, quantitative agreement was less good than in the homogeneous systems. Indeed, the characteristic lengths and times as well as the amplitudes of the correlation provided by the model were smaller than the ones found in the experiments, thus suggesting that the interplay of the interactions might be slightly different in the real system.
Discussion
The work reported here presents a simple physical model that explains the striking spatio-temporal patterns exhibited by an F-pilus over-expressing E. coli strain as it builds a biofilm under constant flow of nutrients. The model also correctly predicted the distinct arrangements formed by wild type cells and other variants upon the same genetic background. In addition, the model qualitatively explains that F-bearing cells enroll their WT kindred into their typical pattern when mixed together, thereby further validating our theoretical description.We thus deciphered the original spatial self-organization of a non-motile E. coli strain growing under flow. This situation involves a constant division rate and no resource depletion within the first hours of biofilm development, when the patterns emerge. Importantly, these conditions – relevant in many real-life immersed adherent microbial communities, e.g. in animal vascular systems, as well as in artificial devices or natural hydrologic networks – substantially differ from those of the standard agar Petri dish extensively employed in previous studies dedicated to microbial spatio-temporal patterns (e.g.ref. 24–26 and 29).
The theoretical model we proposed is based on phase separation driven by cell–cell interactions, with kinetics that depend on the cell–surface adhesion, in a context of constant cell division. Note that our minimal model was implemented in a discrete way through agent-based simulations, thus mimicking the discrete nature of cells.
Our model captures most experimental observations, with good quantitative agreement in single-strain cases and qualitative agreement for mixtures. It demonstrates that sophisticated collective cellular behaviors such as ordered spatial organization can be obtained by combining very few and universal physical driving forces. In particular, chemotaxis, which is an important biological mechanism known to induce pattern formation in microbes,24–26 is not involved in our study, which concerns non-motile cells. From this perspective, our model is close to the one recently developed by Cates and coworkers, which involves a phase separation induced by density-dependent motility, independently of chemotaxis.30 In their model, the phase transition was arrested by logistic growth. Xavier and colleagues also proposed a model for higher-order structures in biofilms which does not involve chemotaxis but nevertheless requires resource limitation.29
The model we proposed here can be summarized as a non-arrested phase separation driving a transient spatio-temporal organization, where the continuing growth finally disrupts the pattern as cells take over the whole surface. The fact that pattern coarsening essentially arise from cell division explains the large values of the power law exponent compared to phase separation in fluids.39,41,45 Consistently, a stationary ordered spotted pattern exhibiting a characteristic length was observed in the absence of cell division.
Our results reveal that the values of the cell–cell and cell–surface interaction energies determine the pattern morphology, as demonstrated using a set of variants of the same genetic background expressing distinct surface appendages. The cell–cell interaction also allows F cells to recruit WT cells into the patterns. This provides an example where co-aggregation, already recognized essential for the development of multispecies biofilm,46 confers to kindred the potential advantage to grow in patterns.
We thus elucidated the conditions supporting the formation of ordered spatio-temporal organization in a bacterial biofilm growing under a mild flow of nutrients, an experimental design not known so far to promote such patterned spatial distribution although three-dimensional regular ripples have already been observed in Xylella fastidiosa biofilm (bacteria isolated from plant fluid) grown under flow. This specific system has motivated a multiphase mathematical model47 more complex than ours. In our case, the mechanism underlying pattern formation is remarkably simple and generic, which makes it likely to occur in various natural systems. This is reminiscent of spatiotemporal self-organization present at very different scales such as vegetation patterns48 or animal ecosystems18,49 which have been shown to improve system resilience to catastrophic shift.50–52 Whether this is also the case for microbial patterns deserves further investigation. So far, the impact of pattern