Open Access Article
Open Access ArticleIn situ visualization of peroxisomal viscosity in the liver of mice with non-alcoholic fatty liver disease by near-infrared fluorescence and photoacoustic imaging†
Yongqing
Zhou
,
Ping
Li
 *,
Xin
Wang
,
Chuanchen
Wu
,
Nannan
Fan
,
Xiaoning
Liu
,
Lijie
Wu
,
Wei
Zhang
,
Wen
Zhang
*,
Xin
Wang
,
Chuanchen
Wu
,
Nannan
Fan
,
Xiaoning
Liu
,
Lijie
Wu
,
Wei
Zhang
,
Wen
Zhang
 ,
Zhenzhen
Liu
and
Bo
Tang
,
Zhenzhen
Liu
and
Bo
Tang
 *
*
College of Chemistry, Chemical Engineering and Materials Science, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Collaborative Innovation Center of Functionalized Probes for Chemical Imaging in Universities of Shandong, Institutes of Biomedical Sciences, Shandong Normal University, Jinan 250014, People's Republic of China. E-mail: tangb@sdnu.edu.cn; lip@sdnu.edu.cn
First published on 6th October 2020
Abstract
Non-alcoholic fatty liver disease (NAFLD) can gradually develop into hepatic failure, and early diagnosis is crucial to improve treatment efficiency. The occurrence of NAFLD is closely related to lipid metabolism. Peroxisomes act as the first and main site for lipid metabolism in the hepatocytes, so abnormal lipid metabolism might directly affect peroxisomal viscosity. Herein, we developed a new near-infrared fluorescence (NIRF) and photoacoustic (PA) imaging probe (PV-1) for the real-time visualization of peroxisomal viscosity in vivo. This PV-1 encompasses the malononitrile group as the rotor, which emits strong NIRF (at 705 nm) and PA (at 680 nm) signals when rotation is hindered as viscosity increases. Through dual-mode imaging, we discovered distinctly higher viscosity in the liver of NAFLD mice for the first time. We further found the remarkable amelioration of NAFLD upon treatment with N-acetylcysteine (NAC). Therefore, we anticipate that the PV-1 imaging method is promising for the early diagnosis and prognostic evaluation of NAFLD.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most common causes of abnormal liver function. The prevalence of NAFLD is gradually increasing, with serious consequences to health.1,2 If the condition is not diagnosed and treated promptly, NAFLD can gradually develop into non-alcoholic steatohepatitis, cirrhosis, and even hepatocellular carcinoma, resulting in liver death.3–6 Therefore, timely and accurate diagnosis is crucial to prevent the deterioration of NAFLD. At present, the existing methods of diagnosing NAFLD mainly include liver biopsy,7 serum biochemistry,8 histopathological examination,9 and ultrasonic examination.10 However, most of these technologies have drawbacks, such as a high risk, false positives, and complicated operation steps.11,12 Therefore, the development of a real-time, reliable, and non-destructive method of early detection may help the timely treatment of NAFLD, and prevent further development.Peroxisomes, as the main site of lipid metabolism, play essential roles in maintaining intracellular morphology and hepatocellular function.13,14 Excessive free fatty acids accumulate in the peroxisomes when the peroxisomal fatty acid β-oxidation is disordered.15,16 These events can cause a disequilibrium in lipid homeostasis, eventually resulting in NAFLD.17,18 Notably, along with lipid accumulation, the peroxisomal microenvironment (including viscosity) can change.19–24 It is speculated that the peroxisomal viscosity in liver tissues with NAFLD might be different from that in normal liver tissues.25,26 Therefore, establishing a method of accurately monitoring peroxisomal viscosity may help the early identification of the possible onset of NAFLD.
