Open Access Article
Open Access ArticleA supramolecular aggregation-based constitutional dynamic network for information processing†
Xiao
Lin
a,
Shu
Yang
b,
Dan
Huang
a,
Chen
Guo
a,
Die
Chen
b,
Qianfan
Yang
 *a and
Feng
Li
*a and
Feng
Li
 *a
*a
aCollege of Chemistry, Sichuan University, Chengdu, 610064, China. E-mail: yangqf@scu.edu.cn; fli@brocku.ca
bWest China School of Pharmacy, Sichuan University, Chengdu, 610041, China
First published on 21st August 2020
Abstract
Concepts and strategies offered by constitutional dynamic chemistry (CDC) hold great promise for designing molecular computing systems adaptive to external environments. Despite demonstrable success in storing and processing chemical information using CDC, further employment of such constitutional dynamic networks (CDNs) for processing more complex digital information has not been realized yet. Herein, we introduced a supramolecular CDN based on the aggregation of cyanine MTC (Agg-CDN), which is composed of four reversibly interconvertible constituents, i.e. monomers, dimers, J-aggregates, and H-aggregates. We demonstrated that the equilibrated Agg-CDN is reconfigurable through constituent exchange in response to well-defined chemical inputs. More importantly, the equilibrated states of the Agg-CDN are spectroscopically distinguishable because of the unique optical properties of MTC. We further tuned the Agg-CDN to at least nine unique states for transforming the chemical inputs into digital outputs, and successfully employed it for encoding and encrypting complex digital information, such as multi-pixel images.
Introduction
Molecular computing may serve as a promising alternative for information processing.1–4 Compared to silicon-based computer technologies, molecular computing may facilitate in developing smaller and more efficient information devices.5–7 Moreover, molecular computers are composed of reaction networks made of diverse chemicals or biomolecules. Therefore, they may respond directly to physical,8 chemical,9–11 or biological stimuli,12 and lead to the development of intelligent computing systems which are adaptive to external stimuli.13,14The adaptive nature of molecular computing systems echoes the emerging field of constitutional dynamic chemistry (CDC).15,16 CDC can be realized using either reconfigurable chemical bonding,17,18 a subfield known as dynamic covalent chemistry (DCC), or component-exchange among supramolecular entities.19,20 In recent years, numerous constitutional dynamic networks (CDNs) operated using either DCC or supramolecular systems have been designed and adapted for processing chemical information.21–23 For example, Lehn et al. constructed a CDN based on a dynamic covalent library of imines and acylhydrazones, which can store, recall and erase chemical information, i.e. the presence/absence of Zn2+ and Cu2+.24 In order to improve the information capacity, a CDN based on acylhydrazones was further constructed, with the ability to memorize diversified information including Zn2+, Et3N and light irradiation.25 Moreover, to expand the realizable functions, a heterogeneous imine-based CDN was designed to implement pattern generation and information transformation in response to ten metal cations.26
Despite the success in using CDNs for storing and processing chemical information, further transformation to binary digital information for subsequent logic functions witnesses very limited efforts. Thus motivated by this, we introduced a supramolecular CDN based on cyanine aggregation (Agg-CDN), which is composed of four reversibly interconvertible constituents, i.e. monomers, dimers, J-aggregates, and H-aggregates (Fig. 1A). We demonstrated that the equilibrated Agg-CDN is reconfigurable through constituent exchange in response to well-defined chemical inputs. More importantly, the equilibrated states of the Agg-CDN are spectroscopically distinguishable because of the unique optical properties of cyanines thus allowing at least nine distinguishable Agg-CDN states for transforming the chemical inputs into digital outputs. By doing so, we successfully employed the Agg-CDN for encoding and encrypting complex digital information, such as multi-pixel images.
