Open Access Article
Open Access ArticleA bipedal DNA nanowalker fueled by catalytic assembly for imaging of base-excision repairing in living cells†
Meng-Mei
Lv
a,
Jin-Wen
Liu
 *b,
Ru-Qin
Yu
*b,
Ru-Qin
Yu
 a and
Jian-Hui
Jiang
a and
Jian-Hui
Jiang
 *a
*a
aState Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China. E-mail: jianhuijiang@hnu.edu.cn; Fax: +86-731-88822872; Tel: +86-731-88822577
bThe First Affiliated Hospital of Guangxi Medical University, Guangxi Collaborative Innovation Center for Biomedicine, School of Preclinical Medicine, Center for Translational Medicine, Guangxi Medical University, Nanning 530021, P.R. China. E-mail: jinwenliu@hnu.edu.cn
First published on 14th August 2020
Abstract
DNA nanowalkers moving progressively along a prescribed DNA track are useful tools in biosensing, molecular theranostics and biosynthesis. However, stochastic DNA nanowalkers that can perform in living cells have been largely unexplored. We report the development of a novel stochastic bipedal DNA walker that, for the first time, realizes direct intracellular base excision repair (BER) fluorescence activation imaging. In our design, the bipedal walker DNA was generated by BER-related human apurinic/apyrimidinic endonuclease 1 (APE1)-mediated cleavage of DNA sequences at an abasic site in the intracellular environment, and it autonomously travelled on spherical nucleic acid (SNA) surfaces via catalyzed hairpin assembly (CHA). Our nanomachine outperforms the conventional single leg-based DNA walker with an improved sensitivity, kinetics and walking steps. Moreover, in contrast to the single leg-based DNA walker, the bipedal DNA walker is capable of monitoring the fluorescence signal of reduced APE1 activity, thus indicating amplified intracellular imaging. This bipedal DNA-propelled DNA walker presents a simple and modular amplification mechanism for intracellular biomarkers of interest, providing an invaluable platform for low-abundance biomarker discovery leading to the accurate identification and effective treatment of cancers.
Introduction
The exquisite programmability of DNA has enabled construction of DNA nanowalkers, one of the sophisticated DNA machines that move progressively along confined tracks. The progressive movement of DNA nanowalkers provides the possibility of executing multiple tasks or signal amplification, affording valuable platforms in molecular transportation, biosensors and biosynthesis.1–3 Current DNA nanowalkers have been engineered to travel in landscapes of different dimensionalities with varying driving energy fueled by nuclease,4–6 DNAzymes,7–9 or toehold-mediated strand displacement.10–12 DNA walkers traversing three-dimensional (3D) tracks confer increased capacity in mimicking complicated biological systems and achieving efficient signal amplification. This advantage has spurred the interest in DNA walkers with 3D-tracks along surfaces of nano- and micro-materials, including carbon nanotubes,13 microparticles,14,15 and gold nanoparticles (GNPs or AuNPs).16–18 A prominent example is a DNA walker that uses catalytic hairpin assembly (CHA) to drive walking on DNA-coated microparticle surfaces, allowing amplified microRNA (miRNA) detection.14 Very recently, DNA nanomachines with 3D tracks have been exploited for live cell applications.19–21 A unipedal DNAzyme nanomachine, which is activated by a specific intracellular target and initiates autonomous walking on AuNPs, has been, for the first time, engineered for miRNA imaging in living cells.19 Alternatively, a unipedal entropy-driven DNA nanomachine has been rationally engineered with three-dimensional tracks on AuNPs for intracellular miRNA imaging.20 However, unipedal DNA nanowalkers are challenging because they can only swing around a specific area near its foothold, which limits their walk distance and amplification efficiency. Bipedal DNA-based walkers that can traverse stochastically on a 3D surface in living cells remain largely unexplored.Here we report for the first time a stochastic bipedal DNA walker that autonomously traverses on GNPs via the CHA reaction, enabling highly sensitive, activatable fluorescence imaging of intracellular base excision repair (BER) activity. Human apurinic/apyrimidinic endonuclease 1 (APE1) is an essential DNA repair protein involved in maintaining the genome integrity and fidelity.22 It plays a key role in the BER pathway by incising DNA adjacent to the abasic (AP) sites.23 Imaging of APE1 activity is critical for understanding the BER pathway in complicated intracellular circumstances as well as for screening the drug candidates for BER. We envision that the CHA cascade has the potential to drive a bipedal DNA walker to traverse the surface on individual nanoparticles in the live cell surroundings. Motivated by this hypothesis, we develop a bipedal DNA walker for live-cell imaging of APE1 activity. Cleavage of a DNA substrate by APE1 generates a bipedal DNA catalyst that forms a DNA walker traversing GNPs through the catalytic assembly. The DNA nanowalker progressively activates the fluorescence of hairpin probes on GNP surfaces for signal-amplified imaging of the APE1 activity. We demonstrated that the bipedal DNA walker exhibits improved sensitivity, kinetics and walking steps as compared to the unipedal DNA nanomachine. The DNA nanowalker is also shown to have high sensitivity for monitoring APE1 activity in living cells. This bipedal DNA walker can provide a simple and modular platform for developing signal amplification strategies for high-contrast imaging and discovery of low-abundance biomarkers in living cells.