Near-infrared fluorescence (NIRF) imaging technology has inherent advantages such as high resolution and a non-invasive nature, and has been widely used in the monitoring of bioactive molecules and the intracellular microenvironment.27–30 NIRF probes based on molecular motors have been developed to detect viscosity changes inside cells and organelles.31–34 However, limited by their penetration depth, most NIRF probes fail to detect changes of viscosity in the deep tissue. Photoacoustic (PA) imaging can successfully overcome this shortcoming, owing to its greater tissue penetration depth (up to 10.0 cm).35,36 In recent years, PA probes have been used for the in situ and non-destructive visualization of various active molecules in deep tissues of animals.37–39 Therefore, NIRF and PA dual-mode imaging may accurately detect peroxisomal viscosity in liver deep tissue, promoting the early diagnosis of NAFLD. To date, no PA probes have been reported for the detection of viscosity in living animals.
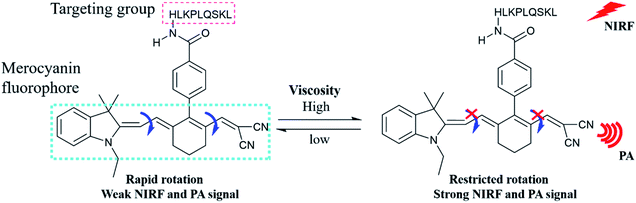
For this purpose, we designed a two-mode (NIRF/PA) small molecule probe (PV-1) for peroxisomal viscosity detection. As shown in Scheme 1, PV-1 is composed of a malononitrile group as a rotor, a HLKPLQSKL peptide chain as a peroxisomal-targeting group,40–43 and a merocyanine dye as a fluorophore. In the non-viscous or low viscosity environment, the rotor rotates freely around the large conjugate structure of the merocyanine dye, which dissipates the excited state energy of the dye by non-radiation.44–48 This process could quench fluorescence of PV-1. As the viscosity increases, the free rotation of the rotor is specifically restricted, thereby reducing the possibility of non-radiative. Therefore, fluorescence of PV-1 rises at 705 nm. At the same time, with the enhanced viscosity, the near-infrared absorbance of PV-1 at about 680 nm is also gradually elevated. Moreover, PV-1 exhibits a high extinction coefficient and low fluorescence quantum yield in glycerol.49 Based on these properties, PV-1 could be responsive to viscosity via changes in PA signals. Therefore, PV-1 could emit strong NIRF and PA signals when the viscosity increases. Given that the PAS8P receptor binds and transports HLKPLQSKL on the peroxisomal membrane surface, PV-1 could accumulate in the peroxisomes. Our experimental results show that PV-1 possesses good specificity and sensitivity for viscosity by dual modes. Fluorescence imaging proves that PV-1 could accurately locate in the peroxisomes and indicate changes in viscosity. More importantly, by using PV-1, we observed a significant increase in peroxisomal viscosity in the liver of NAFLD mouse models.
Results and discussion
Preparation and optical properties of PV-1
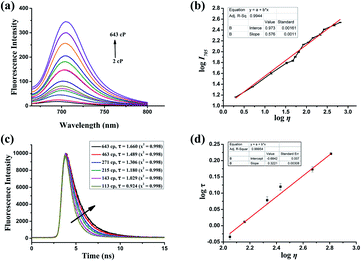
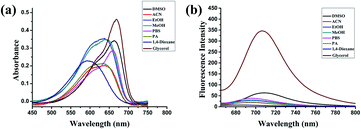
The synthesis and characterization of PV-1 are described in the ESI.† First, we studied the photophysical properties of PV-1 under various environmental viscosities. As shown in the absorption spectra (Fig. 1a and S1†), it could be seen that PV-1 exhibited different NIR absorption peaks in different solvents. Further, PV-1 (5.0 µM) exhibited bright fluorescence in glycerol (Fig. S2†). To precisely evaluate the relationship between fluorescence intensity of PV-1 and environmental viscosity, we gradually increased the media viscosity by adding glycerin into the methanol–glycerol system, and recorded the fluorescence responses of PV-1 in different viscosities. As shown in Fig. S3,† the main absorption peak of PV-1 at about 680 nm gradually increased with increasing viscosity. As shown in Fig. 2a, the fluorescence intensity of PV-1 at 705 nm enhanced continually as the ratio of glycerin increased, and reached the highest value (about 25 times) in pure glycerol. There was a good linear relationship (R2 = 0.994) between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I705 and log
