Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Hajime
Wakui
a,
Yoshikazu
Tanaka
b,
Toyoyuki
Ose
c,
Isamu
Matsumoto
c,
Koji
Kato
d,
Yao
Min
c,
Taro
Tachibana
e,
Masaharu
Sato
f,
Kentaro
Naruchi
f,
Fayna Garcia
Martin
a,
Hiroshi
Hinou
a and
Shin-Ichiro
Nishimura
*a
aField of Drug Discovery Research, Faculty of Advanced Life Science, Graduate School of Life Science, Hokkaido University, N21 W11, Kita-ku, Sapporo, 001-0021, Japan. E-mail: shin@sci.hokudai.ac.jp
bGraduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai 980-8577, Japan
cField of X-ray Structural Biology, Faculty of Advanced Life Science, Graduate School of Life Science, Hokkaido University, N10 W8, Kita-ku, Sapporo 060-0810, Japan
dResearch Institute for Interdisciplinary Science and Graduate School of Natural Science and Technology, Okayama University, 3-1-1, Tsushima-naka, Kita-ku, Okayama 700-8530, Japan
eDepartment of Bioengineering, Graduate School of Engineering, Osaka City University, Sumiyoshi-ku, Osaka 558-8585, Japan
fMedicinal Chemistry Pharmaceuticals, Co., Ltd., N9 W15, Chuo-ku, Sapporo 060-0009, Japan
First published on 18th November 2020
Abstract
Correction for ‘A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes’ by Hajime Wakui et al., Chem. Sci., 2020, 11, 4999–5006, DOI: 10.1039/D0SC00317D.
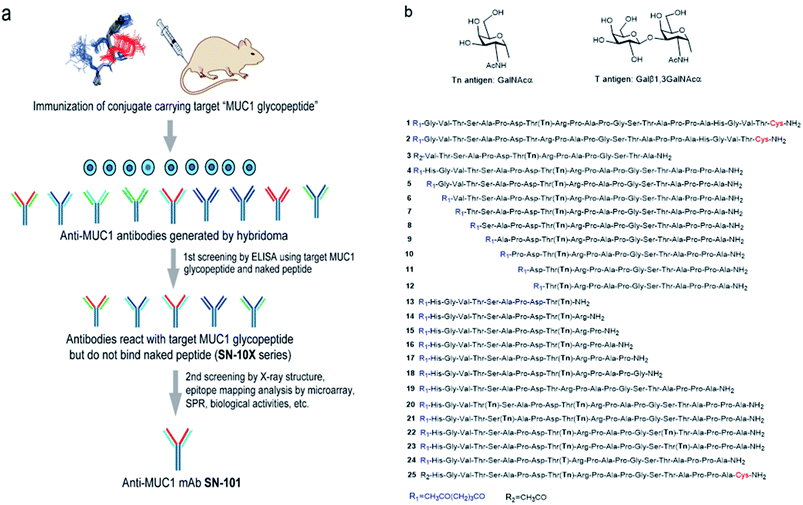
The authors regret that part Fig. 1b was missing from the version of Fig. 1 shown in the original article. The correct version of Fig. 1 is presented below.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2020 |