Results and discussion
Principles of the BER-related DNA walker
To develop a bipedal DNA walker for imaging BER activity of APE1, we construct a bipedal substrate for APE1 and a 3D track on GNPs (Scheme 1). To obtain a bipedal catalyst DNA, we design a DNA substrate hairpin H0 with a stem region that blocks two DNA catalyst sequences flanking an AP site. The hairpin structure masks the bipedal catalyst domains, precluding their hybridization with the hairpin probe H1 on GNPs. A long single-stranded spacer sequence is used to link the bipedal catalyst domain, allowing the two catalyst “legs”, once released, to independently hybridize with two foothold H1 hairpins on the 3D track. Hairpin probe H1 is designed in virtue of the CHA reaction to have a toehold sequence complementary to the catalyst domain and a 3′-end Cy5 tag as well as 5′ terminal thiol moiety. The GNP-supported 3D-track is obtained by self-assembly of hairpin probe H1 on GNPs via the terminal thiol moiety. This design endows the GNP-supported 3D-track with quenched Cy5 fluorescence due to energy transfer between Cy5 and GNPs.24 The other hairpin probe H2 involved in the CHA reaction is supplemented in the GNP-supported 3D-track solution as the fuel. With this GNP-supported H1-assembled 3D-track, the H2 fuel and the bipedal substrate for APE1, we obtain the bipedal DNA walker for APE1 detection and imaging. In the presence of APE1, the AP site is cleaved and the stem region of the substrate H0 is exposed, releasing a bipedal catalyst. The bipedal catalyst is hybridized on two hairpin probes H1 on a single GNP, forming a bipedal DNA nanowalker with the GNP-supported H1-assembled 3D-track. The bipedal DNA nanowalker is capable of walking progressively on the 3D tracks on an individual GNP through the CHA cascade with the aid of the fuel hairpin probe H2. Stochastically, the fuel probes release one of the catalyst legs from the track at a time, while the other remains tethered on the track. The released leg is able to walk and stay on another foothold H1 probe via hybridization. Such reaction cycles can repeat with new fuel hairpin probes, driving the nanowalker to progressively travel along the track on an individual GNP. Progressive walking of the nanomachine results in a cascade of assembly of the fuel probe H2 on the surface-immobilized H1 probe, stretching the H1 probes with the Cy5 labels drawn away from GNP surface. This reaction thus activates an amplified fluorescence signal on GNPs in response to a single bipedal catalyst released by APE1, allowing highly sensitive detection and imaging of APE1 activity. We anticipate that the bipedal catalyst DNA can walk for more steps on individual GNPs with a faster kinetics than a unipedal catalyst, therefore affording improved amplification efficiency. While the unipedal catalyst is also capable of hybridizing to another H1-modified GNP after release from the once-tethered GNP, diffusion of the catalyst to another GNP is a rate-limiting step and thus reduces the amplification efficiency and kinetics. To our knowledge, this is the first time that the CHA reaction has been explored for developing a bipedal DNA nanowalker in living cells. This bipedal DNA walker can afford higher sensitivity and faster kinetics for intracellular detection and imaging. Therefore, it may provide a useful platform for low-abundance biomarker detection in cell biology and disease diagnostics.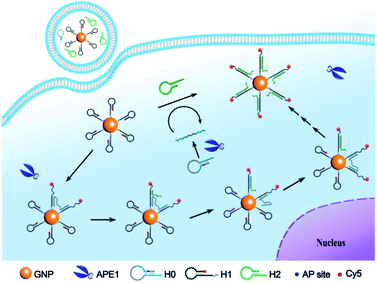 | ||
| Scheme 1 Illustration of the bipedal DNA walker along the 3D track on GNPs for APE1 detection and imaging. | ||
In vitro response of the DNA walker