I705 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) η according to the Förster–Hoffmann equation (Fig. 2b).50 In addition, the fluorescence quantum yield of PV-1 also increased gradually from 0.007 to 0.13 (Table S1†), according to fluorescence quantum yield equation.51 Considering that fluorescence lifetime is an important parameter to study the fluorescence profile of PV-1, the fluorescence lifetimes of PV-1 in different viscosity media were measured. Fig. 2c shows that the fluorescence lifetime of PV-1 has a similar growth tendency to that of solvent viscosity. Furthermore, PV-1 possessed an excellent linear relationship (R2 = 0.999) between log
η according to the Förster–Hoffmann equation (Fig. 2b).50 In addition, the fluorescence quantum yield of PV-1 also increased gradually from 0.007 to 0.13 (Table S1†), according to fluorescence quantum yield equation.51 Considering that fluorescence lifetime is an important parameter to study the fluorescence profile of PV-1, the fluorescence lifetimes of PV-1 in different viscosity media were measured. Fig. 2c shows that the fluorescence lifetime of PV-1 has a similar growth tendency to that of solvent viscosity. Furthermore, PV-1 possessed an excellent linear relationship (R2 = 0.999) between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) τ and log
τ and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) η, increasing from 2.0 cp to 643.0 cp (Fig. 2d). Moreover, compared with the strong fluorescence of PV-1 in glycerol, its fluorescence in solvents with different polarities was negligible. Therefore, the fluorescence intensity of PV-1 was insensitive to polarity changes. These results suggest that PV-1 can detect viscosity with outstanding sensitivity. To further prove the detection mechanism, we also confirmed that the fluorescence responses of precursor 3 were also susceptible to viscosity (Fig. S4†).
η, increasing from 2.0 cp to 643.0 cp (Fig. 2d). Moreover, compared with the strong fluorescence of PV-1 in glycerol, its fluorescence in solvents with different polarities was negligible. Therefore, the fluorescence intensity of PV-1 was insensitive to polarity changes. These results suggest that PV-1 can detect viscosity with outstanding sensitivity. To further prove the detection mechanism, we also confirmed that the fluorescence responses of precursor 3 were also susceptible to viscosity (Fig. S4†).
 | ||
| Fig. 1 Ultraviolet spectra (a) and fluorescence spectra (b) of PV-1 (5.0 µM) in different solvents, respectively. λex = 650 nm. | ||
To clarify the selectivity of PV-1, we studied its fluorescence response to peroxisomal substrates. We first tested the PV-1 fluorescence response to viscosity in the presence of constituents, including reactive oxygen species (ROS), reactive nitrogen species (RNS), reactive sulfur species (RSS), metal ions, and proteins. Experimental results showed that competing species and proteins had a negligible impact on the fluorescence response of PV-1 (Fig. S5†). The fluorescence intensity of PV-1 was investigated in different pH conditions. As demonstrated in Fig. S6,† the fluorescence intensity of PV-1 remained almost unchanged in 30% glycerol solution under pH 5.0 to 9.0. To examine the photostability of PV-1, we recorded the fluorescence spectra of PV-1 in 30% glycerol and phosphate-buffered saline (PBS) for 1.0 h. Fig. S7† suggested that the fluorescence intensity of PV-1 in 30% glycerol was significantly stronger than that in PBS. These results indicate that the fluorescence response of PV-1 remains stable in a complex and varied viscosity environment. Furthermore, these results confirm that PV-1 has potential applications in sensing viscosity changes within a multicomponent biological environment.