Results and discussion
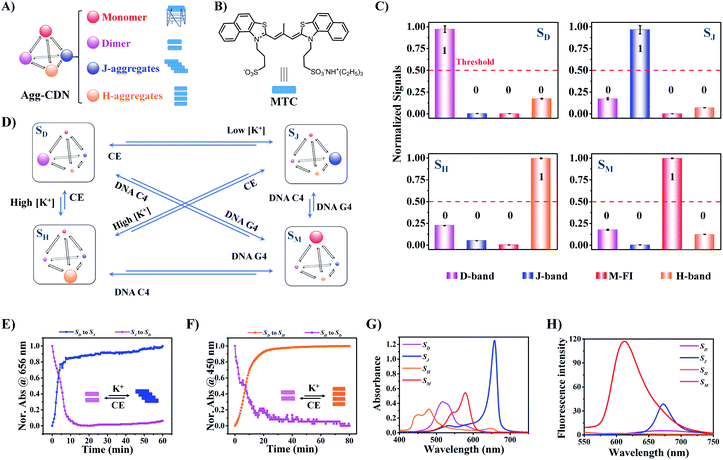
Design of the Agg-CDN
The Agg-CDN was designed using a cyanine MTC [3,3′-di(3-sulfopropyl)-4,5,4′,5′-dibenzo-9-methyl-thiacarbocyanine triethylammonium salt] (the molecular formula is shown in Fig. 1B) as the building block. MTC has the tendency to self-assemble into various supramolecular aggregates in aqueous solutions, the concentration distribution of which is responsive to external chemical inputs.27–30 Specifically, metal cations with various species and concentrations can dramatically regulate the aggregation distribution of MTC because of the metal-specific interactions and the solvent's dielectric constant perturbation due to ionic strength.31–33 As is shown in Fig. S9,† in pH 8.5 Tris–Ac buffer solution with a very low K+ concentration (less than 2 mM), MTC exists mainly as dimers, exhibiting a primary absorption peak at 515 nm (termed D-band). This state of the Agg-CDN is defined as SD. At higher K+ concentration (like 10 mM), the Agg-CDN is dominated by J-aggregates (defined as SJ), showing a sharp and strong absorption peak around 656 nm (termed J-band). Upon further increasing the concentration of K+, the dominant constituent can shift to H-aggregates, evidenced by the gradual decrease of the MTC J-band and an increase in absorbance in the range of 420 nm to 480 nm (termed H-band). The CDN dominated by H-aggregates is defined as SH. In addition to K+, the Agg-CDN is also responsive to DNA G-quadruplexes (G4s). We have previously found that parallel/hybrid G4s could disassemble MTC dimers and J- and H-aggregates to monomers, where a characteristic fluorescence enhancement at 612 nm (termed M-Fl) could be observed (Fig. S10–S12†).34 The Agg-CDN dominated by monomers was then defined as SM.Despite the fact that both J- and H- aggregates contain mixtures of aggregates with various sizes,31 we found that SJ and SH remain spectroscopically distinguishable (Fig. 1C). Therefore, we were able to quantify the concentrations of each constituent in any state through external calibration (Fig. S8 and Table S3†).
More importantly, all the states of the Agg-CDN are reversible (Fig. 1D). For example, the transition from SD to SJ was completed within 10 min by increasing the K+ concentration, whereas the reverse transition was rapidly achieved by chelating K+ using 18-crown-6 (CE) (Fig. 1E and S13†). A rapid kinetic profile was also observed for the interconversion between SD and SH (Fig. 1F). Similarly, G4s could promote the transformation of SJ to SM while complementary cytosine-rich DNAs (C4s) can suppress the formation of G-quadruplexes and thus reverse this process (Fig. S14 and S15†). To ensure that all the reactions in the Agg-CDN reach equilibrium upon measurement, a 90 min incubation was set for all the subsequent reactions throughout the study.