For APE1 detection, we did not require a specific sequence for the bipedal catalyst DNA except for its biorthogonality. In this case, we designed the hairpin probe pairs H1 and H2 using an arbitrarily selected catalyst sequence with biorthogonality (Table S1 in the ESI†). The hairpin substrate H0 was designed using the catalyst sequence with a long single-stranded linker (Fig. S1 in the ESI†). Before construction of the nanowalker, we first investigated the fuel reaction of CHA in response to the APE1-cleavage product using gel electrophoresis analysis (Fig. 1a). We observed that incubation of tethering hairpin probe H1 and fuel hairpin probe H2 did not give a new band, suggesting low leakage of the CHA reaction. Incubation of substrate hairpin probe H0 with H1 or a mixture of H1 and H2 also did not deliver new bands, implying that the catalyst domain was effectively blocked in the hairpin structure of H0. In contrast, incubation of APE1 with H0 followed by reaction with H1 resulted in a new band with remarkable shifts from those for these two probes. This observation evidenced that cleavage of substrate H0 by APE1 facilitated its efficient hybridization with H1. Moreover, a new band was found after incubation of the reaction product between APE1 and H0 with H1 and H2, implying the catalytic assembly of tethering probe H1 and fuel probe H2. These results validated the fact that the CHA cascade was specifically catalyzed by APE1-mediated cleavage of substrate probe H0.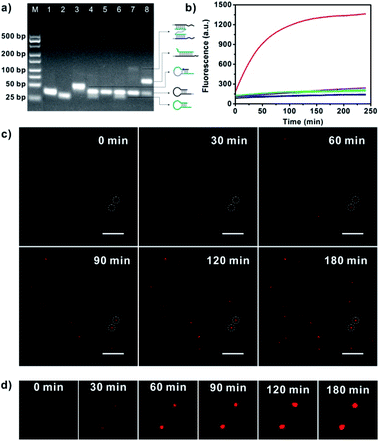 | ||
| Fig. 1 (a) Gel electrophoresis for the CHA reaction. Lane 1, H1; Lane 2, H2; Lane 3, H0; Lane 4, H1 and H2; Lane 5, H0 and H1; Lane 6, H0 plus H1 and H2; Lane 7, H0 plus H1 and 20 U mL−1 APE1; Lane 8, H0 plus H1, H2 and 20 U mL−1 APE1; M, 25 bp DNA size marker. (b) Real-time fluorescence responses of GNP-H1 (black); GNP-H1 and H2 (cyan); H0 and GNP-H1 (blue); HT plus GNP-H1, H2 and APE1 (orange); H0 plus GNP-H1 and H2 (violet); H0 plus GNP-H1 and APE1 (green); H0 plus GNP-H1, H2 and APE1 (red). (c) Real-time TIRF images for APE1 triggered DNA walkers moving on single SNA-based 3D tracks. Scale bar = 5 μm. (d) Detail time-lapse images of the circled dots in Fig. 1c. | ||
The DNA walker triggered by APE1-mediated cleavage was further studied using real-time fluorescence analysis (Fig. 1b). After characterization and evaluation of the GNP-bonded H1 number,25 the ratio of H0 to GNP-H1 and H2 was determined to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (Fig. S2 and S3 in the ESI†). In the mixture of H1-GNPs and H2, a very weak fluorescence signal was observed through incubation for 2.5 h, indicating a low fluorescence background for the CHA system and the nanowalker. Similar findings were obtained with addition of substrate probe H0, testifying that the substrate probe did not initiate the catalytic assembly in the absence of target APE1. However, addition of APE1 in the mixture of H0, H1-GNP and H2 delivered a rapidly increased fluorescence signal up to saturation after 2.5 h. Such a desirable signal-to-background ratio (∼6.5-fold) suggested that H0 was effectively cleaved by APE1 and then acted as an initiator of the DNA walker. In a control experiment, a DNA hairpin (HT) which has the same sequence as H0 except that the AP site was replaced by a thymine was incubated with GNP-H1, H2 and APE1, and an insignificant fluorescence signal was observed. Gel electrophoresis analysis also demonstrated that non-abasic HT could not initiate the APE1-activated DNA walker (Fig. S4 in the ESI†). These data further showed that APE1-induced structure transformation of H0 was essential for activation of the walking behaviour. To validate signal amplification of this approach, APE1 was incubated with H0 and GNP-H1. Theoretically, in the absence of H2, the walker DNA activated by the target could only hybridize with H1 and no further reaction occurred, which did not yield signal amplification. As estimated, obviously weaker fluorescence intensity was observed in the H2-absent experiment, implying that substantial fluorescence signal amplification was achieved in our approach. In addition, the movement behaviour of the APE1-triggered DNA walker on SNAs was monitored by total internal reflection fluorescence (TIRF) microscopy.16,26 The real-time TIRF images revealed that in the imaging area of interest, upon the addition of APE1, the fluorescent counts and fluorescence intensity increased gradually with increasing time and some of them reached a plateau in 120 min (Fig. 1c and d). These observations disclosed that the bipedal walker DNA produced by APE1 cleavage is capable of walking progressively on the individual 3D track, confirming the intermolecular movement behaviour on single given GNPs.