PA properties of PV-1
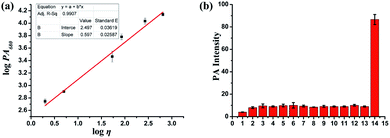
To study the PA imaging properties of the PV-1, the PA signals of PV-1 in a series of methanol–glycerol mixed solvents were measured subsequently. As Fig. 3a suggested, with increasing glycerol ratio, the PA intensity gradually strengthened at about 680 nm, and a good linear relationship (R2 = 0.991) was revealed by fitting the Förster–Hoffmann equation. Moreover, we investigated the PA intensity of PV-1 in the presence of some reactive species under physiological conditions. Our results also revealed that PV-1 displayed a specific PA signal in glycerol at 680 nm compared with other species (Fig. 3b). Altogether, these results prove that PV-1 is capable of PA detection of viscosity changes in complex conditions.Subcellular localization of PV-1
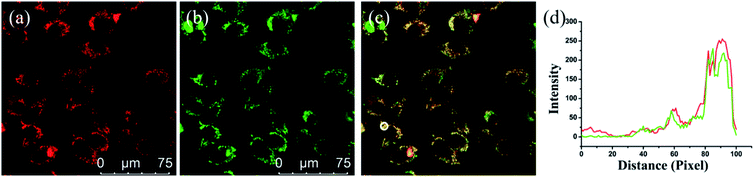
To determine the applicability of PV-1 in a bioimaging experiment, we first tested the cytotoxicity of PV-1 in HL-7702 cells using standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay.52 Fig. S8† showed that the IC50 value of PV-1 was 45 µM, which demonstrates its minimal cytotoxicity and good biocompatibility. Next, we tested its toxicity in C57 mice.53,54 Mice experiments suggested no obvious depilation or inflammation in two mice groups treated with different concentrations of PV-1, proving a low toxicity of PV-1 (Fig. S9†). These results indicate that PV-1 has fascinating biocompatibility, and is suitable for biological imaging. Then, we validated the peroxisomal localization ability of PV-1 in HL-7702 cells and SMMC-7721 cells.55,56 In Fig. 4, the red fluorescence of PV-1 significantly coincident with the green fluorescence of peroxisome-GFP was observed. The Pearson's colocalization coefficient between PV-1 and peroxisome-GFP was 0.91. We also obtained similar results in HL-7702 cells, with a Pearson's colocalization coefficient of 0.90. Further, the PV-1 hardly accumulated in the Golgi apparatus, endoplasmic reticulum, mitochondria, or lysosomes (Fig. S10†). The above results indicate that PV-1 can precisely target and tune in on the peroxisomes of living cells.Fluorescence imaging of peroxisomal viscosity during lipid abnormal metabolism in living cells
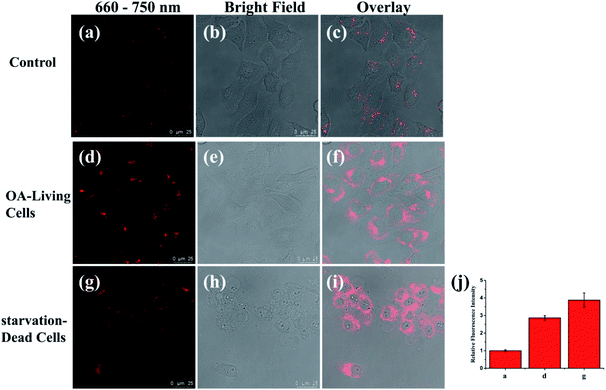
Previous studies have shown that when long free fatty acids are in excess, they impair peroxisomal β-oxidation capacity, causing lipid accumulation.57,58 Thus, we evaluated the viscosity detecting capability of PV-1 during abnormal lipid metabolism in living cells. Oleic acid (OA), a long chain fatty acid which metabolizes in cells, especially in liver cells, was chosen as an initiator of lipid abnormal metabolism in HL-7702 cells.59,60 As shown in Fig. 5, compared with control cells, the OA-simulated cells exhibited brighter fluorescence in peroxisomes shaped like droplet-sized dots. These data verify that PV-1 can respond to peroxisomal viscosity increments in living cells during abnormal lipid metabolism. Moreover, we found that unhealthy cells evoked bright fluorescence due to a higher uptake of the probe. However, unlike droplet-size dots, the fluorescence signals were located in the entire cytoplasm rather than the peroxisomes.61 Therefore, these results demonstrated that the increased fluorescence in the peroxisomes of OA-simulated cells was caused by the higher viscosity in cells under abnormal lipid metabolism. Subsequently, time-dependent fluorescence imaging (from 0–60.0 min) of HL-7702 cells treated with OA was carried out after the addition of PV-1. It was found that the fluorescence intensity of PV-1 could remain stable for as long as 60.0 min (Fig. S11†). Taken together, these results certify that a PV-1-based viscosity detection method can distinguish liver cells with abnormal lipid metabolism from normal cells.NIRF imaging in livers of mice with NAFLD
