Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Subcellular localised small molecule fluorescent probes to image mobile Zn2+
Le
Fang
 a and
Michael
Watkinson
a and
Michael
Watkinson
 *b
*b
aThe Joseph Priestley Building, School of Biological and Chemical Science, Queen Mary University of London, Mile End Road, London, E1 4NS, UK
bThe Lennard-Jones Laboratories, School of Chemical and Physical Science, Keele University, ST5 5BG, UK. E-mail: m.watkinson@keele.ac.uk
First published on 9th October 2020
Abstract
Zn2+, as the second most abundant d-block metal in the human body, plays an important role in a wide range of biological processes, and the dysfunction of its homeostasis is related to many diseases, including Type 2 diabetes, Alzheimer's disease and prostate and breast cancers. Small molecule fluorescent probes, as effective tools for real-time imaging, have been widely used to study Zn2+ related processes. However, the failure to control their localisation in cells has limited their utility somewhat, as they are generally incapable of studying individual processes in a specific cellular location. This perspective presents an overview of the recent developments in specific organelle localised small molecule fluorescent Zn2+ probes and their application in biological milieu, which could help to extend our understanding of the mechanisms that cells use to respond to dysfunction of zinc homeostasis and its roles in disease initiation and development.
Introduction
Zn2+ is the second most abundant d-block metal ion in the human body, with a total content of about 2 g, most of which is bound to proteins. It is estimated to exist in over three thousand proteins and is widely used for catalytic, regulatory, and structural roles.1,2 Zinc plays an important part in a wide range of biological processes, such as brain function and pathology, immune function, gene transcription, and mammalian reproduction. Due to this, it is unsurprising that problems with zinc homeostasis are associated with many diseases, including Alzheimer's disease,3 prostate cancer,4 type 2 diabetes,5 and immune dysfunction and infection.6 Whilst the majority of zinc is found in bound forms, there exists a pool of ‘mobile’ or ‘free’ zinc, which initiates transient signals that stimulate various physiological processes, though its concentration in the cytosol is only in the picomolar range.7 The thiol-rich metallothioneins (MTs) and zinc transporters are normally involved in the processes to maintain cellular zinc homeostasis.7 MTs acting as Zn2+ buffers, can bind a large amount of Zn2+ and release it under conditions of oxidative stress due to the antioxidant role it plays. There are two main families of zinc transporters, ZIP and ZNT, which control the import and export of cytosolic zinc to intracellular organelles or extracellular space (Fig. 1).8 Mobile Zn2+ is associated with the regulation of gene expression, insulin secretion, and is also considered as a signalling ion for intra- and intercellular communication, such as neurotransmission.9 Therefore, effective methods are required to image mobile Zn2+ at the cellular or subcellular level in order to understand these processes.10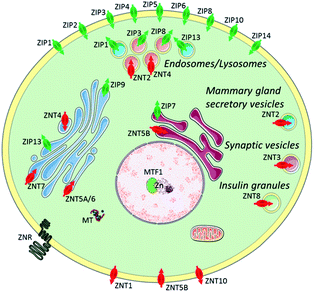 | ||
| Fig. 1 Zinc transporters (ZIP importers and ZNT exporters) and metallothioneins (MTs) to control cellular and subcellular zinc homeostasis. Reproduced from ref. 8 with permission from the Royal Society of Chemistry, Copyright [2014]. | ||
Since Zn2+ has a d10 configuration, it is redox inert in biology and is rendered spectroscopically silent for most of the commonly used photo-spectroscopic techniques.11 As a result, the development of fluorescent chemosensors has become popular for non-invasive real-time Zn2+ imaging in which a change in the fluorescence intensity or wavelength occurs due to analyte binding, which leads to signal output. The most commonly used probes utilise a switch on fluorescent mechanism in which photon-induced electron transfer (PET)12–14 is prevented upon zinc binding (Scheme 1). Typically, such fluorescent chemosensors contain a fluorophore (the signal source), a short spacer unit and a receptor (the recognition site). In addition, fluorescent probes utilising intermolecular charge transfer (ICT)15,16 and Förster Resonance Energy Transfer (FRET)17,18 mechanisms have also been widely applied in biology. In the biological environment, fluorescent chemosensors are generally involved in a competitive exchange equilibrium with endogenous ligands and proteins and thus detect changes of mobile Zn2+ pools rather than total cellular zinc levels. Therefore probes displaying high sensitivity with low detection limit, high specificity and selectivity to distinguish Zn2+ from competing metal ions are preferred.
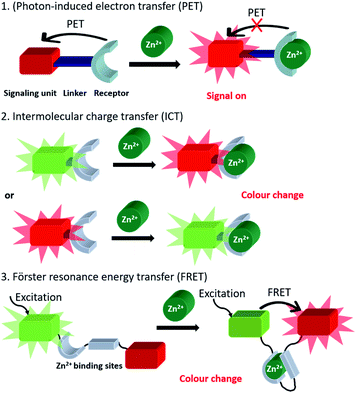 | ||
| Scheme 1 Generic representation of the three main probe classes utilised in the fluorescent imaging of mobile Zn2+. | ||
There are three main types of fluorescent sensors for biological zinc imaging: small molecule probes, protein-based biosensors and peptide targeting fluorescent sensors. Protein-based biosensors and peptide targeting fluorescent sensors can localise to specific organelles by introducing genetically encoded site-directing proteins and selecting specific amino acid sequences, which have high affinity for the particular targets.10 However, they are not without some limitations. The genetically encoded protein-based sensors cannot be transfected into all cell lines, have a small range of excitation and emission wavelengths available, as well as low photochemical stability and brightness. The peptide-based sensors are sensitive to proteases in vivo and cell internalization can be difficult, except for specific peptide sequences. In contrast, small molecule fluorescent probes can display high sensitivity and selectivity, low toxicity, and good photophysical properties. However, the failure to control the small molecule probes' cellular or subcellular location can limit their utility somewhat.
In the last two decades, there have been considerable efforts to overcome this problem. At the subcellular level, organelles require zinc for their normal function and the dysfunction of zinc homeostasis results in pathological processes, such as cellular stress, and these can induce cell apoptosis. Therefore, the development of Zn2+ probes capable of predictable and reliable subcellular localisation is required for biological applications. Reviews exist that summarize a range of subcellularly localised fluorescent probes, for cations, anions, or small molecules,19–22 however, a summary of the recent developments in subcellularly localised Zn2+ probes has, to our knowledge, not yet appeared. In this perspective, an overview of the recent development in specific organelle localised small molecule fluorescent Zn2+ probes is presented together with their applications in biological systems.
Subcellular localised small molecule fluorescent Zn2+ probes
Plasma membrane targeting
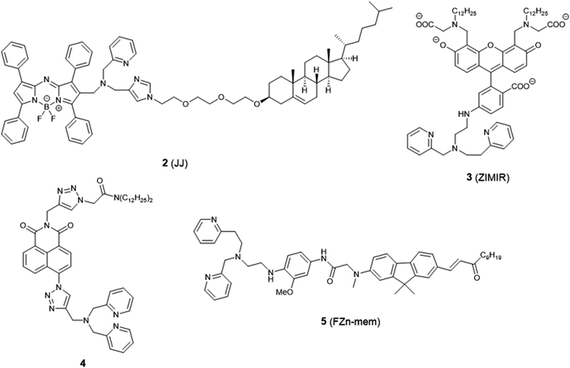
The plasma membrane, also known as the cell membrane, is mainly formed by a lipid bilayer to protect cells from their external environment. The membrane controls the import and export of substances and the selective uptake of ions and organic molecules. Membrane transporters, a class of membrane protein, play an important role in these processes. As shown in Fig. 1, ZIP 1–6, ZIP8, ZIP10 and ZIP14 are zinc transporters that control cellular zinc uptake, while ZNT1, ZNT5B and ZNT10 are responsible for its efflux through the membrane. Some data have suggested that when zinc is present in sufficiently high concentration it can act as a stabilizer of the cell membrane.23 It has also been reported that zinc deficiency in membranes causes a defect in calcium channels, which also impairs the uptake of Ca2+.24There are relatively few small molecule fluorescent probes reported to image Zn2+ in the cell membrane. Due to the phospholipid bilayer nature of the membrane, a highly hydrophobic group is normally used as a membrane targeting unit. In 2011 Yamamoto et al. reported that cholesterol could be applied as a cell membrane targeting unit.25 With fluorescein as the fluorophore and an o-aminophenol-N,N,O-triacetic acid-based zinc-chelating moiety, probe 1 (LF-Chol) showed good cell membrane localisation and fluorescence response when Zn2+ was added or removed (Fig. 2). More recently, cholesterol was also applied by You et al. in a deep-red fluorescent probe 2 (JJ, Fig. 3) to image Zn2+.26 Probe 2 displayed good photophysical properties and tight Zn2+ binding with a low dissociation constant (4 pM). Co-localisation experiments in HeLa cells revealed that 2 was rapidly internalised to intracellular spaces, including the lysosome and endoplasmic reticulum (ER), but the localisation in the membrane was very low (the Pearson's coefficient was reported to be 0.12). Meanwhile, the authors found that probe 2 facilitated the influx of exogenous Zn2+ in the absence of the pyrithione ionophore, while a control probe without cholesterol could not, thus suggesting that the membrane targeting cholesterol unit permeabilised the cell membrane (Table 1).