50 (Fig. S2 and S3 in the ESI†). In the mixture of H1-GNPs and H2, a very weak fluorescence signal was observed through incubation for 2.5 h, indicating a low fluorescence background for the CHA system and the nanowalker. Similar findings were obtained with addition of substrate probe H0, testifying that the substrate probe did not initiate the catalytic assembly in the absence of target APE1. However, addition of APE1 in the mixture of H0, H1-GNP and H2 delivered a rapidly increased fluorescence signal up to saturation after 2.5 h. Such a desirable signal-to-background ratio (∼6.5-fold) suggested that H0 was effectively cleaved by APE1 and then acted as an initiator of the DNA walker. In a control experiment, a DNA hairpin (HT) which has the same sequence as H0 except that the AP site was replaced by a thymine was incubated with GNP-H1, H2 and APE1, and an insignificant fluorescence signal was observed. Gel electrophoresis analysis also demonstrated that non-abasic HT could not initiate the APE1-activated DNA walker (Fig. S4 in the ESI†). These data further showed that APE1-induced structure transformation of H0 was essential for activation of the walking behaviour. To validate signal amplification of this approach, APE1 was incubated with H0 and GNP-H1. Theoretically, in the absence of H2, the walker DNA activated by the target could only hybridize with H1 and no further reaction occurred, which did not yield signal amplification. As estimated, obviously weaker fluorescence intensity was observed in the H2-absent experiment, implying that substantial fluorescence signal amplification was achieved in our approach. In addition, the movement behaviour of the APE1-triggered DNA walker on SNAs was monitored by total internal reflection fluorescence (TIRF) microscopy.16,26 The real-time TIRF images revealed that in the imaging area of interest, upon the addition of APE1, the fluorescent counts and fluorescence intensity increased gradually with increasing time and some of them reached a plateau in 120 min (Fig. 1c and d). These observations disclosed that the bipedal walker DNA produced by APE1 cleavage is capable of walking progressively on the individual 3D track, confirming the intermolecular movement behaviour on single given GNPs.
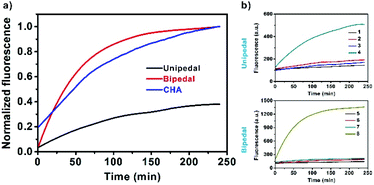
Kinetic analysis of unipedal and bipedal DNA walkers
Next, we systematically investigated the kinetic effects of DNA walkers and CHA in solutions (Fig. 2a). First, as illustrated in Scheme S1,† we employed unipedal and bipedal catalysts to drive CHA on SNAs, and compared them with standard CHA in solutions. Real-time fluorescence monitoring showed that all the catalytic reactions exhibited fluorescence increases along with the separation of fluorophores from the quenchers (BHQ3 for standard CHA; GNPs for SNAs). Moreover, we observed that the bipedal DNA walker displayed faster kinetics than that of the unipedal walker and doubly catalysed CHA in solutions. This result indicated that bipedal catalyst-driven CHA on SNAs provided an improved speed for the developed DNA walker. Such a faster kinetics was understood to be a consequence of the binding of the bipedal catalyst on SNAs, which could speed up branch migration, facilitating its hybridization with neighbouring probes immobilized on the same SNAs, and thus accelerate the CHA reaction. Further investigation of walking efficiency of unipedal and bipedal DNA walkers was performed by comparing the number of steps.The number of walking steps was estimated with the reported method from previous reports.14 The results displayed that the unipedal and bipedal catalysts took 12 and 35 steps on SNA surfaces within 120 min, respectively (Fig. 2b, the detailed illustration is shown in Scheme S2 in the ESI†), suggesting that the walking efficiency of the bipedal catalyst is higher than that of the unipedal catalyst. Presumably, this may be ascribed to the fact that the binding of the bipedal catalyst and H1 is more stable when using two legs, which makes it more likely to stay on the surface and move along the DNA tracks.