formation at the level of microbial communities has been considered from the perspective of the competition of genotypes52–54 and in the general context of the impact of population structure on evolution.55–59 In addition, heterogeneity has long been recognized as a major trait of the biofilm lifestyle14,54,60 but knowledge about the direct impact of defined patterning on biofilm properties progresses slowly.61,62 However, the formation of these non-uniform cell-density distributions is expected to drive important outcomes for the biofilm. Creating local enrichment and boundaries, they generate gradients of density and metabolites14 likely to trigger local cell responses such as cell–cell communication – in E. coli for instance, aggregation has been shown to enhance AI-2-mediated signaling, biofilm formation and stress resistance.43 Other specific functions15,63–65 are also expected to occur with a timing significantly differing from uniform distributions in patterned populations, thereby altering community fate. Also worthy of interest is the specific succession of figures associated with phase separation-based mechanisms such as ours. Initially, they form islands potentially offering microniches favorable for variant emergence; next they combine into a labyrinth of continuous material which might be important for signal transmission efficiency, as suggested by the recent findings of Larkin and co-workers showing that transmission becomes possible when the adherent community is organized near a critical phase transition between a disconnected and a fully connected conduit of signaling cells.66
To summarize, we demonstrated that the interplay of physical interactions and spontaneous diffusion of bacteria on a surface are sufficient to explain a whole zoology of surface colonization figures, including regular patterns. The mechanism does not involve initial biological signaling or sensing, in contrast with the hypothesis of the biofilm as a developmental differentiated entity.9,10 Our model rather advocates that physical forces can initially drive the formation of the biofilm towards a precise spatial organization, which can in turn shape biological response according to local density and biofilm-induced physicochemical alterations of the environment. In other words, we propose here a mechanism where specific biological processes such as chemotaxis and quorum sensing occur under conditions that are physically predefined.
Our work gives clues to think about functional biofilm control strategies amenable to profoundly remodel the biofilm settlement by tuning simple physical processes such as adhesion and diffusion. This opens new avenues to elucidate the role of initial spatial patterning in biofilm development.
Conflicts of interest
NH, the corresponding author declares no financial and non-financial competing interests on behalf of all authors.Acknowledgements
The work was supported in part by a grant from French ‘Agence Nationale pour la Recherche’ (ANR-15-CE02-0001-01 ACToP). The authors thank Claude Loverdo and Raphaël Voituriez for helpful discussions.References
- J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, Annu. Rev. Microbiol., 1995, 49, 711–745 CrossRef CAS PubMed.
- L. Hall-Stoodley and P. Stoodley, Cell. Microbiol., 2009, 11, 1034–1043 CrossRef CAS PubMed.
- H. Vlamakis, Y. Chai, P. Beauregard, R. Losick and R. Kolter, Nat. Rev. Microbiol., 2013, 11, 157–168 CrossRef CAS PubMed.
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Science, 1999, 284, 1318–1322 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- T. J. Battin, W. T. Sloan, S. Kjelleberg, H. Daims, I. M. Head, T. P. Curtis and L. Eberl, Nat. Rev. Microbiol., 2007, 5, 76–81 CrossRef CAS PubMed.
- P. G. Falkowski, T. Fenchel and E. F. Delong, Science, 2008, 320, 1034–1039 CrossRef CAS PubMed.
- H. Koo, R. N. Allan, R. P. Howlin, P. Stoodley and L. Hall-Stoodley, Nat. Rev. Microbiol., 2017, 15, 740–755 CrossRef CAS PubMed.
- G. O'Toole, H. B. Kaplan and R. Kolter, Annu. Rev. Microbiol., 2000, 54, 49–79 CrossRef PubMed.
- R. D. Monds and G. A. O'Toole, Trends Microbiol., 2009, 17, 73–87 CrossRef CAS PubMed.
- C. Beloin and J. M. Ghigo, Trends Microbiol., 2005, 13, 16–19 CrossRef CAS PubMed.
- J. Domka, J. Lee, T. Bansal and T. K. Wood, Environ. Microbiol., 2007, 9, 332–346 CrossRef CAS PubMed.
- M. A. Schembri, K. Kjaergaard and P. Klemm, Mol. Microbiol., 2003, 48, 253–267 CrossRef CAS PubMed.
- P. S. Stewart and M. J. Franklin, Nat. Rev. Microbiol., 2008, 6, 199–210 CrossRef CAS PubMed.
- K. T. Schiessl, F. Hu, J. Jo, S. Z. Nazia, B. Wang, A. Price-Whelan, W. Min and L. E. P. Dietrich, Nat. Commun., 2019, 10, 762 CrossRef PubMed.
- F. Bocci, Y. Suzuki, M. Lu and J. N. Onuchic, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4288–4293 CrossRef CAS PubMed.
