Open Access Article
Open Access ArticleIntegrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA†
Shuang
Peng
,
Zhen
Tan
,
Siyu
Chen
,
Chunyang
Lei
 and
Zhou
Nie
and
Zhou
Nie
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology, Hunan University, Changsha 410082, P. R. China. E-mail: niezhou.hnu@gmail.com
First published on 19th June 2020
Abstract
CRISPR-based diagnostics (CRISPR-Dx) has shown great promise in molecular diagnostics, but its utility in the sensing of microRNA (miRNA) biomarkers is limited by sensitivity, cost and robustness. Here, we describe a CRISPR-Dx method for the sensitive and cost-effective detection of miRNAs by rationally integrating CRISPR-Cas12a with DNA circuits. In this work, a modular catalytic hairpin assembly (CHA) circuit is designed to convert and amplify each target into multiple programmable DNA duplexes, which serve as triggers to initiate the trans-cleavage activity of CRISPR-Cas12a for further signal amplification. Such rational integration provides a generic assay for the effectively amplified detection of miRNA biomarkers. By simply tuning the variable regions in the CHA modules, this assay achieves sub-femtomolar sensitivity for different miRNA biomarkers, which improves the detection limit of CRISPR-Dx in the analysis of miRNA by 3–4 orders of magnitude. With the usage of the proposed assay, the sensitive assessment of miR-21 levels in different cancer cell lines and clinical serum samples has been achieved, providing a generic method for the sensitive detection of miRNA biomarkers in molecular diagnosis.
Introduction
A generic, accurate and sensitive nucleic acid detection method can aid point-of-care pathogen detection, genotyping, and disease monitoring, exhibiting great value in clinical diagnostics.1–3 Recently, the CRISPR-Cas system has attracted much attention in the field of nucleic acid diagnostics since the discovery of the collateral cleavage activity (indiscriminate hydrolysis of ssDNA or RNA) of class II type V and VI Cas proteins,4–6 including Cas13a, Cas12a, Cas12b and Cas14.7–9 Such a collateral cleavage activity can be readily deployed to cleave synthetic fluorescent nucleic acid reporters with thousands of turnovers per second, which endow these Cas proteins with efficient self-signal amplification and reporting capacities. To date, CRISPR-based diagnostics (CRISPR-Dx) methods, such as SHERLOCK (Cas13a),10,11 DETECTR (Cas12a),12 CDetection (Cas12b)13 and Cas14-DETECTR,14 have been exploited for the isothermal detection of genomic DNA and RNA with excellent sensitivity and specificity, and are partly revolutionizing the field of molecular diagnostics.15 As a class of small non-coding RNAs composed of about 19–23 nucleotides, microRNAs (miRNAs) play a pivotal role in numerous biological processes by regulating post-transcriptional gene expression,16,17 and are becoming biomarkers for many diseases in molecular diagnostics. However, CRISPR-Dx methods for miRNAs are yet under development, and only a few Cas13a-based assays have been mainly developed with picomolar sensitivity.18–20 Aside from the unsatisfactory sensitivity, the usage of expensive RNA reporters with unstable nature results in inherent drawbacks in cost and stability. Therefore, a versatile and robust CRISPR-Dx method that allows sensitive analysis of miRNAs is still highly desired in biomedical research and clinical diagnosis.On account of the programmability of Watson–Crick base pairing, nucleic acids, especially DNA, have been deployed as circuits capable of executing algorithms.21,22 DNA circuits serve as programmable intermediates between inputs and outputs, which can perform various tasks, such as logical operation, feedback and signal-transducing amplification isothermally without the participation of protein enzymes.23–25 In a typical DNA circuit, such as the hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA), the ssDNA inputs initiate toehold-mediated strand displacement, resulting in programmable duplex DNA outputs.26–29 Because the functions of the nucleic acid circuits depend solely on the Watson–Crick complementarity, these DNA circuits are also able to work comparably with RNA inputs, including mRNA, long non-coding RNA and miRNA, as with ssDNA inputs.30–36 Moreover, the combination of DNA circuits with nucleases has been demonstrated to be effective in the development of highly sensitive assays for miRNA.37–40 Encouraged by current progress, we are interested in expanding the application scope of DNA-targeting Cas proteins (e.g., Cas12a) to miRNA sensing using a DNA circuit as the converter. Given the modularity and scalability of DNA circuits, the coupling of CRISPR-Cas and DNA circuits might provide a new paradigm for the development of CRISPR-Dx.