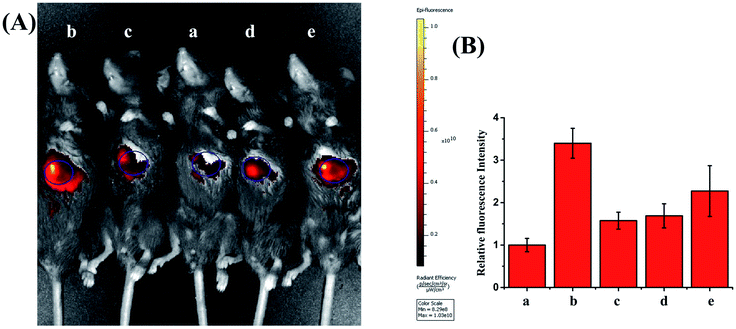
With this viscosity-sensitive tool, we next investigated whether PV-1 could distinguish NAFLD mice from normal mice. Considering that NAFLD is caused by abnormal lipid accumulation, NAFLD mice models were constructed by introducing a high-fat diet (60 kcal% fat) for 6 weeks.62,63 Excess lipid accumulation can trigger a series of stress responses and produce excessive amounts of ROS, which result in serious secondary liver damage in the process of NAFLD.64,65 Therefore, we used NAC (10 mg kg−1) to reduce ROS and alleviate the NAFLD of mice.66,67 Fig. S12† shows that the body weight of mice with NAFLD was significantly higher than that of the other group mice. Hematoxylin and eosin staining experiments demonstrated that the liver of NAFLD mice showed massive hepatic steatosis compared with that of the treatment group and normal group (Fig. S13†). NIRF imaging was performed after intravenous injection of PV-1, (100 µL, 10−4 M). As shown in Fig. 6 and S14,† the livers of normal mice evoked dim fluorescence, while the livers of NAFLD mice emitted a conspicuous fluorescence signal. This shows that the mice with NAFLD had a higher viscosity (about 3.4-times) than the normal mice. In contrast, the fluorescence intensity of livers in the NAC-treated mice was clearly weaker compared with that of the NAFLD mice. Fascinatingly, NAC, as a mucolytic reagent, could also reduce viscosity in vivo due to a mucolytic effect (Fig. S15†).68,69Therefore, we further investigated whether NAFLD can be treated by scavenging ROS. We used antioxidants and drugs to treat and alleviate mice with NAFLD. Vitamin E as an antioxidant protected against scavenging ROS to alleviate NAFLD.70–72 To better evaluate the therapeutic effect of NAC, we also used the pioglitazone drug for treating NAFLD.73–76 Pioglitazone is a highly selective peroxisomal proliferative receptor (PPAR) agonist that regulates lipid metabolism and transport by activating PPAR, thereby reducing fat storage in the liver, ultimately alleviating NAFLD. The fluorescence intensity of livers in the antioxidant- and drug-treated mice was clearly weaker compared with that of NAFLD mice. These results further confirm that the enhanced liver fluorescence is induced from elevated peroxisomal viscosity in the NAFLD mice, and that the NAC can alleviate the NAFLD with mice as well as pioglitazone and vitamin E. Altogether, these results highlight the fact that scavenging ROS can alleviate NAFLD. Moreover, as shown in Fig. S16,† we observed that organs such as the heart, spleen, lung, and kidney showed almost no obvious fluorescence, whereas the liver emitted obvious fluorescence. Therefore, we believe that PV-1 mostly gathers in the mouse liver. Overall, this method of peroxisomal viscosity imaging can be used not only for the early diagnosis of NAFLD, but also for screening drugs for NAFLD.