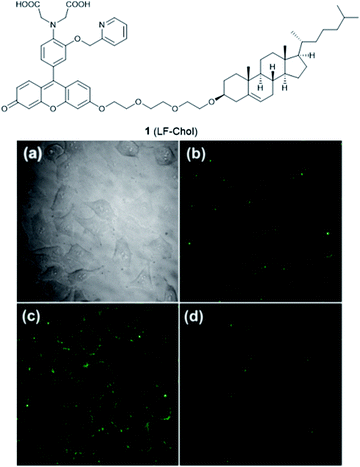 | ||
| Fig. 2 The structure of 1 (LF-Chol). (a) Bright-field transmission image of HeLa cells; (b) the fluorescence of 1 in HeLa cells; (c) the fluorescence response in the presence of 20 μM Zn2+ and (d) the fluorescence response after the subsequent addition of 100 μM EDTA. Reproduced from ref. 25 with permission from the American Chemical Society, Copyright [2011]. | ||
| Probe | Solvent | Max λex/nm | Max λem/nm | ε /M−1 cm−1 | Φ | K d | LODd | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No Zn2+ | With Zn2+ | No Zn2+ | With Zn2+ | No Zn2+ | With Zn2+ | No Zn2+ | With Zn2+ | ||||
a Molar extinction coefficient.
b Fluorescence quantum yield.
c Dissociation constant.
d The limit of detection.
e Not reported.
f
λ
ex = 466 nm.
g The parameters of donor.
h The parameters of acceptor.
i
λ
ex = 565 nm.
j
λ
ex = 400 nm.
k All results shown here were recorded at lysosomal pH 5.2.
l All results shown here were recorded at lysosomal pH 5.0.
m Calculated from turn on fluorescence emission at wavelength 578 nm.
n Calculated from the ratio of F578 nm/F680 nm.
o The pH of the buffer is 5.0.
p
K
d1 = 3.5 ± 0.1 mM, Kd2 = 150 ± 100 μM.
q
K
d1 = 220 ± 30 μM, Kd2 = 160 ± 80 μM, Kd3 = 9 ± 6 μM.
r The results were obtained in solution EtOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MOPS buffer (1 MOPS buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). 1, v/v).
|
|||||||||||
| 1 | HEPES buffer | 470 | 470 | 517 | 517 | NRe | NR | <0.005 | 0.18 | 126 nM | NR |
| 2 | PIPES buffer | 643 | 637 | 663 | 662 | 3.68 × 104 | 3.62 × 104 | 0.038 | 0.21 | 4 pM | NR |
| 3 | HEPES buffer | 493 | 493 | 515 | 515 | 7.3 × 104 | NR | 0.0032 | 0.225 | 0.45 μM | NR |
| 4 | HEPES buffer | 347 | 347 | 420 | 443 | NR | NR | NR | NR | NR | NR |
| 5 | MOPS/DPPC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
400 | 415 | 506 | 520 | 1.64 × 104 | NR | 0.19 | NR | 20 ± 0.4 nM | NR |
| 6 | MOPS buffer | 545 | NR | 575 | 575 | NR | NR | NR | NR | 65 ± 10 nM | NR |
| 7 | PIPES buffer | 497 | 490 | 522 | 517 | 3.3 × 104 | 4.2 × 104 | 0.36 | 0.80 | 1.5 ± 0.2 μM | NR |
| 8 | PIPES buffer | 569 | 573 | 629 | 631 | 2.84× 104 | 3.67 × 104 | 0.0355 | 0.5025 | 3.25 ± 0.12 nM | NR |
| 9 | MOPS buffer | 388 | 375 | 500 | 493 | 1.87 × 104 | 2.31 × 104 | 0.15 | 0.92 | 3.1 nM | NR |
| 10 | HEPES buffer | 413 | 395 | 536 | 536 | 1.85 × 104 | 2.10 × 104 | 0.0048 | 0.33 | 1.4 nM | NR |
| 11 | MOPS buffer | 392 | NR | 559 | NR | 1.32 × 104 | NR | 0.023 | NR | 17 ± 2 nM | NR |
| 12 | HEPES buffer | 453 | 391 | 625f | 542f | 6.2 × 103 | 4.2 × 103 | NR | 0.03 | 8.2 ± 0.2 nM | NR |
| 13 | HEPES buffer | 420 | 370 | 550 | 504 | NR | NR | 0.11 | 0.22 | 0.45 ± 0.01 nM | NR |
| 14 | CH3CN | 347g | 363g | 633 | 633 | 81 × 103g | 67 × 103g | 0.28i | 0.29j | 9.1 μM | NR |
| 602h | 602h | 23 × 103h | 21 × 103h | ||||||||
| 15 | PIPES buffer | 510 | 510 | 535 | 529 | NR | NR | <0.001 | 0.75 ± 0.03 | 0.6 ± 0.03 nM | NR |
| 16 | HEPES buffer | 496 | 469 | 550 | 534 | 2.10 × 104 | 1.40 × 104 | 0.003 | 0.046 | 1.0 ± 0.1 nM | NR |
| 17 | MES buffer | 285 | 261 | 542 | 495 | 4.30 × 103 | 1.66 × 103 | 0.11 | 0.17 | 16 ± 1.1 nM | NR |
| 18 | MES buffer | 435 | 444 | 528 | 536 | 1.55 × 104 | 1.81 × 104 | 0.03 | 0.23 | 8.5 μM | 0.18 μM |
| 19 | EtOH/Tris–HCl (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
597 | 568 | 647 | 578 | NR | NR | NR | 0.23 | 68 ± 4 μMm | 0.48 μM |
| 123 ± 6 μMn | |||||||||||
| 20 | MES buffero | 500 | 490 | 536 | 536 | NR | NR | NR | NR | NR | 0.477 μM |
| 21a | HEPES buffer/CH3CN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
350 | 365 | 504 | 504 | NR | NR | 0.25 | 0.24 | 1 nM2 | NR |
| 21b | 350 | 359 | 504 | 504 | NR | NR | 0.32 | 0.33 | 0.17 nM2 | NR | |
| 22a | HEPES buffer | 456 | 454 | 515 | 515 | NR | NR | 0.0027 | 0.0482 | 1.54 nM | NR |
| 22b | HEPES buffer | 454 | 451 | 509 | 515 | NR | NR | 0.0064 | 0.2606 | 2.57 nM | NR |
| 22c | HEPES buffer | 453 | 453 | 512 | 517 | NR | NR | 0.0077 | 0.3283 | 1.91 nM | NR |
| 23a | PIPES buffer | 478 | 530 | 625 | 628 | 1.93 × 104 | 2.64 × 104 | 0.067 | 0.41 | 0.69 nM | NR |
| 23b | PIPES buffer | 480 | 524 | 630 | 630 | 1.69 × 104 | 2.56 × 104 | 0.069 | 0.22 | 0.70 nM | NR |
| 23c | PIPES buffer | 480 | 535 | 623 | 628 | 1.33 × 104 | 1.93 × 104 | 0.342 | 0.60 | <0.001 nM | NR |
| 24 | EtOH | 406 | NR | 490 | NR | NR | NR | <0.01 | 0.12 | 5.5 nM | 5.8 nM |
| 25 | HEPES buffer | 346 | 346 | 414 | 414 | NR | NR | 0.041 | 0.25 | 3.5 nM | 47 pM |
| 26 | HEPES buffer | 346 | 346 | 414 | 414 | NR | NR | 0.013 | 0.041 | 4.7 nM | 0.71 nM |
| 27 | HEPES buffer | 346 | 346 | 414 | 414 | NR | NR | 0.05 | 0.28 | 2.83 ± 0.11 nM | 48 pM |
| 28 | PIPES buffer | 515 | 507 | 530 | 525 | 7.95 × 104 | 8.40 × 104 | 0.39 | 0.87 | 0.7 ± 0.1 nM | NR |
| 29a | PIPES buffer | 499 | 496 | 519 | 514 | 2.72 × 104 | 1.60 × 104 | 0.004 | 0.73 | NR | |
| 29b | PIPES buffer | 498 | 492 | 515 | 515 | 6.41 × 104 | 4.00 × 104 | 0.012 | 0.51 | NR | |
| 30 | HEPES buffer | 302 | 360 | 532 | 532 | 420 | 4680 | 0.015 | 0.055 | 1.8 ± 0.1 pM | 0.042 pM |
| 32a | MOPS buffer | 384 | 388 | 518 | 518 | 2.89 × 104 | NR | 0.12l | 0.93l | 1.7 nMr | NR |
| 33 | HEPES buffer | 453 | 442 | 546 | 536 | NR | NR | NR | 0.19 | 4 nM | 57 nM |
| 34 | HEPES buffer | 346 | 346 | 414 | 414 | NR | NR | 0.02 | 0.05 | 18.8 nM | NR |
Besides cholesterol, long alkyl chains have also been widely used for membrane targeting. Rutter, Li et al. reported the fluorescent probe 3 (ZIMIR) in which a pair of dodecyl alkyl chains were used as the membrane targeting unit as a Zn2+ indicator to image dynamic insulin release.27 The probe was quenched based on the PET mechanism and displayed a robust fluorescence increase after binding with Zn2+ and showed low toxicity and membrane labelling in a wide range of cell lines. Using this probe, the authors demonstrated exocytotic activity at subcellular resolution from pancreatic β cells in intact islets and found that the sites of Zn2+/insulin release are mainly in small groups of adjacent β cells. Similar results were observed by Watkinson et al. with the plasma membrane targeting Zn2+ probe 4.28 The di-dodecylamide motif as the membrane targeting unit was introduced through a one-pot modular ‘click-SNAr-click’ approach, which was shown to be more efficient in the synthesis. Probe 4 showed a significant switch on fluorescence response to Zn2+ due to aggregation phenomena, and in cellulo experiments in mouse pancreatic islets demonstrated its localisation to the exterior of the plasma membrane. This probe was subsequently used to measure dynamic insulin secretion by Hodson et al. since zinc is co-released from insulin-containing granules.29
Similarly, Cho et al. synthesized the two-photon (TP) probe, 5 (FZn-mem), to image near-membrane Zn2+ by introducing a long alkyl chain to 2-amino-7-(3-oxo-1-dodecen-1′-yl)-9.9-dimethylfluorene (ADF), a two-photon fluorophore with a large TP cross section.30 With the same metal-binding motif as 3, probe 5 was shown to be highly selective for Zn2+ over competing cations, and its dissociation constant Kd was determined to be 20 ± 0.4 nM and 19 ± 0.2 nM by one-photon and two-photon spectroscopy, respectively. After TP excitation by 820 nm femtosecond laser pulses, the fluorescence of 5 could be collected in the emission wavelength range of 450–600 nm, and near-membrane Zn2+ could be detected in living cells and tissue at a depth of 110 μm.