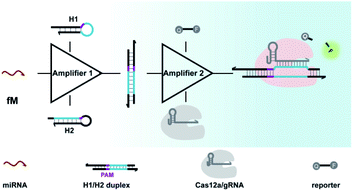
Aimed at this goal, we here integrated CRISPR-Cas with a DNA circuit to develop a generic CRISPR-Dx method for the sensitive analysis of miRNA biomarkers. In this work, the CRISPR-Cas12a system was chosen because the collateral cleavage of ssDNA reporters by activated Cas12a protein is highly efficient, with approximately 1250 turnovers per second.12,41 As shown in Scheme 1, with the introduction of CHA, miRNA targets are transduced into amplified outputs of programmable DNA duplexes, in which the protospacer adjacent motif (PAM) and protospacer sequence can be recognized by Cas12a/gRNA complexes. The collateral cleavage activity toward fluorescent ssDNA reporters is triggered along with the binding and cleavage of the DNA duplexes by the Cas12a/gRNA complexes, thereby leading to the generation of appreciable fluorescence signals. As a result, a CRISPR-Cas12a and CHA-based assay (CRISPR-CHA) is established for the effectively amplified detection of miRNA biomarkers. In CRISPR-CHA, self-signal amplification of the CRISPR-Cas12a system and CHA-mediated signal conversion and amplification are rationally coupled, which not only expands the application scope of CRISPR-Cas12a to miRNA sensing, but also greatly enhances the signal gain compared with the sole CHA.
Moreover, the proposed CRISPR-CHA possesses two important engineering novelties: (1) by introducing unpaired sites into the DNA duplex outputs, the seed sequence-dependent DNA targeting mechanism of CRISPR-12a is exploited to enhance the collateral cleavage-mediated signal amplification. (2) The modular design of CHA components allows the use of the same gRNA to probe the DNA duplex outputs from different miRNA targets, which provides a universal strategy for miRNA probing. The versatility of CRISPR-CHA has been demonstrated in the isothermal detection of miRNA biomarkers, including miR-21, miR-141 and miR-155 with sub-femtomolar sensitivity, which is significantly better than that of Cas13a-based methods.18,19 As the gold standard for miRNA assay, the quantitative polymerase chain reaction requires multiple enzymes, complicated primer design and thermal cycles.42 In contrast, CRISPR-CHA achieves the isothermal detection of miRNA with only one enzyme. Therefore, the proposed CRISPR-CHA enables versatile and sensitive sensing of miRNA biomarkers, providing a helpful tool for clinical diagnosis.