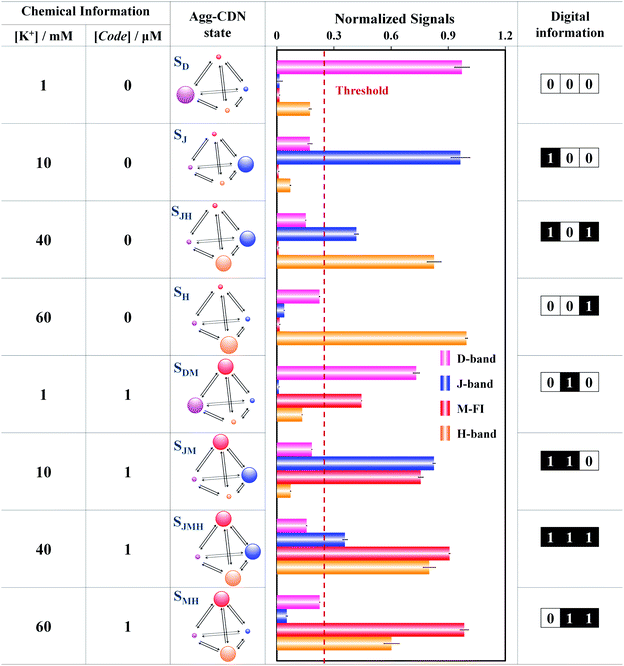
As shown in Fig. 1G and H, all four Agg-CDN states, SM, SD, SJ and SH, are spectroscopically distinguishable, because of the exciton delocalization effect of MTC supramolecular aggregation. In addition, the transition between any two Agg-CDN states may also be monitored by resolving a mixed spectrum. By doing so, we have achieved as many as nine spectroscopically distinguishable CDN states which store specific combinations of two external chemical inputs, including K+ and a G4 oligonucleotide named Code (Fig. 2). The further transformation of the chemical information stored in the Agg-CDN to binary signals was achieved via setting a threshold (red dashed lines in Fig. 1C) for the normalized MTC spectral signals. We defined the normalized signals greater than the threshold as “1”, otherwise as “0”. By assigning the binarized J-band, M-FI and H-band as the 1st, 2nd and 3rd bit values, respectively, we were able to finally transform the chemical information into three-bit digital information though the Agg-CDN (Fig. 2).
Molecular computing of the Agg-CDN
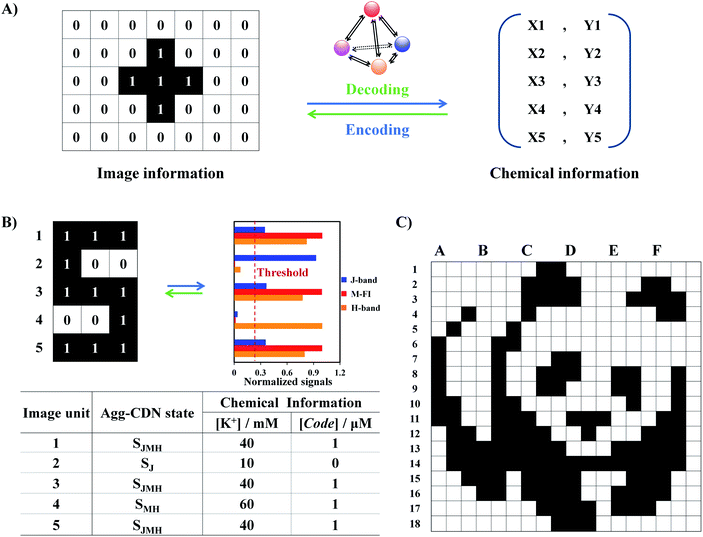
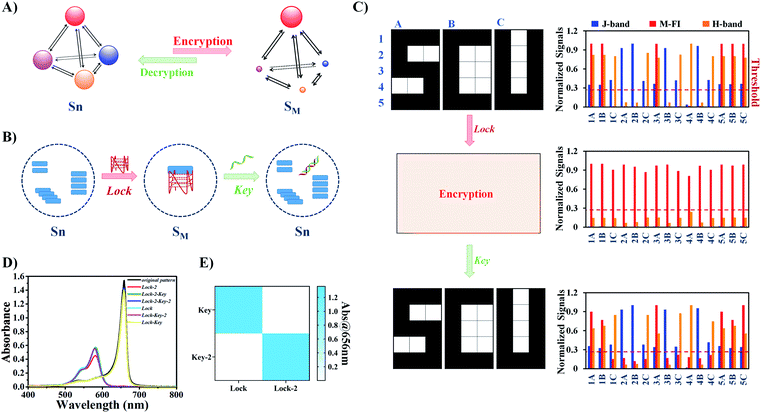
The power of the Agg-CDN for molecular computing was next demonstrated by performing image encoding and decoding operations, the two-way conversions between black-and-white pixels and chemical information (Fig. 3A). In a typical workflow (Fig. 3B), a graphic of letter “S” was first divided into units of three-pixel patterns. As defined in Fig. 2, each pattern corresponds to one specific state of the Agg-CDN, which also stores a specific combination of K+ and Code inputs. Through the Agg-CDN, chemical information can be converted to a specific image and vice versa. A similar principle can be scaled up to encode/decode patterns with a much larger footprint and complexity. For example, we demonstrated that the panda logo for the World Wide Fund for Nature (WWF) with 108 units could also be encoded by arranging varying states of the Agg-CDN in the same manner (Fig. 3C and S16–S21†).Having established the functions of encoding and decoding, we further applied the Agg-CDN to more complicated image processing operations, encryption and decryption (Fig. 4A and S22†). As shown in Fig. 4B and S23,† the encryption is achieved by reconfiguring all abovementioned eight CDN states (termed Sn) into a unified SM state by adding a 12 μM concentration of G4 strand Lock as a molecular lock. Spectroscopically, all the CDN states for image encoding are masked by the Lock strand, evidenced by a unified signal readout. Moreover, this encryption process is reversible. Upon the addition of a Key strand that is complementary to Lock, all the CDN states were restored without any loss of the original chemical information, allowing the accurate identification of the original image (Fig. 4C and S24†).