Mitochondria targeting
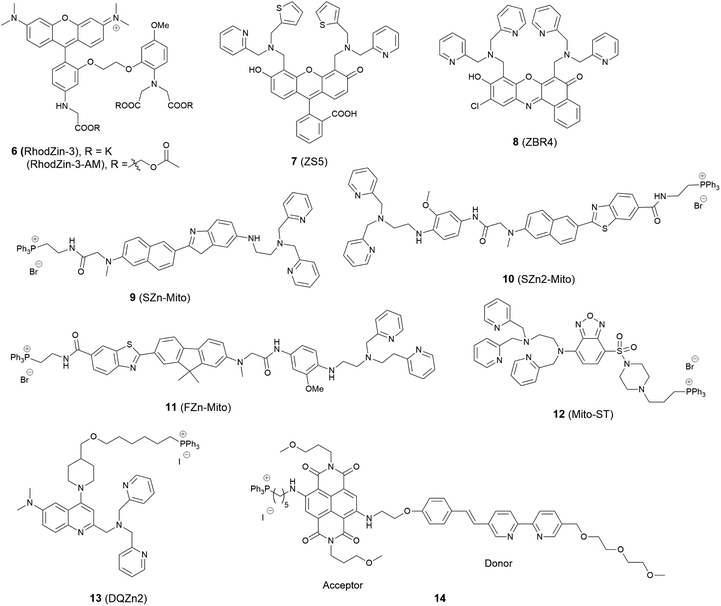
The mitochondrion is an important organelle in eukaryotic organisms. It generates most of the chemical energy supply of adenosine triphosphate (ATP) in all cells, and takes part in many biological processes, such as signalling, the cell cycle, cell growth and cell death.31 Mitochondria are involved in intracellular Zn2+ storage and as a co-factor for a wide range of enzymatic reactions and Zn2+ is closely related to the mitochondrial respiratory chain.32 It was also reported that zinc can reduce mitochondrial damage under stress conditions, which can protect cells from oxidatively-induced apoptosis.33 In contrast, mitochondrial dysfunction may cause rapid Zn2+ entry, which is one of the major contributors to neuronal injury.34 Therefore, to understand these processes better, mitochondria localised zinc imaging is in demand.The first example of a mitochondrial fluorescent Zn2+ probe was reported by Gee et al. in 2003.35 Probe 6 (RhodZin-3, Fig. 4) displayed a 75-fold fluorescence increase after binding Zn2+ and good selectivity over competing cations. The ester form was loaded into cells due to its membrane permeability and it showed co-localisation with a mitochondrial marker MitoTracker Green and Zn2+ response in cortical neurons.
In 2006, Lippard et al. synthesized a series of probes and found 7 (ZS5), displaying green fluorescence, could localise to mitochondria36 although this was the only probe to display mitochondrial localisation within a larger series of structurally similar probes synthesized. Another probe 8 (ZBR4), a red fluorescent probe, was subsequently found to spontaneously accumulate in mitochondria in 2014 by the same group.37 However, the localisation was again apparently serendipitous as this was the only probe in another large family of probes that failed to localise to the endoplasmic reticulum (ER).
Delocalised lipophilic cations, such as the triphenylphosphonium salt (TPP), are known as effective mitochondrial targeting groups, since they can cross hydrophobic membranes and accumulate in the mitochondria in living cells. By introducing this group Kim and Cho et al. obtained a mitochondria-targeting two-photon probe 9 (SZn-Mito) for Zn2+ imaging (Fig. 5).38 The probe showed a 7-fold increase of two-photon excited fluorescence after being bound with Zn2+ and was able to detect zinc in living tissues to a depth of 100–200 μm through two-photon microscopy. Just one year later, the same group developed a similar mitochondria targeting probe 10 (SZn2-Mito) which had a 70-fold two-photon excited fluorescence increase in response to Zn2+ and displayed similar properties.39 Besides these, another two-photon Zn2+ probe 11 (FZn-Mito) was synthesized with a different fluorophore, but it had similar mitochondria targeting behaviour and two-photon emission response to Zn2+.40
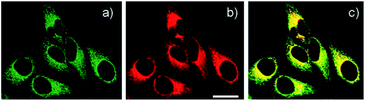 | ||
| Fig. 5 (a) TPM and (b) OPM images of HeLa cells co-labelled with (a) 9 (0.5 μM) and (b) Mitotracker Red FM (1 μM) for 30 min; (c) colocalised image. Reproduced from ref. 38 with permission from the American Chemical Society, Copyright [2011]. | ||
In 2012 Guo et al. synthesized the ratiometric probe 12 (Mito-ST) which was capable of imaging zinc ions in the mitochondria.41 With Zn2+ associated, the probe showed a blue shift in both excitation and emission wavelength. Interestingly, they observed the concentration of Zn2+ in mitochondria started to increase immediately upon H2O2 (10 mM) stimulation and increased to 1.6 nM (from 0.6 nM) with the ratiometric imaging result based on the method reported by Tsien et al.42 However, the Zn2+ release stimulated by S-nitrosocysteine (SNOC, 10 mM), a precursor of the NO radical, experienced a lag phase, and the concentration was much higher (76 nM) compared to that stimulated by H2O2. In the same year Jiang et al. reported the quinoline-based ratiometric probe 13 (DQZn2) with similar properties.43 The emission of the probe showed a 46 nm blueshift, with about a 5-fold change of the emission intensity ratio after binding with Zn2+, which allowed the concentration of mitochondrial free Zn2+ to be quantitatively measured in NIH3T3 cells.
The mitochondria localised Zn2+ probe 14 utilising a FRET mechanism was reported by Zhu et al.44 The FRET donor fluorophore displayed a bathochromic shift of emission when coordinated with Zn2+. The spectral overlap between the emission of the donor and the absorption of the acceptor increased upon Zn2+ binding, which enhanced the FRET efficiency. In HeLa cells, when the probe was excited at 405 nm, red fluorescence with emission wavelength 580–680 nm could be observed after addition of ZnCl2.
In 2014, a reaction-based fluorescent probe 15 (DA-ZP1-TPP) was reported to investigate mobile Zn2+ in mitochondria.45 As shown in Fig. 6, the phenolic oxygen atoms of the xanthene ring in the fluorescein moiety was protected with an acetyl group, which rendered it non-fluorescent in metal-free media. In the presence of Zn2+, the ester group was hydrolysed, and the PET mechanism quenched by Zn2+ association, giving a strong fluorescence response (more than a 140-fold increase). Interestingly, using this probe, the authors found that tumorigenic epithelial prostate cells could not accumulate mobile Zn2+ in their mitochondria, compared to that of healthy epithelial prostate cells, which could.