Results and discussion
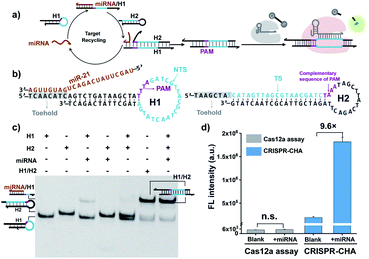
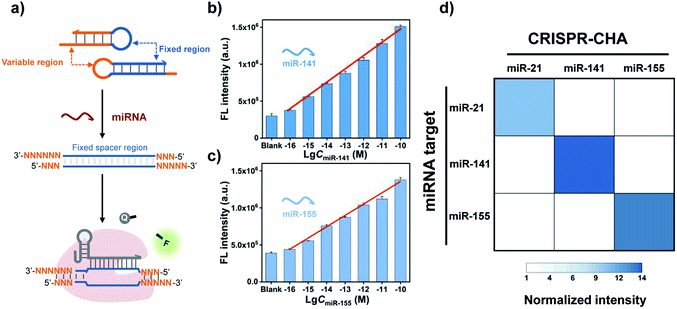
Cas12a from a Lachnospiraceae bacterium (LbCas12a) is the most widely used Cas protein in CRISPR-Dx, and thus LbCas12a fused with a maltose-binding protein (MBP) tag was expressed and purified in this work according to a previously reported procedure (Fig. S1 and S2†).12,43 The collateral cleavage activity of LbCas12a was assessed using a FQ reporter (FAM-TTATT-BHQ1). After the incubation of the LbCas12a protein and its guide RNA (gRNA) with DNA triggers, both ssDNA and dsDNA triggers induced significant fluorescence signals, suggesting the efficient digestion of FQ reporters by the activated LbCas12a proteins. Alternatively, the collateral cleavage activity initiated by the dsDNA trigger is much higher than that by the ssDNA trigger (Fig. S3†), which is consistent with the findings of Doudna's group.12 The dsDNA trigger at a low concentration of 100 pM could induce detectable fluorescence signals (Fig. S4†), indicating the good sensitivity of LbCas12a toward dsDNA targets.In CRISPR-CHA, the efficient coupling of CRISPR-Cas12a and a DNA circuit primarily depends on the recognition of the DNA duplex outputs in CHA by LbCas12a proteins (Fig. 1a). Therefore, the DNA duplex outputs in CHA should include the following components, a protospacer adjacent motif (PAM) and a protospacer sequence targeted by the spacer of LbCas12a gRNA. Based on the previously reported design principle of CHA, we here devised a CHA circuit composed of two DNA hairpins (H1 and H2) for coupling with CRISPR-Cas12a (Fig. 1b). Both H1 and H2 consist of a single stem-loop DNA with an 8-nt 5′-overhang. In H1, the 5′-overhang serves as the toehold for the target miRNA, and the loop region contains the 5′-TTTA-3′ PAM of LbCas12a as well as the non-target strand (NTS). Meanwhile, the 5′-overhang of H2 acts as the toehold for the 3′-overhang of the miRNA/H1 duplex, and the target strand (TS) is blocked in the stem region followed by the complementary sequence of PAM in the loop region. In this scenario, only the H1/H2 duplex can activate the LbCas12a/gRNA complex, because neither of the hairpins can be recognized by the LbCas12a/gRNA complex.
To test the feasibility of CRISPR-CHA assay, we selected miR-21 as the model target. Many studies have revealed that miR-21 plays a pivotal role in numerous biological processes by regulating the post-transcription of gene expression.44–46 The aberrant level of miR-21 is considered to be associated with the etiology, progression, and prognosis of many tumour types, including brain, liver, prostate, and lung cancers.47–50 In order to verify whether miR-21 can be converted into DNA duplex outputs by the proposed CHA circuit, miR-21 was incubated with H1 and H2, and the mixture was analysed by gel shift assay. As shown in Fig. 1c, an intense band of the H1/H2 duplex in the lane of the mixture (lane 7) appeared, suggesting the effective initiation of the CHA circuit by miR-21. Moreover, a negligible band of the H1/H2 duplex was observed in the absence of miR-21 (line 5), which indicated low background leakage in the CHA circuit. The activation of LbCas12a/gRNA by the generated H1/H2 duplex was confirmed using fluorophore-labelled H1 (Fig. S6†). Furthermore, the activated LbCas12a proteins exhibited strong collateral cleavage toward ssDNA reporters (Fig. S7†). Then, the fluorescence response to miR-21 in CRISPR-CHA was examined (Fig. S8†), and miR-21 was able to induce a dramatic fluorescence signal, which is 9.6-fold over that of the control without miR-21 (Fig. 1d). In contrast, LbCas12a alone is incapable of sensing miR-21. Collectively, by integrating CRISPR-Cas12a with the CHA circuit, a CRISPR-CHA for miRNA assay was developed.