Having successfully demonstrated the encryption and decryption of one image using the pair of Lock/Key, we further demonstrated that other G4/C4 DNA pairs can be used to perform the same encrypting/decrypting operations to the same system (Fig. 4D, E and S25†). The multiple orthogonality of lock and key pairs provides a higher level of security in practice.
Conclusions
In conclusion, we have introduced a novel constitutional dynamic network (Agg-CDN) based on supramolecular aggregates. At least nine spectroscopically distinguishable states of the Agg-CDN can be finely tuned, which allows the high-density storage of specific chemical information, including varying combinations of K+ and DNA G4 strands. Because of the unique design and spectroscopic properties, the Agg-CDN enables further encoding/decoding and encrypting/decrypting operations on complex digital information, such as multi-pixel images. Our study has demonstrated the possibility and potential for devising novel molecular computing systems using the principle of CDC. The success of designing the Agg-CDN by manipulating supramolecular aggregates may also inspire the design of new CDC systems.Experimental section
Chemicals and materials
DNA oligonucleotides (listed in Table S1†) were purchased from Sangon Biotechnology Co. Ltd. (Shanghai, China). The cyanine dye MTC was synthesized in our lab according to the methods of the methods of Hamer35 and Ficken,36 and the purity was proved by high resolution mass spectrometry (HRMS), high performance liquid chromatography (HPLC), nuclear magnetic resonance (1H NMR and 13C NMR) and absorption spectroscopy. All the analytical reagent grade chemicals were purchased from Aldrich Ltd. (Shanghai, China) and used as received without further purification. Ultrapure water prepared using an ULUPURE (Chengdu, China) ultrapure water system and was used throughout the experiments.Instruments and equipment
The absorption spectra were acquired using a 10 mm quartz cuvette on an EVOLUTION 201 spectrophotometer (Thermo Scientific, USA) at room temperature in a 10 mm quartz cell, and the scan range was 400–750 nm. The fluorescence spectra were collected with an F-4600 spectrophotometer (HITACHI, Japan) in a 10 mm quartz cell at room temperature, and a xenon arc lamp was used as the excitation light source. Both the excitation and emission slits were 5 nm, and the PMT voltage was 400 V. The excitation wavelength was 530 nm and emission wavelength was 530–750 nm.Sample preparation
The Code stock solution was dissolved in 10 mM, pH 6.5 Tris–Ac buffer solution and the other DNA stock solutions were dissolved in 10 mM, pH 8.5 Tris–Ac buffer solution. The concentrations of DNA were determined by measuring absorbance at 260 nm. A 150 μM MTC stock solution was prepared by dissolving 7.67 mg MTC powder in 50 mL methanol and stored in the dark. The concentration of the MTC stock solution was corrected by diluting a 50 μL stock solution to 1000 μL with methanol, measuring its absorbance at 573 nm and calculating the actual concentration via Lambert-beer's law. The stock solution of KCl was diluted to 1 M using ultrapure water.Experimental operations