Sensitivity and specificity of the DNA walker
After confirming the APE1-triggered bipedal walking behaviour on the SNA surface, we then explored the dependence of the bipedal DNA walker on target concentration. The walker was found to produce fluorescence signals linearly correlated with the concentrations of APE1 in the range from 0.02 to 1 U mL−1 (Fig. S4 in the ESI†), with a detection limit estimated to be 0.01 U mL−1, which was sensitive enough to sense endogenous APE1 activity in living systems.27 Additionally, we found that the bipedal DNA walker would not be activated by other potential interfering substances such as Exo III, DNase I, λ-exo, T7 exo and complex cell growth media (Fig. S5 in the ESI†).This finding suggested the specificity of the developed bipedal DNA walker to target APE1, supporting the potential of this bipedal DNA walker for high specific detection of APE1 activity in living cells.Intracellular imaging of the DNA walker for APE1 activity
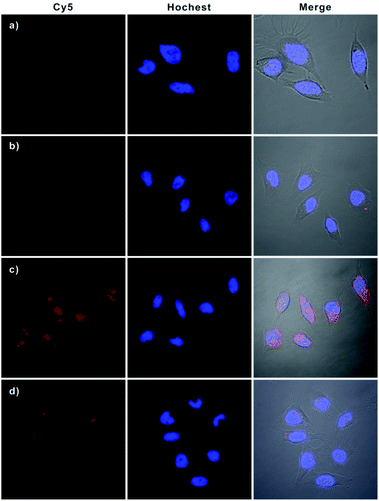
Having demonstrated the excellent APE1-driven walking efficiency of the bipedal DNA walker in vitro, we then investigated its imaging ability in living cells (Fig. 3). After transfection into HeLa cells via lipofectamine 3000, the bipedal DNA walker (H0, GNPs-H1, and H2) was incubated with cells at 37 °C for 3 h. An intense fluorescence signal from the bipedal DNA walker was observed, which was ascribed to the fact that the H0 probe delivered in cytosol reacted with APE1 in the HeLa cells, and the APE1-activated bipedal walker DNA moved along the GNPs-H1, resulting in the formation of the GNPs-H1-H2 duplex and accompanied by the enhancement of fluorescence. This result provided direct evidence for the successful realization of the bipedal DNA walker for the detection of APE1 in living cells. A control experiment with the AP-site absent hairpin probe (HT) instead of H0 revealed that no fluorescence activation was observed for incubation of HeLa cells with the probes GNPs-H1 and H2. This result manifested the specificity of the bipedal DNA walker for APE1 imaging. On the other hand, when HeLa cells were incubated with only H0 and GNPs-H1, we obtained a low-contrast red fluorescence image. In this case, APE1 can activate H0 to produce a bipedal walker DNA and then form a duplex conformation with GNPs-H1; however, the CHA cascade cannot occur without the aid of the fuel hairpin probe H2. Therefore, this result indeed indicated a much lower fluorescence signal in the absence of bipedal walker DNA-triggered amplification. Moreover, in order to compare the sensitivity enhancement performance between unipedal and bipedal DNA walkers, we then performed an additional experiment by incubating HeLa cells with GNPs-H1, H0 as well as HS, a hairpin with only a single catalytic domain. The confocal microscopy imaging results revealed that the unipedal DNA walker generated a weaker fluorescence signal than the bipedal DNA walker, but more intense than the cells only incubated with probes GNPs-H1 and H0. This finding was consistent with those of the aforementioned results that the bipedal DNA walker produced a much higher amplified signal than the unipedal DNA walker14 and also clearly demonstrated that the bipedal DNA walker was a prominent strategy for live cell imaging with more efficient signal amplification. Further inspection using flow cytometric assay also verified that the bipedal DNA walker-based strategy offered remarkable enhancement in sensitivity for fluorescence detection of living cells (Fig. S7 in the ESI†), and the relative fluorescence intensities of different groups of cells are consistent with the trend analysed from the confocal fluorescence images (Fig. S8 in the ESI†).To further verify that this bipedal DNA walker could be used to monitor low activity levels of the APE1 target, we used 7-nitroindole-2-carboxylicacid (NCA), a commonly used inhibitor of APE1,23 to reduce APE1 activity levels. Cell extract analysis revealed that APE1 activity significantly reduced ∼70% compared with that of normal HeLa cells after treatment with 2 μM NCA (Fig. S9 in the ESI†). Therefore, it was challenging to image intracellular APE1 activity at such a low level. Surprisingly, for cells pretreated with 2 μM NCA, we found that the bipedal DNA walker still produced an observable fluorescence signal in HeLa cells. However, only a negligible fluorescence signal was observed from cells transfected with the unipedal DNA walker under the same conditions (Fig. 4 and S10 in the ESI†). These results not only clearly testified that the bipedal DNA walker is capable of monitoring a low level of APE1 activity, but also further manifested that the bipedal DNA walker had higher sensitivity than the unipedal DNA walker for live cell imaging.