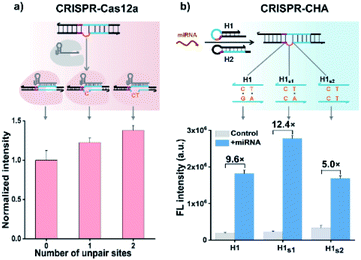
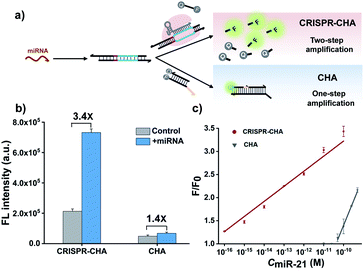
Previous structural studies suggested that the DNA targeting of the Cas12a/gRNA complex depends on the presence of a seed sequence in the gRNA.51–53 As shown in Fig. S9,† after the PAM recognition by the Cas12a/gRNA complex, local strand unwinding in the vicinity of the PAM allows the TS to base pair to the seed sequence in gRNA, thereby leading to the formation of an R-loop as well as the allosteric activation of specific and collateral cleavage activities. Given this seed sequence-dependent mechanism of DNA targeting, we envisioned to facilitate seed binding by introducing unpaired sites into the TS/NTS duplex adjacent to the PAM, which might result in the enhanced activation of Cas12a proteins. To validate this hypothesis, we tested the collateral cleavage activities of the Cas12a/gRNA complex triggered by the target DNA duplex with one or two unpaired sites adjacent to the PAM, respectively (Fig. 2a). As displayed in Fig. 2a, the fluorescence signals of the target DNA containing unpaired sites were slightly higher than that of control DNA without unpaired sites. Moreover, the enhancement in fluorescence signals showed a positive correlation with the number of unpaired sites. Based on these findings, we sought to increase the sensitivity of CRISPR-CHA by introducing unpaired sites into the H1/H2 duplex. Specifically, H1 containing one or two substitutes (H1s1 and H1s2) in the loop region was designed, and then the fluorescence signals of miR-21 in CRISPR-CHA with different H1 were measured. The ratios of signal to background (S/B) in the groups H1, H1s1 and H1s2 were 9.6, 12.4 and 5.0, respectively (Fig. 2b). The H1s1-based CRISPR-CHA had the highest S/B value, which might be attributed to the compromise between the enhancement of collateral cleavage activity and the decline in CHA efficiency caused by the unpaired sites. In CRISPR-CHA, the CHA-mediated signal conversion and self-signal amplification of CRISPR-Cas12a are rationally coupled (Fig. 3a). Each miRNA target is converted into hundreds of H1/H2 duplex outputs by the CHA circuit. Meanwhile, each H1/H2 duplex output could initiate the efficient cleavage of FQ reporters, because of the rapid turnover of LbCas12a (1250 turnover per s).12 As a result, a two-stage signal amplification for the sensitive detection of miRNA biomarkers is achieved. In contrast, each H1/H2 duplex could only restore the emission of a fluorophore in the conventional CHA assay (Fig. S10†). Therefore, the proposed CRISPR-CHA has a much better detection sensitivity compared to classical CHA assay in theory. Then, the fluorescence signals of CRISPR-CHA and CHA in response to different concentrations of miR-21 were investigated. Here, F/F0 was defined as the fluorescence response signal, where F and F0 are the fluorescence intensities of each assay with and without the presence of miR-21, respectively. The fluorescence response signals of 100 pM miR-21 from CRISPR-CHA and CHA assay were determined to be 3.4 and 1.4, respectively (Fig. 3b), suggesting an efficient signal gain in CRISPR-CHA. For CRISPR-CHA, a linear relationship between the fluorescence response signal and the logarithmic value of miR-21 concentration was obtained in the concentration range from 0.1 fM to 100 pM (R2 = 0.99) with a limit of detection (LOD) of 0.07 fM (3S/N, Fig. 3c). In contrast, the linear range of the CHA assay was 0.05–0.5 nM with a LOD of 0.04 nM (Fig. 3c). Therefore, CRISPR-CHA was able to decrease the LOD by approximately 6 orders of magnitude compared with the classical CHA assay. Moreover, the LOD of CRISPR-CHA is 3–4 orders of magnitude lower than that of Cas13a-based miRNA assays,18,19 providing an ultrasensitive method for the analysis of miRNA biomarkers.