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 at 90 °C for 5 min. The Lock–Key (Lock-2 and Key-2) complex was added to the samples in which decryption operation has been implemented. After incubation at 37 °C in the dark for 90 min, absorption and fluorescence spectra were collected. The flow chart for the encryption/decryption operations is shown in Fig. S18.†
1.2 at 90 °C for 5 min. The Lock–Key (Lock-2 and Key-2) complex was added to the samples in which decryption operation has been implemented. After incubation at 37 °C in the dark for 90 min, absorption and fluorescence spectra were collected. The flow chart for the encryption/decryption operations is shown in Fig. S18.†
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The kind assistance of Prof. Peng Wu and Dr Yani Xie from the Analytical & Testing Center at Sichuan University is greatly appreciated.Notes and references
- L. M. Adleman, Science, 1994, 266, 1021–1024 CrossRef CAS PubMed.
- H. Tian, Angew. Chem., Int. Ed., 2010, 49, 4710–4712 CrossRef CAS PubMed.
- M. Amelia, L. Zou and A. Credi, Coord. Chem. Rev., 2010, 254, 2267–2280 CrossRef CAS.
- K. M. Cherry and L. Qian, Nature, 2018, 559, 370–376 CrossRef CAS PubMed.
- M. Lv, W. Zhou, D. Fan, Y. Guo, X. Zhu, J. Ren and E. Wang, Adv. Mater., 2020, 32, e1908480 CrossRef PubMed.
- D. Fan, J. Wang, E. Wang and S. Dong, Chem. Sci., 2019, 10, 7290–7298 RSC.
- R. Orbach, S. Lilienthal, M. Klein, R. D. Levine, F. Remacle and I. Willner, Chem. Sci., 2015, 6, 1288–1292 RSC.
- S. M. Chirieleison, P. B. Allen, Z. B. Simpson, A. D. Ellington and X. Chen, Nat. Chem., 2013, 5, 1000–1005 CrossRef CAS PubMed.
- D. Huang, C. Yang, Y. Yao, J. Li, C. Guo, J. Chen, Y. Zhang, S. Yang, Q. Yang and Y. Tang, Chem.–Eur. J., 2019, 25, 6996–7003 CrossRef CAS PubMed.
- D. Huang, C. Guo, J. Miao, Y. Zhang, X. Lin, D. Chen, S. Yang, Q. Yang and Y. Tang, Nanoscale, 2019, 11, 16241–16244 RSC.
- J. Chen, S. Yang, C. Yang, J. Li, D. Huang, X. Lin, C. Guo, Q. Zhou, Q. Yang and Y. Tang, Chem.–Eur. J., 2019, 25, 5691–5697 CrossRef CAS PubMed.
- Y. Benenson, Nat. Rev. Genet., 2012, 13, 455–468 CrossRef CAS PubMed.
- P. Zhang, D. Gao, K. An, Q. Shen, C. Wang, Y. Zhang, X. Pan, X. Chen, Y. Lyv, C. Cui, T. Liang, X. Duan, J. Liu, T. Yang, X. Hu, J. J. Zhu, F. Xu and W. Tan, Nat. Chem., 2020, 12, 381–390 CrossRef CAS PubMed.
- C. Liu, Z. Huang, W. Jiang, X. Liu, J. Li, X. Han, H. Tu, L. Qiu and W. Tan, Anal. Chem., 2020, 92, 3620–3626 CrossRef CAS PubMed.
- P. T. Corbett, J. Leclaire, L. Vial, K. R. West, J.-L. Wietor, J. K. M. Sanders and S. Otto, Chem. Rev., 2006, 106, 3652–3711 CrossRef CAS PubMed.