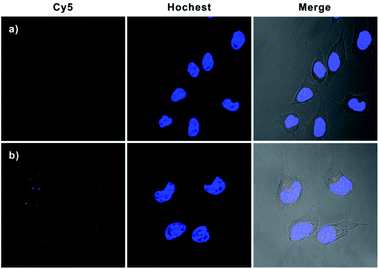 | ||
| Fig. 4 Confocal fluorescence images of HeLa cells with reduced APE1 activity. Cells were pretreated with 2 μM NCA and then incubated with Hs, GNP-H1 and H2 (a); H0, GNP-H1 and H2 (b). | ||
In addition, real-time fluorescence imaging for the bipedal DNA walker showed that the intracellular fluorescence gradually increased and reached a plateau after 3 h (Fig. S11† in the ESI), implying that the developed bipedal DNA walker could be adopted for monitoring the APE1 activation events and shedding light on the related kinetics in living cells. Subsequently, a series of Z-stack imaging through the entire HeLa cells manifested that the operation of the bipedal DNA walker was mainly in the cytoplasm (Fig. S12 in the ESI†). Furthermore, to testify the universality of our developed strategy, we further applied our bipedal DNA walker for imaging of APE1 in another cell line (MCF-7). Notably, a relative stronger fluorescence signal in MCF-7 cells was observed in comparison with HeLa cells (Fig. 5 and S13 in the ESI†), which is in good agreement with flow cytometric assay results (Fig. S14 in the ESI†) and previous reports that APE1 activity in MCF-7 was higher than in HeLa cells.28
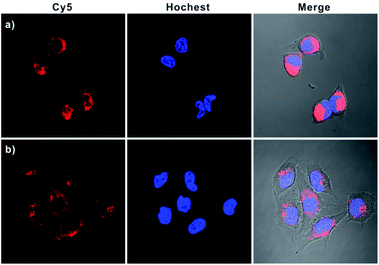 | ||
| Fig. 5 Confocal microscopy images of MCF-7 (a) and HeLa cells (b) incubated with H0, GNP-H1, and H2. | ||
Conclusions
In summary, we have developed a novel stochastic bipedal DNA walker and applied it specifically to BER-related APE1 activation imaging inside living cells. The bipedal walker DNA produced by APE1 cleavage is capable of moving progressively on the SNAs through the fuel-aided CHA cascade, resulting in the amplified fluorescence signal for highly sensitive detection and imaging of APE1 activity. We demonstrated that the bipedal DNA walker exhibits improved sensitivity, kinetics and walking steps as compared to the unipedal DNA nanomachine for APE1 activity detection. To our knowledge, this is the first time that the design of a target-generated bipedal DNA walker for live cell imaging and, at the same time, the first bipedal DNA walker for imaging intracellular APE1 activity have been produced. In virtue of these advantages, our developed strategy might broaden insights into the design of bipedal DNA walkers and expand the application field to living cells, which holds great promise for low-abundance biomarker discovery and for related biomedical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NSFC (21527810 and 21705041).Notes and references
- P. Yin, H. Yan, X. G. Daniell, A. J. Turberfield and J. H. Reif, Angew. Chem., Int. Ed., 2004, 43, 4906–4911 CrossRef CAS PubMed.