PA imaging in livers of mice with NAFLD
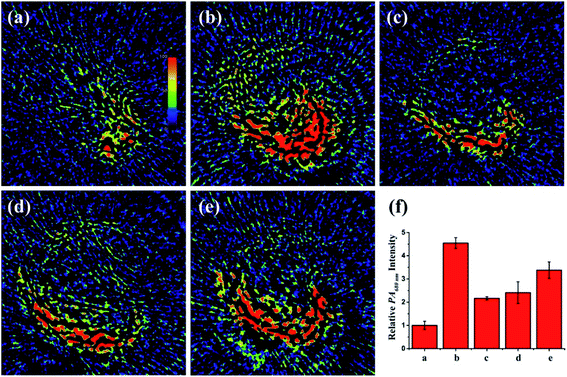
Since the liver is located deep in the body, NIRF imaging has insufficient penetration depth for the liver in vivo. Fortunately, with favorable near-infrared absorbance/emission, PV-1 might be appropriate for peroxisomal viscosity detection in mouse liver by a PA imaging technique. In an PA experiment, we discovered stronger PA signals (about 4.4-fold) at 680 nm in NAFLD mice livers compared with normal mice. In contrast, after intragastric administration of a dose of therapeutic reagents in NAFLD mice, such as pioglitazone, vitamins E and NAC, the intensity of the PA signals decreased. These results revealed that the peroxisomal viscosity was enhanced in mice with NAFLD, and could be reduced by NAC treatment (Fig. 7 and S17†). The above results suggest that the PA signal variation tendency is consistent with that of in vivo fluorescence imaging. Thus, the PV-1-based PA imaging technique can accurately reflect the peroxisomal viscosity changes and indicate the occurrence of NAFLD. Furthermore, the content of alanine aminotransferase (ALT) is an important index to measure the degree of liver lesions in NAFLD.77,78 To further make sure of the accuracy of the NIRF and PA imaging results, the liver ALT levels of five mice groups were determined by enzyme-linked immunosorbent assay (ELISA). The ELISA results showed that the ALT in NAFLD mice liver was higher than that of normal mice (Fig. S18†). All the data further illustrate that the imaging method can effectively realize the early diagnosis of NAFLD by monitoring liver viscosity change in NAFLD mice. Currently, the reported fluorescence viscosity probes are mostly applied to superficial tissue imaging, while single-channel optical imaging is not reliable for tracking viscosity fluctuations. Hence, PV-1 can monitor viscosity in the deep liver tissue by superior PA imaging. Importantly, the dual model imaging viscosity method is used for the early accurate diagnosis and clinical drug evaluation of NAFLD.Conclusions
In summary, we designed and synthesized the first dual-mode (NIRF/PA) peroxisomal viscosity probe PV-1. The PV-1 exhibited high specificity and selectivity to viscosity. By HLKPLQSKL peptide chain, PV-1 can solely gather in peroxisomes. Moreover, the detection method of PV-1 could indicate the peroxisomal viscosity differences in OA stimulated and normal cells. Using this PV-1, we found that the liver viscosities of NAFLD mice were significantly higher than those of normal mice and three groups of treated NAFLD mice. Therefore, the PV-1-based dual-mode detection viscosity method can not only be utilized for indicating the occurrence and development of NAFLD, but can also be used for evaluating the efficacy of drugs for NAFLD.Live subject statement
All of the animal experiments were performed strictly in accordance with the relevant laws and guidelines issued and approved by the Ethical Committee of Shandong Normal University for the Care and Use of Laboratory Animals.Conflicts of interest
There are no conflicts to declare.Note added after first publication
This article replaces the version published on 19th October 2020, which contained a duplication of one of the figures. Fig. 3 and Fig. 6 have now been updated correctly.Acknowledgements
This work was supported by the National Natural Science Fund of China (21675105, 21535004, 21927811) and the Key Research and Development Program of Shandong Province (2018YFJH0502), National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2017ZX09301030004).Notes and references
- L. A. Adams, P. Angulo and K. D. Lindor, CMAJ, 2015, 172, 899–905 Search PubMed.