- J. M. Lehn, Chem. Soc. Rev., 2007, 36, 151–160 RSC.
- S. P. Black, J. K. Sanders and A. R. Stefankiewicz, Chem. Soc. Rev., 2014, 43, 1861–1872 RSC.
- Y. Jin, C. Yu, R. J. Denman and W. Zhang, Chem. Soc. Rev., 2013, 42, 6634–6654 RSC.
- S. Wang, L. Yue and I. Willner, Nano Lett., 2020, 20, 5451–5457 CrossRef CAS PubMed.
- L. Yue, S. Wang, V. Wulf and I. Willner, Nat. Commun., 2019, 10, 4774 CrossRef PubMed.
- P. Zhang, L. Yue, M. Vazquez-Gonzalez, Z. Zhou, W. H. Chen, Y. S. Sohn, R. Nechushtai and I. Willner, ACS Nano, 2020, 14, 1482–1491 CrossRef CAS PubMed.
- L. Yue, S. Wang and I. Willner, Sci. Adv., 2019, 5, eaav5564 CrossRef CAS PubMed.
- Z. Zhou, X. Liu, L. Yue and I. Willner, ACS Nano, 2018, 12, 10725–10735 CrossRef CAS PubMed.
- J. Holub, G. Vantomme and J. M. Lehn, J. Am. Chem. Soc., 2016, 138, 11783–11791 CrossRef CAS PubMed.
- G. Men and J. M. Lehn, Chem. Sci., 2019, 10, 90–98 RSC.
- A. Osypenko, S. Dhers and J. M. Lehn, J. Am. Chem. Soc., 2019, 141, 12724–12737 CrossRef CAS PubMed.
- C. Yang, S. Yang, L. Song, Y. Yao, X. Lin, K. Cai, Q. Yang and Y. Tang, Chem. Commun., 2019, 55, 8005–8008 RSC.
- C. Yang, L. Song, J. Chen, D. Huang, J. Deng, Y. Du, D. Yang, S. Yang, Q. Yang and Y. Tang, NPG Asia Mater., 2018, 10, 497–508 CrossRef CAS.
- C. Yang, D. Zou, J. Chen, L. Zhang, J. Miao, D. Huang, Y. Du, S. Yang, Q. Yang and Y. Tang, Chem.–Eur. J., 2018, 24, 4019–4025 CrossRef CAS PubMed.
- D. Chen, S. Yang, Z. Wu and X. Xu, Chem.–Eur. J., 2020 DOI:10.1002/chem.202001240.
- H. X. Sun, J. F. Xiang, Q. F. Yang, Q. A. Shang, Q. J. Zhou, Y. X. Zhang, G. Z. Xu and Y. L. Tang, Appl. Phys. Lett., 2011, 98, 031103 CrossRef.
- L. Wang, J. Xiang, H. Sun, Q. Yang, L. Yu, Q. Li, A. Guan and Y. Tang, Dyes Pigm., 2015, 122, 382–388 CrossRef CAS.
- H. Sun, J. Xiang, W. Gai, Y. Liu, A. Guan, Q. Yang, Q. Li, Q. Shang, H. Su, Y. Tang and G. Xu, Chem. Commun., 2013, 49, 4510–4512 RSC.
- Q. Yang, J. Xiang, S. Yang, Q. Li, Q. Zhou, A. Guan, L. Li, Y. Zhang, X. Zhang, H. Zhang, Y. Tang and G. Xu, Anal. Chem., 2010, 82, 9135–9137 CrossRef CAS PubMed.
- F. M. Hamer, The cyanine dyes and related compounds, Interscience Publishers, New York, 1964 Search PubMed.
- G. E. Ficken, The Chemistry of Synthetic Dyes, Academic Press, New York, 1971 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: All the DNA sequences used in this study; synthesis and characterization of MTC; detailed spectrogram in the experiment. See DOI: 10.1039/d0sc03392h |
| This journal is © The Royal Society of Chemistry 2020 |