A major advantage of CRISPR-CHA, aside from the good sensitivity, is its versatility originated from the high predictability and programmability of the CHA circuit. By tuning the variable regions in H1 and H2, CRISPR-CHAs for other miRNA targets are facile to construct using the same gRNA as in the CRISPR-CHA for miR-21 (Fig. 4a). In contrast, the gRNA has to be adjusted according to different miRNA targets in the Cas13a-based assays. Additionally, the use of a fluorescence RNA reporter with unstable nature requires extra precautions to avoid false-positive results. Therefore, CRISPR-CHA exhibits merits in cost and robustness over the Cas13a-based assays. To evaluate the versatility of CRISPR-CHA, miRNA biomarkers including miR-141 and miR-155 were chosen as the targets. miR-141 and miR-155 are the biomarkers of many cancers, so the development of a novel invasive diagnostic tool for detecting these biomarkers is important for risk assessment of cancers.54–56 CHA circuits were designed for the amplified conversion of the two miRNA targets into the outputs of H1/H2 duplexes, based on which CRISPR-CHAmiR-141 and CRISPR-CHAmiR-155 were fabricated for the analysis of miR-141 and miR-155, respectively (Fig. S11 and S12†). Here, the DNA mimics of miR-141 and miR-155 were used based on the theoretical computation using Oligo Analyzer 3.1 (Fig. S13†). Then, the fluorescence response signals of the mimics of miRNA targets with different concentrations were tested (Fig. S14–S16†). As shown in Fig. 4b and c, there was a linear correlation between the fluorescence signal and the logarithmic value of miR-141 concentration ranging from 0.1 fM to 100 pM (R2 = 0.99), and the LOD of CRISPR-CHAmiR-141 was calculated to be 0.14 fM. A similar linear relationship was observed in CRISPR-CHAmiR-155 with a LOD of 0.15 fM. To investigate the specificity, we assessed the responses of a panel of CRISPR-CHAs toward different miRNA targets, and the result is shown as a heatmap in Fig. 4d. Each CRISPR-CHA selectively responded to its target miRNA with a low cross-response (Fig. 4d), indicating the high specificity of CRISPR-CHA. Since the target recognition in CRISPR-CHA is based on the Watson–Crick base pair of CHA, its ability to discriminate a single-nucleotide mutation might be worse than that of the CRISPR-Dx methods utilizing CRISPR-Cas for target recognition. Taken together, these results have demonstrated the potential of CRISPR-CHA as a generic sensing platform for sensitive and specific detection of miRNA.