- X. Yang, Y. Tang, S. D. Mason, J. Chen and F. Li, ACS Nano, 2016, 10, 2324–2330 CrossRef CAS PubMed.
- Y. He and D. R. Liu, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
- J. Bath, S. J. Green and A. J. Turberfield, Angew. Chem., Int. Ed., 2005, 44, 4358–4361 CrossRef CAS PubMed.
- K. Yehl, A. Mugler, S. Vivek, Y. Liu, Y. Zhang, M. Fan, E. R. Weeks and K. Salaita, Nat. Nanotechnol., 2016, 11, 184–190 CrossRef CAS PubMed.
- Y. Li, G. A. Wang, S. D. Mason, X. Yang, Z. Yu, Y. Tang and F. Li, Chem. Sci., 2018, 9, 6434–6439 RSC.
- Y. Tian, Y. He, Y. Chen, P. Yin and C. Mao, Angew. Chem., Int. Ed., 2005, 117, 4429–4432 CrossRef.
- T. G. Cha, J. Pan, H. Chen, H. N. Robinson, X. Li, C. Mao and J. H. Choi, J. Am. Chem. Soc., 2015, 137, 9429–9437 CrossRef CAS PubMed.
- J. Jiang, P. Zhang, Y.-m. Nie, K.-f. Peng, Y. Zhuo, Y. Q. Chai and R. Yuan, Chem. Sci., 2020, 11, 2193–2199 RSC.
- L. Wang, R. Deng and J. Li, Chem. Sci., 2015, 6, 6777–6782 RSC.
- C. Jung, P. B. Allen and A. D. Ellington, ACS Nano, 2017, 11, 8047–8054 CrossRef CAS PubMed.
- J. Li, A. Johnson-Buck, Y. R. Yang, W. M. Shih, H. Yan and N. G. Walter, Nat. Nanotechnol., 2018, 13, 723–729 CrossRef CAS PubMed.
- T. G. Cha, J. Pan, H. Chen, J. Salgado, X. Li, C. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39–43 CrossRef CAS PubMed.
- C. Jung, P. B. Allen and A. D. Ellington, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed.
- M. Q. He, K. Wang, W. J. Wang, Y. L. Yu and J. H. Wang, Anal. Chem., 2017, 89, 9292–9298 CrossRef CAS PubMed.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- M. Oishi and K. Saito, ACS Nano, 2020, 14, 3477–3489 CrossRef CAS PubMed.
- W. Li, L. Wang and W. Jiang, Chem. Commun., 2017, 53, 5527–5530 RSC.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- C. P. Liang, P. Q. Ma, H. Liu, X. Guo, B. C. Yin and B. C. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed.
- F. Chen, J. Xue, M. Bai, J. Qin and Y. Zhao, Chem. Sci., 2019, 10, 3103–3109 RSC.
- C. Vascotto, D. Fantini, M. Romanello, L. Cesaratto, M. Deganuto, A. Leonardi, J. P. Radicella, M. R. Kelley, C. D'Ambrosio, A. Scaloni, F. Quadrifoglio and G. Tell, Mol. Cell. Biol., 2009, 29, 1834–1854 CrossRef CAS PubMed.
- G. Tell, F. Quadrifoglio, C. Tiribelli and M. R. Kelley, Antioxid. Redox Signaling, 2009, 11, 601–620 CrossRef CAS PubMed.
- C. Jing, Z. Gu, T. Xie and Y. T. Long, Chem. Sci., 2016, 7, 5347–5351 RSC.
- J. S. Lee, S. I. Stoeva and C. A. Mirkin, J. Am. Chem. Soc., 2006, 128, 8899–8903 CrossRef CAS PubMed.
- C. Y. Zhang, H. C. Yeh, M. T. Kuroki and T. H. Wang, Nat. Mater., 2005, 4, 826–831 CrossRef CAS PubMed.
- J. Zhai, Y. Liu, S. Huang, S. Fang and M. Zhao, Nucleic Acids Res., 2017, 45, e45 CrossRef PubMed.
- F. Chen, M. Bai, K. Cao, Y. Zhao, J. Wei and Y. Zhao, Adv. Funct. Mater., 2017, 27, 1702748 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc03698f |
| This journal is © The Royal Society of Chemistry 2020 |