- T. Karlas, R. Physician, J. Wiegand, S. Physician and T. Berg, Best Pract. Res., Clin. Endocrinol. Metab., 2013, 27, 195–208 CrossRef CAS.
- E. Hashimoto, M. Taniai and K. Tokushige, J. Gastroenterol. Hepatol., 2013, 28, 64–70 CrossRef CAS.
- L. Vigan, A. Lleo and A. Aghemo, Hepatobiliary Surg. Nutr., 2018, 7, 130–133 CrossRef.
- O. Massoud and M. Charlton, Clin. Liver Dis., 2018, 22, 201–211 CrossRef.
- C. H. Ma, A. Kesarwala, T. Eggert, J. M. Echeverz, D. E. Kleiner, P. Jin, D. F. Stroncek, M. Terabe, V. Kapoor, A. M. ElGindi, M. Han, A. Thornton, H. B. Zhang, M. Egger, J. Luo, D. Felsher, D. McVicar, A. Weber, M. Heikenwalder and T. F. Greten, Nature, 2016, 531, 253–257 CrossRef CAS.
- P. Golabi, M. Sayiner, Y. Fazel, A. Koenig, L. Henry and Z. M. Younossi, Expet Rev. Gastroenterol. Hepatol., 2015, 10, 1–9 Search PubMed.
- E. Cleveland, A. Bandy and L. VanWagner, Clin. Liver Dis., 2018, 11, 98–104 CrossRef.
- E. Hashimoto, K. Tokushige and J. Ludwig, Hepatol. Res., 2015, 45, 20–28 CrossRef.
- S. R. Mehta, E. L. Thomas, J. D. Bell, D. G. Johnston and S. T. Robinson, World J. Gastroenterol., 2008, 14, 3476–3483 CrossRef.
- Y. Sumida, A. Nakajima and Y. Itoh, World J. Gastroenterol., 2014, 20, 475–485 CrossRef.
- J. D. Browning and J. D. Horton, J. Clin. Invest., 2004, 114, 147–152 CrossRef CAS.
- I. J. Lodhi and C. F. Semenkovich, Cell Metab., 2014, 19, 380–392 CrossRef CAS.
- T. B. Dansen and K. A. Wirtz, IUBMB Life, 2001, 51, 223–230 CrossRef CAS.
- N. C. Tapia, N. Rosso and C. Tiribelli, BMC Gastroenterol., 2012, 12, 20–30 CrossRef.
- N. Latruffe, M. C. Maki, V. N. Frances, M. C. Clemencet, B. Jannin and J. P. Berlot, Biochem. Pharmacol., 2000, 60, 1027–1032 CrossRef CAS.
- P. Tessari, A. Coracina, A. Cosma and A. Tiengo, Nutr. Metabol. Cardiovasc. Dis., 2009, 19, 291–302 CrossRef CAS.
- O. Cheung and A. J. Sanyal, Semin. Liver Dis., 2008, 28, 351–359 CrossRef CAS.
- S. D. Kohlwein, M. Veenhuis and J. V. Klei, Lipid Droplets and Peroxisomes: Key Players in Cellular Lipid Homeostasis or A Matter of Fat—Store ‘em Up or Burn ’em Down, 2012 Search PubMed.
- I. J. Lodhi and C. F. Semenkovich, Cell Metab., 2014, 19, 380–392 CrossRef CAS.
- T. C. Walther and R. V. Farese, Annu. Rev. Biochem., 2012, 81, 1–28 CrossRef.
- G. Mannaerts, P. Veldhoven and M. Casteels, Cell Biochem. Biophys., 2000, 32, 73–87 CrossRef CAS.
- B. L. Parker, A. C. calkin, M. M. Seldin, M. F. Keating, A. Vallim and B. G. Drew, Nature, 2019, 567, 187–193 CrossRef CAS.
- A. D. Barbosa, D. B. Savage and S. Siniossoglou, Curr. Opin. Cell Biol., 2015, 35, 91–97 CrossRef CAS.