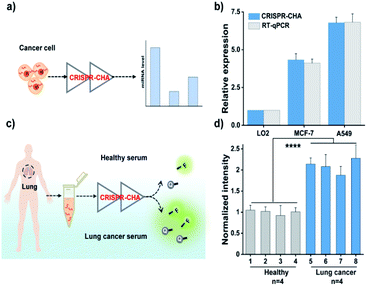
Finally, given the excellent sensitivity and specificity of the proposed CRISPR-CHA, its practical detection performance for the miRNA biomarker in clinical samples was assessed. To this end, the utility of CRISPR-CHA in the analysis of miR-21 levels in different cell lines was first verified. Total RNA extracted from human breast cancer cells (MCF-7), human non-small cell lung cancer cells (A549) and human hepatocyte cells (LO2) were analysed by CRISPR-CHA (Fig. 5a), and the result revealed different expression levels of miR-21 in these cell lines (Fig. 5b, Table S4 and Fig. S17†), which is consistent with the result quantified by RT-qPCR using a commercially available kit (Fig. 5b). Next, the performance of CRISPR-CHA in the detection of miR-21 in serum samples was further investigated. Serum from a healthy donor spiked with synthetic miR-21 at different concentrations was used as the standard sample. It was found that the serum only has a slight matrix effect on CRISPR-CHA (Fig. S18†). A good linear relationship between the fluorescence signal and logarithmic value of miR-21 concentration was obtained in the range from 1.0 fM to 100 pM, and as low as 1.0 fM miR-21 could be distinguished from the blank sample. Then, the level of miR-21 in serum samples from 4 healthy donors and 4 lung cancer patients was tested by CRISPR-CHA (Fig. 5c). As shown in Fig. 5d, serum samples from lung cancer patients induced much higher fluorescence signals compared to those from healthy donors, suggesting the high expression levels of miR-21 in the lung cancer patients' serum, which agrees with previously reported results.57,58 Moreover, the pre-addition of the miR-21 inhibitor (ssDNA fully complementary to miR-21) in the lung cancer patient's serum samples markedly decreased the fluorescence signals (Fig S19†), demonstrating that the responses indeed originated from miR-21 in the serum samples. These results indicated the promise of CRISPR-CHA in the detection of low-abundance miRNA biomarkers in clinical samples.
Conclusions
In summary, we presented a CRISPR-CHA method for the sensitive detection of miRNA by integrating CRISPR-Cas12a with a CHA circuit. In CRISPR-CHA, the self-signal amplification of CRISPR-Cas12a and CHA-mediated signal conversion and amplification are rationally coupled, resulting in a two-stage amplified detection of miRNA with sub-femtomolar sensitivity. Given the modularity and scalability of the CHA circuit, the generality of CRISPR-CHA toward different miRNA targets is facile to implement without involving any alterations to the CRISPR-Cas components, exhibiting unparalleled merits in cost, robustness and sensitivity over Cas13a-based assays. The versatility of CRISPR-CHA has been demonstrated in the assessment of miR-21 levels in different cancer cells and clinical serum samples. Therefore, this work expands the application scope of CRISPR-Cas12a to miRNA sensing and provides a valuable tool for biochemical research and clinical diagnosis.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21725503 and 21974038), and the Fundamental Research Funds for the Central Universities.Notes and references
- Y. V. Gerasimova and D. M. Kolpashchikov, Chem. Soc. Rev., 2014, 43, 6405–6438 RSC.
- M. Urdea, L. A. Penny, S. S. Olmsted, M. Y. Giovanni, P. Kaspar, A. Shepherd, P. Wilson, C. A. Dahl, S. Buchsbaum, G. Moeller and D. C. H. Burgess, Nature, 2006, 444, 73–79 CrossRef PubMed.
- L. Wu and X. Qu, Chem. Soc. Rev., 2015, 44, 2963–2997 RSC.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- L. A. Marraffini and E. J. Sontheimer, Science, 2008, 322, 1843–1845 CrossRef CAS PubMed.
- S. Shmakov, O. O. Abudayyeh, K. S. Makarova, Y. I. Wolf, J. S. Gootenberg, E. Semenova, L. Minakhin, J. Joung, S. Konermann, K. Severinov, F. Zhang and E. V. Koonin, Mol. Cell, 2015, 60, 385–397 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, P. Essletzbichler, S. Han, J. Joung, J. J. Belanto, V. Verdine, D. B. Cox, M. J. Kellner, A. Regev, E. S. Lander, D. F. Voytas, A. Y. Ting and F. Zhang, Nature, 2017, 550, 280–284 CrossRef PubMed.