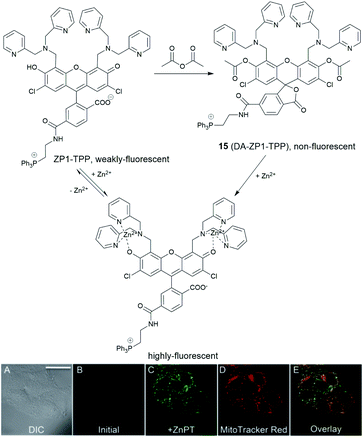 | ||
| Fig. 6 The structure of the reaction-based probe 15 (DA-ZP1-TPP). Its fluorescence response to Zn2+ and co-localisation with Mito-tracker red. Reproduced from ref. 45 with permission from the National Academy of Sciences, Copyright [2014]. | ||
Lysosome targeting
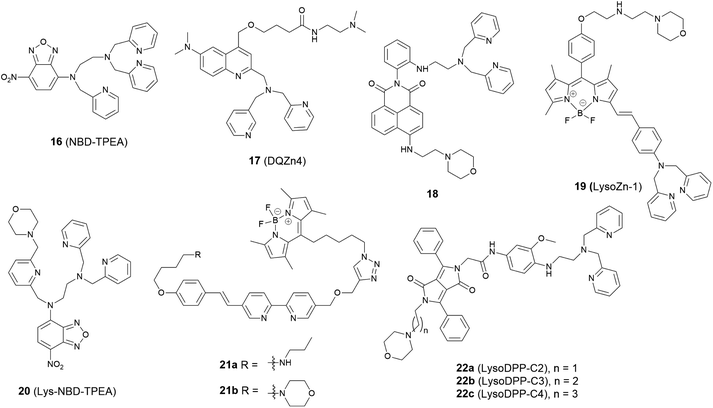
The lysosome acts as the waste disposal system in cells by digesting unwanted materials in the cytoplasm. Zn2+ homeostasis in the lysosome is regulated by zinc transporters, ZIP3, ZIP8 and ZNT2, and its dysregulation is associated with a wide range of pathological processes at the cellular level. For example, oxidative stress induced by H2O2 causes Zn2+ accumulation in the lysosome in hippocampal neurons, which eventually undergo lysosomal membrane permeabilization (LMP) and this may be the mechanism of oxidative neuronal death.46 It was also reported that lysosome-related organelles in intestinal cells of C. elegans (gut granules) are the major site of zinc storage, which promotes detoxification and subsequent mobilization, regulating cellular and organismal zinc metabolism.47 Therefore, it is vital to develop fluorescent probes to image Zn2+ in the lysosomal space to help understand these kinds of biological processes.In 2009 Guo et al. synthesized the intramolecular charge transfer (ICT) based probe 16 (NBD-TPEA, Fig. 7).48 The probe demonstrated a good selectivity for Zn2+, a large Stokes shift, and was also applied for in vivo Zn2+ imaging in zebrafish larva. However, subcellular experiments showed it to not only accumulate in the lysosome but also in the Golgi apparatus, which may perhaps limit its application in lysosomal Zn2+ imaging.
The basic ethylenediamine group has been reported to accumulate in the lysosome since it can be protonated by the acid environment in lysosomal space (pH = 4.5–5.5). The basic dimethylethylamino moiety has therefore been introduced as a lysosome targeting group in the ratiometric probe 17 (DQZn4) based on quinoline and showed good targeting behaviour.49 It performed well in the acidic pH environment, showing a significant turn-on fluorescence response and 47 nm blueshift of emission wavelength on binding Zn2+. With these desirable properties in vitro, it was successfully utilised for imaging lysosomal Zn2+ changes in NIH 3T3 cells.
Another group that has been frequently used for targeting the lysosome is the morpholine unit. One example is the two-photon probe 18 based on a naphthalimide dye with an N,N-di-(2-picolyl)ethylenediamine (DPEN) Zn2+ ligand, which can image intracellular Zn2+ in the lysosome and mouse brain tissues under two-photon excited microscopy.50 Unlike the other subcellular targeting probes, in this case organelle differentiation relied on a concentration gradient of the probe intracellularly. Probe 18 showed a strong fluorescence switch on response to Zn2+ in the lysosomal pH range (pH = 4.5–5.5) but the intensity increase was much smaller at cytosolic pH range (pH = 7.2–7.4), therefore it was able to detect lysosomal Zn2+ specifically.
Using the same targeting unit Peng et al. reported a ratiometric probe 19 (LysoZn-1) to image lysosomal Zn2+ (Fig. 8).51 The authors introduced an electron donor 4-ethoxylphenyl to the meso-position of a styryl-Bodipy-DPA scaffold, which distinguished Zn2+ from Cd2+ very well. They explained this using Hard-Soft Acid–Base theory,52 which was supported by theoretical calculations. Probe 19 exhibited a significant fluorescence increase and ratiometric (F578 nm/F680 nm) changes upon Zn2+ binding and had a good response to Zn2+ in the lysosomal pH range. Using 19 it was observed that lysosomal Zn2+ concentration increased upon H2O2 stimulation in neuronal stem cells and a similar phenomenon was observed by Uvdal et al. with a PET based lysosome localised probe 20 (Lys-NBD-TPEA) in which morpholine was again utilised as the targeting unit.53
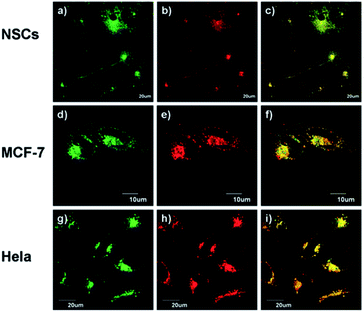 | ||
| Fig. 8 19 (LysoZn-1) co-localises to lysosomes in NSCs (a–c), MCF-7 (d–f) and HeLa (g–i) cells. Cells were stained with LysoSensor Green (1 μM) (a, d and g), and 19 (1 μM) (b, e and h), (c, f and i) Overlap of these two channels. Reproduced from ref. 51 with permission from the Royal Society of Chemistry, Copyright [2014]. | ||
Based on a FRET mechanism, two fluorescent Zn2+ probes 21 were prepared by Zhu et al. with different aliphatic amino groups as the lysosome targeting units.54 A Zn2+-sensitive arylvinylbipyridyl fluorophore was selected as the FRET donor, and through the efficient intramolecular FRET process, its broad emission band was transformed into a strong, narrow emission band of the acceptor Bodipy, which is preferable for multi-colour imaging. With a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry between 21 and Zn2+, the molar absorptivity of the donor was increased upon Zn2+ coordination, leading to the enhancement of acceptor emission. The lysosome localisation was confirmed by confocal microscopy and with the high resolution of structured illumination microscopy (SIM), they found 21b localised to the interior of lysosomes in HeLa cells, rather than anchoring at the lysosomal membranes (Fig. 9). Sessler et al. synthesized a series of probes 22 (LysoDPP-C2-C4), which were designed based on AND logic to detect both Zn2+ and the acidic pH of the lysosome.55 The morpholine moiety served not only as a lysosome targeting unit but also as a pH-responsive marker, since the nitrogen in the morpholine moiety was protonated at low pH and the PET was quenched, which increased the fluorescence of the diketopyrrolopyrrole (DPP) fluorophore in the same way as the PET quenching upon Zn2+ binding. The experiments in cellulo showed 22c was the most effective probe in terms of the initial fluorescence and the fluorescence response to Zn2+. When incubated with 22c, the cancerous prostate cell lines PC3 and DU145 showed no change in fluorescence intensity, while the normal human prostate epithelial cell line RWPE1 displayed a significant increase on the addition of exogenous Zn2+. The authors also demonstrated that the probe was capable of imaging the prostate in vivo nude mice models and discriminating between cancerous and normal prostate tissue through histological studies.
1 stoichiometry between 21 and Zn2+, the molar absorptivity of the donor was increased upon Zn2+ coordination, leading to the enhancement of acceptor emission. The lysosome localisation was confirmed by confocal microscopy and with the high resolution of structured illumination microscopy (SIM), they found 21b localised to the interior of lysosomes in HeLa cells, rather than anchoring at the lysosomal membranes (Fig. 9). Sessler et al. synthesized a series of probes 22 (LysoDPP-C2-C4), which were designed based on AND logic to detect both Zn2+ and the acidic pH of the lysosome.55 The morpholine moiety served not only as a lysosome targeting unit but also as a pH-responsive marker, since the nitrogen in the morpholine moiety was protonated at low pH and the PET was quenched, which increased the fluorescence of the diketopyrrolopyrrole (DPP) fluorophore in the same way as the PET quenching upon Zn2+ binding. The experiments in cellulo showed 22c was the most effective probe in terms of the initial fluorescence and the fluorescence response to Zn2+. When incubated with 22c, the cancerous prostate cell lines PC3 and DU145 showed no change in fluorescence intensity, while the normal human prostate epithelial cell line RWPE1 displayed a significant increase on the addition of exogenous Zn2+. The authors also demonstrated that the probe was capable of imaging the prostate in vivo nude mice models and discriminating between cancerous and normal prostate tissue through histological studies.