- K. Siteur, E. Siero, M. B. Eppinga, J. D. M. Rademacher, A. Doelman and M. Rietkerk, Ecological Complexity, 2014, 20, 81–96 CrossRef.
- M. Rietkerk and J. van de Koppel, Trends Ecol. Evol., 2008, 23, 169–175 CrossRef PubMed.
- S. Lion and M. Baalen, Ecol. Lett., 2008, 11, 277–295 CrossRef PubMed.
- Q.-X. Liu, A. Doelman, V. Rottschäfer, M. de Jager, P. M. J. Herman, M. Rietkerk and J. van de Koppel, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11905–11910 CrossRef CAS PubMed.
- C. A. Klausmeier, Science, 1999, 284, 1826–1828 CrossRef CAS PubMed.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef CAS PubMed.
- R. Hoyle, Pattern Formation: An Introduction to Methods, Cambridge University Press, Cambridge, 2006 Search PubMed.
- E. O. Budrene and H. C. Berg, Nature, 1991, 349, 630–633 CrossRef CAS PubMed.
- E. O. Budrene and H. C. Berg, Nature, 1995, 376, 49–53 CrossRef CAS PubMed.
- D. E. Woodward, R. Tyson, M. R. Myerscough, J. D. Murray, E. O. Budrene and H. C. Berg, Biophys. J., 1995, 68, 2181–2189 CrossRef CAS PubMed.
- E. Ben-Jacob, I. Cohen, O. Shochet, I. Aranson, H. Levine and L. Tsimring, Nature, 1995, 373, 566–567 CrossRef CAS PubMed.
- P. Ghosh, J. Mondal, E. Ben-Jacob and H. Levine, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E2166–E2173 CrossRef CAS PubMed.
- J. B. Xavier, E. Martinez-Garcia and K. R. Foster, Am. Nat., 2009, 174, 1–12 CrossRef PubMed.
- M. E. Cates, D. Marenduzzo, I. Pagonabarraga and J. Tailleur, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11715–11720 CrossRef CAS PubMed.
- N. De Lay and S. Gottesman, Mol. Microbiol., 2012, 86, 524–538 CrossRef CAS PubMed.
- C. Beloin, A. Houry, M. Froment, J. M. Ghigo and N. Henry, PLoS Biol., 2008, 6, e167 CrossRef PubMed.
- P. Thomen, J. Robert, A. Monmeyran, A. F. Bitbol, C. Douarche and N. Henry, PLoS One, 2017, 12, e0175197 CrossRef PubMed.
- A. Nussbaumer, E. Bittner and W. Janke, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2008, 77, 041109 CrossRef CAS PubMed.
- V. A. Shneidman and R. K. P. Zia, Phys. Rev. B: Condens. Matter Mater. Phys., 2001, 63, 085410 CrossRef.
- D. T. Gillespie, J. Comput. Phys., 1976, 22, 403–434 CrossRef CAS.
- J.-L. Barrat and J.-P. Hansen, Basic Concepts for Simple and Complex Liquids, 2003 Search PubMed.
- I. M. Lifshitz and V. V. Slyozov, J. Phys. Chem. Solids, 1961, 19, 35–50 CrossRef.
- D. A. Huse, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 34, 7845–7850 CrossRef PubMed.
- J. G. Amar, F. E. Sullivan and R. D. Mountain, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 196–208 CrossRef PubMed.
- P. W. Voorhees and M. E. Glicksman, Acta Metall., 1984, 32, 2001–2011 CrossRef CAS.
- K. Kjaergaard, M. A. Schembri, H. Hasman and P. Klemm, J. Bacteriol., 2000, 182, 4789–4796 CrossRef CAS PubMed.
- L. Laganenka, R. Colin and V. Sourjik, Nat. Commun., 2016, 7, 12984 CrossRef CAS PubMed.
- J. Geng, C. Beloin, J. M. Ghigo and N. Henry, PLoS One, 2014, 9, e102049 CrossRef PubMed.
- S. J. Mitchell and D. P. Landau, Phys. Rev. Lett., 2006, 97, 025701 CrossRef CAS PubMed.
- S. Katharios-Lanwermeyer, C. Xi, N. S. Jakubovics and A. H. Rickard, Biofouling, 2014, 30, 1235–1251 CrossRef CAS PubMed.