- B. Zetsche, J. S. Gootenberg, O. O. Abudayyeh, I. M. Slaymaker, K. S. Makarova, P. Essletzbichler, S. E. Volz, J. Joung, J. van Der Oost, A. Regev, E. V. Koonin and F. Zhang, Cell, 2015, 163, 759–771 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, S. Konermann, J. Joung, I. M. Slaymaker, D. B. T. Cox, S. Shmakov, K. S. Makarova, E. Semenova, L. Minakhin, K. Severinov, A. Regev, E. S. Lander, E. V. Koonin and F. Zhang, Science, 2016, 353, aaf5573 CrossRef PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- F. Teng, L. Guo, T. Cui, X.-G. Wang, K. Xu, Q. Gao, Q. Zhou and W. Li, Genome Biol., 2019, 20, 132 CrossRef.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839–842 CrossRef CAS.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, J. Am. Chem. Soc., 2020, 142, 207–213 CrossRef CAS PubMed.
- F. J. Slack and A. M. Chinnaiyan, Cell, 2019, 179, 1033–1055 CrossRef CAS PubMed.
- A. Qu, M. Sun, L. Xu, C. Hao, X. Wu, C. Xu, N. A. Kotov and H. Kuang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 3391–3400 CrossRef CAS PubMed.
- Y. Shan, X. Zhou, R. Huang and D. Xing, Anal. Chem., 2019, 91, 5278–5285 CrossRef CAS PubMed.
- Y. Dai, R. A. Somoza, L. Wang, J. F. Welter, Y. Li, A. I Caplan and C. C. Liu, Angew. Chem., Int. Ed., 2019, 58, 17399–17405 CrossRef CAS PubMed.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, Adv. Mater., 2019, 31, 1905311 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–332 CrossRef CAS.
- B. Li, A. D. Ellington and X. Chen, Nucleic Acids Res., 2011, 39, e110 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- Y. S. Jiang, S. Bhadra, B. Li and A. D. Ellington, Angew. Chem., Int. Ed., 2014, 53, 1845–1848 CrossRef CAS PubMed.
- C. Jung and A. D. Ellington, Acc. Chem. Res., 2014, 47, 1825–1835 CrossRef CAS PubMed.
- S. Yue, X. Song, W. Song and S. Bi, Chem. Sci., 2019, 10, 1651–1658 RSC.
- C. Wu, S. Cansiz, L. Zhang, I.-T. Teng, L. Qiu, J. Li, Y. Liu, C. Zhou, R. Hu, T. Zhang, C. Cui and W. Tan, J. Am. Chem. Soc., 2015, 137, 4900–4903 CrossRef CAS PubMed.
- S. Wang, W. Song, S. Wei, S. Zeng, S. Yang, C. Lei, Y. Huang, Z. Nie and S. Yao, Anal. Chem., 2019, 91, 8622–8629 CrossRef CAS PubMed.
- D. Y. Zhang and E. Winfree, Nucleic Acids Res., 2010, 38, 4182–4197 CrossRef CAS PubMed.
- M.-H. Zhu, X.-M. Mu, H.-M. Deng, X. Zhong, R. Yuan and Y.-L. Yuan, Chem. Commun., 2019, 55, 9622–9625 RSC.
- Q. Wei, J. Huang, J. Li, J. Wang, X. Yang, J. Liu and K. Wan, Chem. Sci., 2018, 9, 7802–7808 RSC.
- A. R. Chandrasekaran, J. A. Punnoose, L. Zhou, P. Dey, B. K. Dey and K. Halvorsen, Nucleic Acids Res., 2019, 47, 10489–10505 CrossRef CAS PubMed.
- R. Duan, X. Zuo, S. Wang, X. Quan, D. Chen, Z. Chen, L. Jiang, C. Fan and F. Xia, J. Am. Chem. Soc., 2013, 135, 4604–4607 CrossRef CAS.