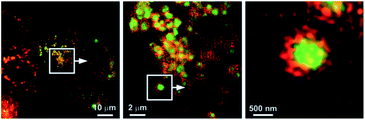 | ||
| Fig. 9 SIM images to show the localisation of compound 21b (green) compared to that of the lysosome-associated membrane protein (LAMP) fused to the fluorescent protein FusionRed (red). Reproduced from ref. 54 with permission from Wiley-VCH, Copyright [2015]. | ||
Endoplasmic reticulum (ER) targeting
The ER, an organelle in eukaryotic cells, serves a number of important cellular roles, such as protein synthesis and transport, protein folding, carbohydrate metabolism, lipid and steroid synthesis.56 Proteins synthesized in the ER are normally properly folded and transported to the Golgi apparatus, however, when there are changes to ER function, resulting from factors such as ageing, genetic mutations, or the environment, unfolded or misfolded proteins are synthesized and accumulate in the ER, causing ER stress, which activates the unfolded protein response (UPR).57It is known the ER acts as an intracellular store for biological mediators, including zinc, which it requires for normal function. For example, it has been found that zinc can be released from thapsigargin- and inositol 1,4,5-trisphosphate (IP3)-sensitive ER storage in cortical neurons.58 ZIP7, ZIP9, ZIP13 and ZNT5-7 are the transporters to regulate Zn2+ inside the ER and the depletion of zinc transporters and zinc deficiency can cause ER stress and upregulate the UPR,59,60 which can result in inflammation61 and a wide range of diseases, such as diabetes62 and neurodegenerative disorders, including Parkinson's and Alzheimer's diseases.63 However, the role of ‘mobile’ zinc in these processes is little understood due to the lack of suitable molecular tools to image this subcellular region that exist.
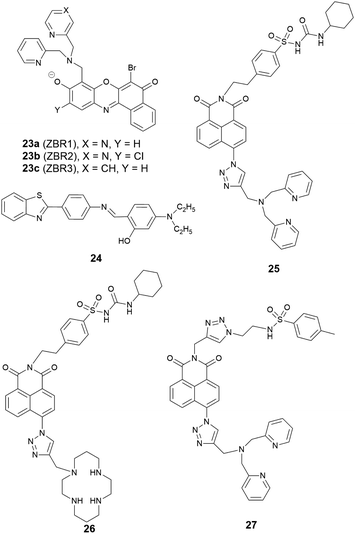
Reports of ER localised small molecule Zn2+ probes are very limited, and all early reports were of systems found to accumulate in the ER adventitiously. In 2013 Lippard et al. reported a series of benzoresorufin based red-emitting fluorescent probes 23 (the ZBR family, Fig. 10) for labile Zn2+.64 The probes displayed a broad absorption band and a bathochromic shift after binding with Zn2+, while the emission showed up to 8.4-fold increase with addition of Zn2+. In cellulo studies revealed all probes accumulated in the ER in a variety of cell lines, as the Pearson's correlation coefficient with ER tracker was much higher the other organelle tracker dyes (Fig. 11). Interestingly, with 23a, the authors observed the depletion of labile zinc in the ER of neural stem cells under ER stress induced by peroxynitrite.
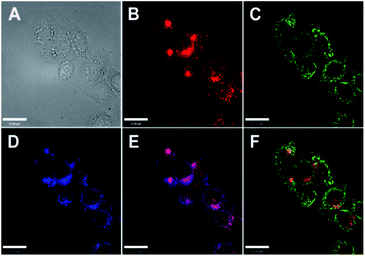 | ||
| Fig. 11 Co-localisation analysis of 23a (ZBR1) with organelle-specific markers in HeLa cells (A) bright-field image; (B) 23a (5 μM); (C) Mito-Tracker Green (0.2 μM); (D) ER-Tracker (1 μM); (E) overlay of 23a and ER-Tracker; (F) overlay of 23a and Mito-Tracker Green. Scale bar = 25 μm. Reproduced from ref. 64 with permission from the American Chemical Society, Copyright [2013]. | ||
The conveniently prepared small molecular fluorescent probe 24 included benzothiazole as the fluorophore and an o-hydroxyl Schiff base as the Zn2+ receptor.65 Upon binding Zn2+, the PET process was blocked, and the fluorescence response had a 65-fold increase with a slight red shift, with a Job's plot showing a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry between 24 and Zn2+. In human liver hepatocellular carcinoma cells, 24 was able to detect intracellular endogenous Zn2+ and its subcellular localisation was found to be the ER with a Pearson's correlation coefficient of 0.88 compared to ER tacker Red. Moreover, 24 was applied to investigate Zn2+ in plant tissues where it was shown that Zn2+ mainly accumulates in vascular tissue and epidermal cells in the cotyledons when treated with exogenous Zn2+.
1 binding stoichiometry between 24 and Zn2+. In human liver hepatocellular carcinoma cells, 24 was able to detect intracellular endogenous Zn2+ and its subcellular localisation was found to be the ER with a Pearson's correlation coefficient of 0.88 compared to ER tacker Red. Moreover, 24 was applied to investigate Zn2+ in plant tissues where it was shown that Zn2+ mainly accumulates in vascular tissue and epidermal cells in the cotyledons when treated with exogenous Zn2+.
Given the ongoing need to develop an effective and reliable strategy for targeting the ER, a number of targeting units for the organelle have been explored. Glibenclamide functions as the ER targeting group in the commercial ER tracker red and ER tracker green, and using a sulfonyl urea analogue, we developed two probes 25 and 26 as ER targeting probes.66 It was shown that both probes accumulated in the ER in a number of cell lines, and displayed a good switch on fluorescence response to Zn2+. Probe 25 was used to demonstrate that a decrease of mobile Zn2+ concentration in the ER occurs under conditions of ER stress induced by tunicamycin and thapsigargin.
In addition to this cyclohexyl sulfonylurea moiety, the recently reported methyl phenyl sulfonamide was also incorporated as an ER targeting group through an alternative modular ‘click-SNAr-click’ approach.67 The probe obtained, 27, was also found to localise to the ER compared to the other organelles tested.
Golgi apparatus targeting
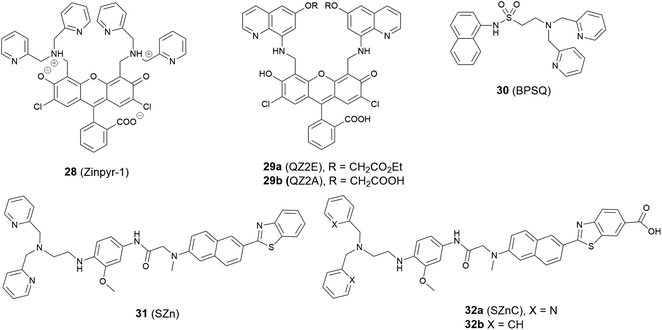
The Golgi apparatus works as a central station in cells, receiving secretory cargoes exported from the ER packing proteins into membrane-bound vesicles and sending them to their intra- and extra-cellular destinations. Zn2+ is integral to these processes for a variety of proteins, functioning in catalytic, regulatory, and structural roles. For example, it was found that zinc takes part in the interaction between the two main Golgi proteins GRASP55 and Golgin45, maintaining the normal morphology of the Golgi apparatus;68 Zn2+ also coordinates with insulin monomers in the trans-Golgi network to package it into secretory granules, which are then released from pancreatic β-cells.69 To regulate Zn2+ homeostasis in the Golgi apparatus, transporters ZNT4-7 and ZIP7, ZIP9, ZIP11, ZIP13 are responsible for Zn2+ import and export, respectively. The breakdown of Zn2+ homeostasis in the Golgi apparatus is likely to be associated with a range of human disorders, such as cancer and neuronal, liver, kidney and eye diseases70 and the development of effective methods for its detection and monitoring are required.The first Zn2+ probe to localise in the Golgi apparatus was reported in 2000 by Lippard, Tsien et al.71 With fluorescein as the signalling unit, probe 28 (Zinpyr-1, Fig. 12) has a large extinction coefficient, high quantum yield, and good membrane permeability. Experiments in Cos-7 cells showed it colocalised well with a galactosyl transferase-enhanced cyan fluorescent protein fusion (GT-ECFP), confirming 28 stains the Golgi apparatus. Some years later Lippard et al. presented two cell-trappable fluorescent Zn2+ probes: a carboxylic ester probe 29a (QZ2E) and its carboxylic acid analogue 29b (QZ2A).72 The probes were poorly emissive but had a significant increase in emission after binding with Zn2+ (120-fold for 29a, 30-fold for 29b). Interestingly, the authors found the 29b was cell membrane-impermeable due to its carboxylic acid moieties, but 29a was membrane permeable and mainly localised to the Golgi apparatus; after 18 hours' incubation time, the ester was hydrolysed to produce 29b, which was trapped inside cells.
Guo et al. modified the 8-sulfonamidoquinoline based commercial probe Zinquin to produce a new fluorescent probe 30 (BPSQ) to image Zn2+ with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry, which was better at discriminating mobile Zn2+.73 Co-localisation experiments showed it to accumulate preferentially in the Golgi apparatus.
1 binding stoichiometry, which was better at discriminating mobile Zn2+.73 Co-localisation experiments showed it to accumulate preferentially in the Golgi apparatus.