- N. G. Cogan, M. R. Donahue, M. Whidden and L. De La Fuente, Biophys. J., 2013, 104, 1867–1874 CrossRef CAS PubMed.
- G. G. Penny, K. E. Daniels and S. E. Thompson, Philos. Trans. R. Soc., A, 2013, 371, 20120359 CrossRef PubMed.
- G. Theraulaz, E. Bonabeau, S. C. Nicolis, R. V. Sole, V. Fourcassie, S. Blanco, R. Fournier, J. L. Joly, P. Fernandez, A. Grimal, P. Dalle and J. L. Deneubourg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9645–9649 CrossRef CAS PubMed.
- H. de Paoli, T. van der Heide, A. van den Berg, B. R. Silliman, P. M. J. Herman and J. van de Koppel, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 8035–8040 CrossRef CAS PubMed.
- Q. X. Liu, P. M. Herman, W. M. Mooij, J. Huisman, M. Scheffer, H. Olff and J. van de Koppel, Nat. Commun., 2014, 5, 5234 CrossRef CAS PubMed.
- J. van Gestel, F. J. Weissing, O. P. Kuipers and A. T. Kovacs, ISME J., 2014, 8, 2069–2079 CrossRef CAS PubMed.
- C. D. Nadell, K. Drescher and K. R. Foster, Nat. Rev. Microbiol., 2016, 14, 589–600 CrossRef CAS PubMed.
- J. Vandermeer and S. Yitbarek, J. Theor. Biol., 2012, 300, 48–56 CrossRef PubMed.
- R. Korona, C. H. Nakatsu, L. J. Forney and R. E. Lenski, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 9037–9041 CrossRef CAS PubMed.
- O. Hallatschek, P. Hersen, S. Ramanathan and D. R. Nelson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19926–19930 CrossRef CAS PubMed.
- A. F. Bitbol and D. J. Schwab, PLoS Comput. Biol., 2014, 10, e1003778 CrossRef PubMed.
- P. Lombardo, A. Gambassi and L. Dall'Asta, Phys. Rev. Lett., 2014, 112, 148101 CrossRef PubMed.
- J. R. Nahum, P. Godfrey-Smith, B. N. Harding, J. H. Marcus, J. Carlson-Stevermer and B. Kerr, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7530–7535 CrossRef CAS PubMed.
- J. W. Costerton, Z. Lewandowski, D. DeBeer, D. Caldwell, D. Korber and G. James, J. Bacteriol., 1994, 176, 2137–2142 CrossRef CAS PubMed.
- M. R. Parsek and T. Tolker-Nielsen, Curr. Opin. Microbiol., 2008, 11, 560–566 CrossRef CAS PubMed.
- A. K. Wessel, T. A. Arshad, M. Fitzpatrick, J. L. Connell, R. T. Bonnecaze, J. B. Shear and M. Whiteley, mBio, 2014, 5, e00992 CrossRef PubMed.
- C. Okegbe, B. L. Fields, S. J. Cole, C. Beierschmitt, C. J. Morgan, A. Price-Whelan, R. C. Stewart, V. T. Lee and L. E. P. Dietrich, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5236–E5245 CAS.
- J. L. Connell, J. Kim, J. B. Shear, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18255–18260 CrossRef CAS PubMed.
- S. E. Darch, O. Simoska, M. Fitzpatrick, J. P. Barraza, K. J. Stevenson, R. T. Bonnecaze, J. B. Shear and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4779–4784 CrossRef CAS PubMed.
- J. W. Larkin, X. Zhai, K. Kikuchi, S. E. Redford, A. Prindle, J. Liu, S. Greenfield, A. M. Walczak, J. Garcia-Ojalvo, A. Mugler and G. M. Suel, Cell Syst., 2018, 7(137–145), e133 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sm01375j |
| ‡ Present address: Université Côte d’Azur, CNRS UMR 7010, Institut de Physique de Nice, Parc Valrose 06108 Nice, France. |
| § Present address: Laboratory for Biointerfaces, Empa, Swiss Federal Laboratories for Materials Science and Technology, Lerchenfeldstrasse 5, 9014 St. Gallen, Switzerland. |
| This journal is © The Royal Society of Chemistry 2020 |