- C.-H. Zhang, Y. Tang, Y.-Y. Sheng, H. Wang, Z. Wu and J.-H. Jiang, Chem. Commun., 2016, 52, 13584–13587 RSC.
- Y. Guo, J. Wu and H. Ju, Chem. Sci., 2015, 6, 4318–4323 RSC.
- H. Wang, C. Li, X. Liu, X. Zhou and F. Wang, Chem. Sci., 2018, 9, 5842–5849 RSC.
- M. A. English, L. R. Soenksen, R. V. Gayet, H. de Puig, N. M. A. Mari, A. S. Mao, P. Q. Nguyen and J. J. Collins, Science, 2019, 365, 780–785 CrossRef CAS.
- D. Duan, K. Zheng, Y. Shen, R. Cao, L. Jiang, Z. Lu, X. Yan and J. Li, Nucleic Acids Res., 2011, 39, e154 CrossRef CAS PubMed.
- S.-Y. Li, Q.-X. Cheng, J.-K. Liu, X.-Q. Nie, G.-P. Zhao and J. Wang, Cell Res., 2018, 28, 491–493 CrossRef CAS.
- S. Wang, W. Song, S. Wei, S. Zeng, S. Yang, C. Lei, Y. Huang, Z. Nie and S. Yao, Anal. Chem., 2019, 91, 8622–8629 CrossRef CAS.
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- E. van Rooij, Circ. Res., 2011, 108, 219–234 CrossRef CAS PubMed.
- M.-L. Si, S. Zhu, H. Wu, Z. Lu, F. Wu and Y.-Y. Mo, Oncogene, 2007, 26, 2799–2803 CrossRef CAS PubMed.
- R. Kumarswamy, I. Volkmann and T. Thum, RNA Biol., 2011, 8, 706–713 CrossRef CAS PubMed.
- V. Jazbutyte and T. Thum, Curr. Drug Targets, 2010, 11, 926–935 CrossRef CAS PubMed.
- G. Zhu, L. Liang and C. Zhang, Anal. Chem., 2014, 86, 11410–11416 CrossRef CAS PubMed.
- D. C. Swarts, J. van der Oost and M. Jinek, Mol. Cell, 2017, 66, 221–233 CrossRef CAS PubMed.
- D. Dong, K. Ren, X. Qiu, J. Zheng, M. Guo, X. Guan, H. Liu, N. Li, B. Zhang, D. Yang, C. Ma, S. Wang, D. Wu, Y. Ma, S. Fan, J. Wang, N. Gao and Z. Huang, Nature, 2016, 532, 522–526 CrossRef CAS PubMed.
- T. Yamano, H. Nishimasu, B. Zetsche, H. Hirano, I. M. Slaymaker, Y. Li, I. Fedorova, T. Nakane, K. S. Makarova, E. V. Koonin, R. Ishitani, F. Zhang and O. Nureki, Cell, 2016, 165, 949–962 CrossRef CAS PubMed.
- A. F. Jou, C.-H. Lu, Y.-C. Ou, S.-S. Wang, S.-L. Hsu, I. Willner and J. A. Ho, Chem. Sci., 2015, 6, 659–665 RSC.
- Q. Liu, D. Wang, M. Yuan, B. He, J. Li, C. Mao, G. Wang and H. Qian, Chem. Sci., 2018, 9, 7562–7568 RSC.
- F. Yang, Y. Cheng, Y. Cao, H. Dong, H. Lu, K. Zhang, X. Meng, C. Liu and X. Zhang, Chem. Sci., 2019, 10, 1709–1715 RSC.
- M. P. Saguer and M. C. Rodicio, Clin. Biochem., 2013, 46, 869–878 CrossRef PubMed.
- S. Volinia, G. A. Calin, C.-G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris and C. M. Croce, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2257–2261 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc03084h |
| This journal is © The Royal Society of Chemistry 2020 |