Following the previous experience on mitochondria targeting Zn2+ probes,38,39 Kim et al. reported a Golgi-localised two-photon probe 32a (SZnC), which had a strong two-photon excited fluorescence enhancement in response to Zn2+ for real-time monitoring of Golgi Zn2+ changes (Fig. 13).74 According to theoretical predictions for Golgi apparatus staining, the lipophilicity value (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value) should be within the range of 3–5.75 The log
P value) should be within the range of 3–5.75 The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values were calculated through measuring the probe's partitioning ratio between n-octanol and buffer, and the value of 32a was found to be 2.9 ± 0.1, which is reasonably matched to the theoretical range whereas it was determined to be 2.5 ± 0.1 for the control compound 31 (SZn), which experiment proved to be non-Golgi targeting. However, the log
P values were calculated through measuring the probe's partitioning ratio between n-octanol and buffer, and the value of 32a was found to be 2.9 ± 0.1, which is reasonably matched to the theoretical range whereas it was determined to be 2.5 ± 0.1 for the control compound 31 (SZn), which experiment proved to be non-Golgi targeting. However, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of another control probe 32b was 3.1 ± 0.1 and this was spread over the entire cell except the nucleus. The authors attributed the Golgi localisation of 32a to the weakly basic pyridyl group, which 32b does not possess. Therefore, both the lipophilicity and pyridyl moiety of 32a appear to contribute to its accumulation in the Golgi apparatus and the probe was applied for imaging Zn2+ in rat hippocampal tissue and could be observed at a depth of more than 100 μm with two-photon microscopy. In addition to the log
P values of another control probe 32b was 3.1 ± 0.1 and this was spread over the entire cell except the nucleus. The authors attributed the Golgi localisation of 32a to the weakly basic pyridyl group, which 32b does not possess. Therefore, both the lipophilicity and pyridyl moiety of 32a appear to contribute to its accumulation in the Golgi apparatus and the probe was applied for imaging Zn2+ in rat hippocampal tissue and could be observed at a depth of more than 100 μm with two-photon microscopy. In addition to the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value, it may be that other factors, such as amphiphilicity, electric charge and pKa values, molecular and ionic weights and conjugated bond number of the probes could also be considered to predict their location more accurately.75
P value, it may be that other factors, such as amphiphilicity, electric charge and pKa values, molecular and ionic weights and conjugated bond number of the probes could also be considered to predict their location more accurately.75
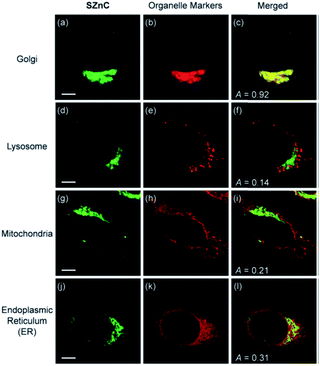 | ||
| Fig. 13 The colocalisation of probe 32a (SZnC) (2 μM) and organelle markers in HeLa cells. Reproduced from ref. 74 with permission from Royal Society of Chemistry, Copyright [2015]. | ||
It appears that all of the probes described above were found to reside in the Golgi apparatus adventitiously, therefore a Golgi targeting unit is still required in order to develop reliable Golgi apparatus localised Zn2+ probes. Ceramide may be an option for Golgi apparatus targeting since it has been employed in commercial stains for the Golgi apparatus such as NBD-ceramide and Bodipy-ceramide. However, it requires multi-step synthesis and also co-localises to the plasma membrane, which may limit future applications to some extent. More recently a phenylsulfonamide-based fluorescent probe was reported to localise to the Golgi apparatus for H2S imaging,76 which may provide a possible Golgi apparatus targeting unit for Zn2+ probes. Furthermore, cysteine has been reported as a Golgi apparatus targeting unit in other systems,77,78 since galactosyltransferase and protein kinase D were found to anchor to the Golgi region via their cysteine residues or cysteine rich domains79,80 and this may also find utility. However, it has been noted that cysteine is too hydrophilic,81 and consequently not membrane permeable in some cases. We therefore incorporated an S-trityl protected cysteine moiety into a new Zn2+ probe through ‘click’ chemistry, and confirmed the probe to be membrane permeable.82 Interestingly, the trityl group appeared to be removed in cellulo within 24 hours, and the deprotected probe accumulated in the Golgi apparatus.
Nucleus targeting
The nucleus, as the control centre of the cell, maintains the integrity of genes and controls the activities of the cell by regulating gene expression. In eukaryotic cells, there is a double membrane which encloses the organelle from the cytoplasm. This nuclear membrane is impermeable to large molecules, and the nuclear pores are the channels for large molecules, which must be actively transported by carrier proteins, while also allowing free movement of small molecules and ions. Among these, ZNT9 and ZIP7 were found to be responsible for zinc influx and efflux. About 30–40% of total intracellular zinc is found in the nucleus, which plays an important role in the regulation of cell proliferation. It stabilises the structures of DNA, RNA and the ribosome, and is involved in DNA and protein synthesis. Nuclear hormone receptors are regulated by zinc finger domains, and zinc deficiency impairs their responsiveness, which also has effects on metabolic processes related to growth.83 Some zinc-dependent proteins have been found in the nucleus functioning as transporters.84 It is therefore necessary to image Zn2+ in the nucleus to better understand zinc influx and distribution and its related biological processes.Nucleus localised Zn2+ probes have been realised by genetically encoded sensors85 or small molecule fusion protein tags,86 however there are no small molecule Zn2+ probes showing specific nucleus targeting, though the commercial probes Newport Green87 and Zinquin88 have been reported to display entire cell distribution, including the nucleus. A fluorescent probe based on naphthalimide, 33 (Naph-BPEA, Fig. 14), reported by Guo et al.89 displayed a 4-fold fluorescence enhancement and blue shift of the ICT absorption band on Zn2+ binding. The intracellular distribution of the probe was studied in HeLa and HepG2 cells and when the cells were stained with 33, only cytosolic fluorescence was observed. However, when exogenous Zn2+ was added, the entire cell became fluorescent, including the nucleus, which was confirmed with the Hoechst stain, a commercial DNA dye (Fig. 14). It demonstrated the ability of the probe to penetrate the nuclear envelope, but the nuclear fluorescence response was small due to the low nuclear concentration of labile Zn2+. The authors speculated that incorporation of the 4-amino-1,8-naphthalimide moiety to be the reason for its nuclear envelope penetrability through positive and negative control experiments. Furthermore, in vivo Zn2+ imaging in zebrafish larva was also performed. In 2018, we developed the biotinylated probe 34 based on 1,8-naphthalimide moiety for imaging Zn2+ in breast cancer MCF-7 cells and found it has nuclear envelope penetrability.90 However, it suffers from the same issue as 33 in that it was distributed over the entire cell and its sensitivity was insufficient to detect endogenous nuclear Zn2+ with the nucleus fluorescence only being switched on after the addition of exogenous Zn2+.
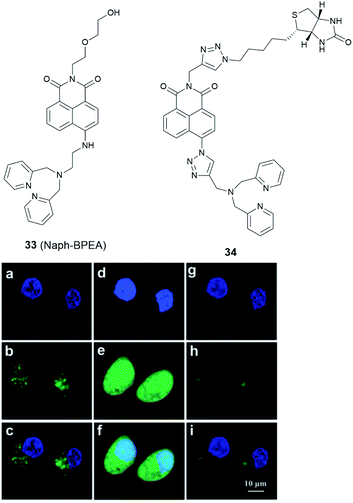 | ||
Fig. 14 The structure of 33 (Naph-BPEA) and 34. Confocal fluorescence images of HeLa cells co-stained with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (blue) and 33 (green). (a–c): images of the co-stained cells; (d–f): images of the co-stained cells followed by Zn2+-pyrithione incubation; (g–i): images of cells which were subsequently treated by TPEN. Reproduced from ref. 90 with permission from the Royal Society of Chemistry, Copyright [2013]. 342 (blue) and 33 (green). (a–c): images of the co-stained cells; (d–f): images of the co-stained cells followed by Zn2+-pyrithione incubation; (g–i): images of cells which were subsequently treated by TPEN. Reproduced from ref. 90 with permission from the Royal Society of Chemistry, Copyright [2013]. | ||
Nonetheless, the challenge remains to develop small molecule probes to image Zn2+ in the nucleus specifically, where the concentration of mobile Zn2+ in this organelle is also very low. In this case, targeting groups that bind to DNA may be possibilities, such as the non-fluorescent pyrrole–imidazole polyamides.91,92
Summary and outlook
Efforts in the development of small molecule fluorescent probes in last two decades have provided a wide range of tools to study Zn2+ related processes at the subcellular level, which are already beginning to be utilised to provide answers to questions around the role of this important cation in biology. However, its role, especially its function in individual cellular processes, remains far from being fully understood, and a number of challenges and requirements endure for the further development of small molecule Zn2+ probes.Reliable and effective small molecule probes to study mobile Zn2+, as well as a number of other analytes, in some specific organelles, such as the Golgi apparatus and nucleus, are still needed. It should be noted that many factors need to be considered to achieve organelle targeting, including their pH,93 ionic strength and temperature of the medium, concentration of the probe and incubation time.75 It is also desirable to demonstrate the organelle targeting efficacy of new probes in a wide range of cell lines in case of false positive results in individual cell lines if they are to find widespread utility in biological applications.
There remains a significant demanded to develop tools to accurately quantify the endogenous Zn2+ in specific locations by developing ratiometric fluorescent probes, since most of the probes discussed herein are based on the typical PET mechanism and are not able to image Zn2+ quantitively due to either no or a small shift in emission wavelength on analyte binding. One viable route towards the development of ratiometric PET probes has been reported by Radford et al. in which a second zinc-insensitive fluorophore was appended to a zinc-sensitive PET based probe using a polyproline helix as linker to provide sufficient rigidity.94 This approach may open possibilities for other ratiometric organelle targeted probes, although this would further complicate their synthesis and it is possible that such derivatisation may affect cell uptake and targeting properties. As an alternative to PET probes, small molecule ICT probes utilising similar organelle targeting vectors may come to the fore, but again their addition to existing systems adds to synthetic complexity and may adversely affect their photochemical properties.
Probes with longer wavelength excitation and emission profiles, such as in the near-infrared range (650–900 nm), which are finding increasing application in vivo, are also required and could be utilised to study Zn2+ in deeper tissue with fewer undesirable effects from autofluorescence of surrounding tissues and less harm to biological samples. More importantly, with the rapid development of high spatiotemporal resolution fluorescent microscopic techniques, especially super-resolution microscopy, a number of new challenges are emerging. Besides the basic requirements for biological applications, probes with excellent photostability under the high photon intensity that these techniques utilise are required, whilst also displaying selective organelle targeting ability and a capability to provide real-time information about the location, dynamics and interactions of Zn2+ in living cells. In this case, small molecule markers are preferable as they have shown higher photostability compared to fluorescent proteins. The cyanine, BODIPYS, and especially the rhodamines and their derivatives have been successfully applied in super-resolution imaging,95,96 which could have potential as fluorophores for organelle specific Zn2+ imaging at the nanometre level. In this case, it may also be necessary to develop routinely performed experiments for the assessment of photostability as part of the characterisation of newly reported probes so that they can be compared more easily.
Additionally, the extension of the application of fusion tags, such as SNAP-tag97 and HaloTag,86 or bioorthogonal reactions could be considered to localise Zn2+ probes to an individual protein, to study spatiotemporal zinc fluctuations in a specific process, or its response to intra- or extracellular environmental changes, which may help better understand the mechanism of disease initiation and development due to the dysfunction of zinc homeostasis.
Thus, whilst many exciting developments in small molecule fluorescent probes have occurred in recent years and many significant advances have been made there is still much work to be done and synthetic chemists, collaborating with biologists and biomedical scientists, are likely to be at the vanguard of future developments in the area.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
LF is grateful to the Chinese Scholarship Council for the provision of a PhD studentship.Notes and references
- W. Maret, Adv. Nutr., 2013, 41, 82–91 CrossRef.
- W. Maret, Int. J. Mol. Sci., 2017, 18, 2285–2296 CrossRef.
- N. M. Hooper, N. T. Watt and I. J. Whitehouse, Int. J. Alzheimers. Dis., 2011, 971021 Search PubMed.
- L. C. Costello and R. B. Franklin, Arch. Biochem. Biophys., 2016, 611, 100–112 CrossRef CAS.
- P. Chabosseau and G. A. Rutter, Arch. Biochem. Biophys., 2016, 611, 79–85 CrossRef CAS.
- A. S. Prasad, Curr. Opin. Clin. Nutr. Metab. Care, 2009, 12, 646–652 CrossRef CAS.
- W. Maret, Metallomics, 2015, 7, 202–211 RSC.
- A. Wedd and W. Maret, Binding, Transport and Storage of Metal Ions in Biological Cells, The Royal Society of Chemistry, 2014 Search PubMed.
- Y. Chen, Y. Bai, Z. Han, W. He and Z. Guo, Chem. Soc. Rev., 2015, 44, 4517–4546 RSC.
- P. Chabosseau, J. Woodier, R. Cheung and G. A. Rutter, Metallomics, 2018, 10, 229–239 RSC.
- Z. Xu, J. Yoon and D. R. Spring, Chem. Soc. Rev., 2010, 39, 1996–2006 RSC.
- B. Daly, J. Ling and A. P. De Silva, Chem. Soc. Rev., 2015, 44, 4203–4211 RSC.
- W. Sun, M. Li, J. Fan and X. Peng, Acc. Chem. Res., 2019, 52, 2818–2831 CrossRef CAS.
- J. V. Jun, D. M. Chenoweth and E. J. Petersson, Org. Biomol. Chem., 2020, 18, 5747–5763 RSC.
- S. H. Park, N. Kwon, J. H. Lee, J. Yoon and I. Shin, Chem. Soc. Rev., 2020, 49, 143–179 RSC.
- Y. Fu and N. S. Finney, RSC Adv., 2018, 8, 29051–29061 RSC.
- H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC.
- L. Wu, C. Huang, B. P. Emery, A. C. Sedgwick, S. D. Bull, X.-P. He, H. Tian, J. Yoon, J. L. Sessler and T. D. James, Chem. Soc. Rev., 2020, 49, 5110–5139 RSC.
- P. Gao, W. Pan, N. Li and B. Tang, Chem. Sci., 2019, 10, 6035–6071 RSC.
- H. Zhu, J. Fan, J. Du and X. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS.
- W. Xu, Z. Zeng, J. H. Jiang, Y. T. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658–13699 CrossRef CAS.
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- W. J. Bettger and B. L. O'Dell, J. Nutr. Biochem., 1993, 4, 194–207 CrossRef CAS.
- B. L. O'Dell, J. Nutr., 2000, 130, 1432S–1436S CrossRef.
- S. Iyoshi, M. Taki and Y. Yamamoto, Org. Lett., 2011, 13, 4558–4561 CrossRef CAS.
- J. J. Kim, J. Hong, S. Yu and Y. You, Inorg. Chem., 2020, 59, 11562–11576 CrossRef CAS.
- D. Li, S. Chen, E. A. Bellomo, A. I. Tarasov, C. Kaut, G. A. Rutter and W. H. Li, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 21063–21068 CrossRef CAS.
- J. Pancholi, D. J. Hodson, K. Jobe, G. A. Rutter, S. M. Goldup and M. Watkinson, Chem. Sci., 2014, 5, 3528–3535 RSC.
- N. R. Johnston, R. K. Mitchell, E. Haythorne, M. P. Pessoa, F. Semplici, J. Ferrer, L. Piemonti, P. Marchetti, M. Bugliani, D. Bosco, E. Berishvili, P. Duncanson, M. Watkinson, J. Broichhagen, D. Trauner, G. A. Rutter and D. J. Hodson, Cell Metab., 2016, 24, 389–401 CrossRef CAS.
- K. Rathore, C. S. Lim, Y. Lee, H. J. Park and B. R. Cho, Asian J. Org. Chem., 2014, 3, 1070–1073 CrossRef CAS.
- H. M. McBride, M. Neuspiel and S. Wasiak, Curr. Biol., 2006, 16, R551–R560 CrossRef CAS.
- A. Atkinson, O. Khalimonchuk, P. Smith, H. Sabic, D. Eide and D. R. Winge, J. Biol. Chem., 2010, 285, 19450–19459 CrossRef CAS.
- D. Rajapakse, T. Curtis, M. Chen and H. Xu, Oxid. Med. Cell. Longevity, 2017, 6926485 Search PubMed.
- S. L. Sensi, D. Ton-That, P. G. Sullivan, E. A. Jonas, K. R. Gee, L. K. Kaczmarek and J. H. Weiss, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 6157–6162 CrossRef CAS.
- S. L. Sensi, D. Ton-That, J. H. Weiss, A. Rothe and K. R. Gee, Cell Calcium, 2003, 34, 281–284 CrossRef CAS.
- E. M. Nolan, J. W. Ryu, J. Jaworski, R. P. Feazell, M. Sheng and S. J. Lippard, J. Am. Chem. Soc., 2006, 128, 15517–15528 CrossRef CAS.
- A. Loas, R. J. Radford and S. J. Lippard, Inorg. Chem., 2014, 53, 6491–6493 CrossRef CAS.
- H. M. Kim, C. S. Lim, G. Masanta, H. J. Kim, B. R. Cho and J. H. Han, J. Am. Chem. Soc., 2011, 133, 5698–5700 CrossRef.
- N. Y. Baek, C. H. Heo, C. S. Lim, G. Masanta, B. R. Cho and H. M. Kim, Chem. Commun., 2012, 48, 4546–4548 RSC.
- K. Rathore, C. S. Lim, Y. Lee and B. R. Cho, Org. Biomol. Chem., 2014, 12, 3406–3412 RSC.
- Z. Liu, C. Zhang, Y. Chen, W. He and Z. Guo, Chem. Commun., 2012, 48, 8365–8367 RSC.
- G. Grynkiewicz, M. Poenie and R. Y. Tsien, J. Biol. Chem., 1985, 260, 3440–3450 CAS.
- L. Xue, G. Li, C. Yu and H. Jiang, Chem.–Eur. J., 2012, 18, 1050–1054 CrossRef CAS.
- K. Sreenath, J. R. Allen, M. W. Davidson and L. Zhu, Chem. Commun., 2011, 47, 11730–11732 RSC.
- W. Chyan, D. Y. Zhang, S. J. Lippard and R. J. Radford, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 143–148 CrossRef CAS.
- J. J. Hwang, S.-J. Lee, T.-Y. Kim, J.-H. Cho and J.-Y. Koh, J. Neurosci., 2008, 28, 3114–3122 CrossRef CAS.
- H. C. Roh, S. Collier, J. Guthrie, J. D. Robertson and K. Kornfeld, Cell Metab., 2012, 15, 88–99 CrossRef CAS.
- F. Qian, C. Zhang, Y. Zhang, W. He, X. Gao, P. Hu and Z. Guo, J. Am. Chem. Soc., 2009, 131, 1460–1468 CrossRef CAS.
- Q. Liu, D. Zhu, H. Jiang, L. Xue and G. Li, Inorg. Chem., 2012, 51, 10842–10849 CrossRef.
- H. J. Lee, C. W. Cho, H. Seo, S. Singha, Y. W. Jun, K. H. Lee, Y. Jung, K. T. Kim, S. Park, S. C. Bae and K. H. Ahn, Chem. Commun., 2016, 52, 124–127 RSC.
- H. Zhu, J. Fan, S. Zhang, J. Cao, K. Song, D. Ge, H. Dong, J. Wang and X. Peng, Biomater. Sci., 2014, 2, 89–97 RSC.
- R. G. Pearson, J. Am. Chem. Soc., 1963, 85, 3533–3539 CrossRef CAS.
- Z. Hu, G. Yang, J. Hu, H. Wang, P. Eriksson, R. Zhang, Z. Zhang and K. Uvdal, Sens. Actuators, B, 2018, 264, 419–425 CrossRef CAS.
- K. Sreenath, Z. Yuan, A. Allen, M. W. Davidson and L. Zhu, Chem.–Eur. J., 2015, 21, 867–874 CrossRef.
- C. Du, S. Fu, X. Wang, A. C. Sedgwick, W. Zhen, M. Li, X. Li, J. Zhou, Z. Wang, H. Wang and J. L. Sessler, Chem. Sci., 2019, 10, 5699–5704 RSC.
- D. S. Schwarz and M. D. Blower, Cell. Mol. Life Sci., 2016, 73, 79–94 CrossRef CAS.
- K. Mori, Cell, 2000, 101, 451–454 CrossRef CAS.
- C. J. Stork and Y. V. Li, J. Mol. Signaling, 2010, 5, 1–6 CrossRef.
- J. Jeong, J. M. Walker, F. Wang, J. G. Park, A. E. Palmer, C. Giunta, M. Rohrbach, B. Steinmann and D. J. Eide, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E3530–E3538 CrossRef CAS.
- T. S. Le Nguyen, K. Kohno and Y. Kimata, Biosci., Biotechnol., Biochem., 2013, 77, 1337–1339 CrossRef CAS.
- K. Zhang and R. J. Kaufman, Nature, 2008, 454, 455–462 CrossRef CAS.
- S. H. Back and R. J. Kaufman, Annu. Rev. Biochem., 2012, 81, 767–793 CrossRef CAS.
- C. Hetz and S. Saxena, Nat. Rev. Neurol., 2017, 13, 477–491 CrossRef CAS.
- W. Lin, D. Buccella and S. J. Lippard, J. Am. Chem. Soc., 2013, 135, 13512–13520 CrossRef CAS.
- X. Gan, P. Sun, H. Li, X. Tian, B. Zhang, J. Wu, Y. Tian and H. Zhou, Biosens. Bioelectron., 2016, 86, 393–397 CrossRef CAS.
- L. Fang, G. Trigiante, R. Crespo-Otero, C. S. Hawes, M. P. Philpott, C. R. Jones and M. Watkinson, Chem. Sci., 2019, 10, 10881–10887 RSC.
- L. Fang, G. Trigiante, R. Crespo-Otero, M. P. Philpott, C. R. Jones and M. Watkinson, Org. Biomol. Chem., 2019, 17, 10013–10019 RSC.
- H. Wu and J. Zhao, Metallomics, 2019, 11, 1984–1987 RSC.
- Y. V. Li, Endocrine, 2014, 45, 178–189 CrossRef CAS.
- M. Aridor and L. A. Hannan, Traffic, 2000, 1, 836–851 CrossRef CAS.
- G. K. Walkup, S. C. Burdette, S. J. Lippard and R. Y. Tsien, J. Am. Chem. Soc., 2000, 122, 5644–5645 CrossRef CAS.
- L. E. Mcquade and S. J. Lippard, Inorg. Chem., 2010, 49, 9535–9545 CrossRef CAS.
- C. Zhang, Y. Zhang, Y. Chen, Z. Xie, Z. Liu, X. Dong, W. He, C. Shen and Z. Guo, Inorg. Chem. Commun., 2011, 14, 304–307 CrossRef CAS.
- H. Singh, H. W. Lee, C. H. Heo, J. W. Byun, A. R. Sarkar and H. M. Kim, Chem. Commun., 2015, 51, 12099–12102 RSC.
- R. W. Horobin and F. Rashid-Doubell, Biotech. Histochem., 2013, 88, 461–476 CrossRef CAS.
- H. Zhu, C. Liu, C. Liang, B. Tian, H. Zhang, X. Zhang, W. Sheng, Y. Yu, S. Huang and B. Zhu, Chem. Commun., 2020, 56, 4086–4089 RSC.
- R. S. Li, P. F. Gao, H. Z. Zhang, L. L. Zheng, C. M. Li, J. Wang, Y. F. Li, F. Liu, N. Li and C. Z. Huang, Chem. Sci., 2017, 8, 6829–6835 RSC.
- W. Zhang, J. Zhang, P. Li, J. Liu, D. Su and B. Tang, Chem. Sci., 2019, 10, 879–883 RSC.
- D. Aoki, N. Lee, N. Yamaguchi, C. Dubois and M. N. Fukuda, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 4319–4323 CrossRef CAS.
- Y. Maeda, G. V. Beznoussenko, J. Van Lint, A. A. Mironov and V. Malhotra, EMBO J., 2001, 20, 5982–5990 CrossRef CAS.
- H. Wang, Z. He, Y. Yang, J. Zhang, W. Zhang, W. Zhang, P. Li and B. Tang, Chem. Sci., 2019, 10, 10876–10880 RSC.
- L. Fang, R. Crespo-Otero, C. R. Jones and M. Watkinson, 2020, submitted for publication.
- S. R. Powell, J. Nutr., 2000, 130, 1488–1492 CrossRef.
- L. M. Green and J. M. Berg, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 6403–6407 CrossRef CAS.
- J. G. Miranda, A. L. Weaver, Y. Qin, J. G. Park, C. I. Stoddard, M. Z. Lin and A. E. Palmer, PLoS One, 2012, 7, e49371–e49380 CrossRef CAS.
- M. L. Zastrow, Z. Huang and S. J. Lippard, ACS Chem. Biol., 2020, 15, 396–406 CrossRef CAS.
- L. M. Canzoniero, D. M. Turetsky and D. W. Choi, J. Neurosci., 1999, 19, RC31 CrossRef CAS.
- H. Haase and D. Beyersmann, Biochem. Biophys. Res. Commun., 2002, 296, 923–928 CrossRef CAS.
- C. Zhang, Z. Liu, Y. Li, W. He, X. Gao and Z. Guo, Chem. Commun., 2013, 49, 11430–11432 RSC.
- L. Fang, G. Trigiante, C. J. Kousseff, R. Crespo-Otero, M. P. Philpott and M. Watkinson, Chem. Commun., 2018, 54, 9619–9622 RSC.
- P. B. Dervan and B. S. Edelson, Curr. Opin. Struct. Biol., 2003, 13, 284–299 CrossRef CAS.
- F. Yang, N. G. Nickols, B. C. Li, G. K. Marinov, J. W. Said and P. B. Dervan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1863–1868 CrossRef CAS.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS.
- D. Y. Zhang, M. Azrad, W. Demark-wahnefried, C. J. Frederickson, S. J. Lippard and R. J. Radford, ACS Chem. Biol., 2015, 10, 385–389 CrossRef CAS.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS.
- N. Li, R. Zhao, Y. Sun, Z. Ye, K. He and X. Fang, Natl. Sci. Rev., 2017, 4, 739–760 CrossRef CAS.
- S. J. Lippard, E. Tomat, E. M. Nolan and J. Jaworski, J. Am. Chem. Soc., 2008, 130, 15776–15777 CrossRef.
| This journal is © The Royal Society of Chemistry 2020 |