- P. Tessari, A. Coracina, A. Cosma and A. Tiengo, Nutr., Metab. Cardiovasc. Dis., 2009, 19, 291–302 CrossRef CAS.
- N. C. Tapia, N. Rosso and C. Tiribelli, BMC Gastroenterol., 2012, 12, 20 CrossRef.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2017, 53, 12822–12825 RSC.
- H. B. Xiao, P. Li, W. Zhang and B. Tang, Chem. Sci., 2016, 7, 1588–1593 RSC.
- Y. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- Y. Z. Min, J. M Li, F. Liu, E. L. Yeow and B. G. Xing, Angew. Chem., Int. Ed., 2014, 53, 1012–1016 CrossRef CAS.
- K. Dou, W. J. Huang, Y. H. Xiang, S. J. Li and Z. H. Liu, Anal. Chem., 2020, 92, 4177–4181 CrossRef CAS.
- R. Guo, J. L. Yin, Y. Y. Ma, G. H. Li, Q. Wang and W. Y. Lin, Sens. Actuators, B, 2018, 271, 321–328 CrossRef CAS.
- Z. G. Yang, Y. X. He, J. H. Lee, W. S. Cha, W. X. Ren, J. H. Lee, C. h. Kang and J. S. Kim, Chem. Commun., 2014, 50, 11672–11675 RSC.
- Y. Liu, C. H. Wolstenholme, G. C. Carter, H. B. Liu, H. Hu, L. S. Grainger, K. Miao, M. Fares, C. A. Hoelzel, H. P. Yennawar, G. Ning, M. Y. Du, L. Bai, X. S. Li and X. Zhang, J. Am. Chem. Soc., 2018, 140, 7381–7384 CrossRef CAS.
- E. Y. Zhou, H. Knox, C. Liu and W. Zhao, J. Am. Chem. Soc., 2019, 141, 17601–17609 CrossRef CAS.
- L. L. Zeng, G. C. Ma, J. Lin and P. Huang, Small, 2018, 14, 1800782 CrossRef.
- J. J. Zhang, X. Zhen, P. K. Upputuri, M. Pramanik, P. Chen and K. Y. Pu, Adv. Mater., 2017, 29, 1604764 CrossRef.
- H. Wang, Y. X. Zhang, Y. Y. Yang, Z. X. He, C. C. Wu, W. Zhang, W. Zhang, J. Liu, P. Li and B. Tang, Chem. Commun., 2019, 55, 9685–9688 RSC.
- S. T. Li, F. Y. Meng, X. L. Liao, Y. M. Wang, Z. X. Sun, F. C. Guo, X. X. Li, M. Meng, Y. Li and C. H. Sun, PLoS One, 2014, 9, e86724 CrossRef.
- J. Y. Bai, P. Y. Lei, C. Y. Zhao, Y. Q. Wang, D. D. Yan and S. Y. Yang, Toxicol. Rep., 2016, 3, 98–104 CrossRef CAS.
- D. M. Xi, M. Xiao, J. F. Cao, L. Y. Zhao, N. Xu, S. Long, J. L. Fan, K. Shao, W. Sun, X. H. Yan and X. J. Peng, Adv. Mater., 2020, 32, 1907855 CrossRef CAS.
- S. R. Terlecky, W. M. Nuttley, D. McCollum, E. Sock and S. Subramani, EMBO J., 1995, 14, 3627–3634 CrossRef CAS.
- A. Jankowski, J. H. Kim, R. F. Collins, R. Daneman, P. Walton and S. Grinstein, J. Biol. Chem., 2001, 276, 48748 CrossRef CAS.
- J. A. McNew, K. Sykes and J. M. Goodman, Mol. Biol. Cell, 1993, 4, 223 CrossRef CAS.
- Y. Q. Zhou, P. Li, N. N. Fan, X. Wang, X. N. Liu, L. J. Wu, W. Zhang, W. Zhang, C. Ma and B. Tang, Chem. Commun., 2019, 55, 6767–6770 RSC.
- M. Haidekker, T. Brady, D. Lichlyter and E. Theodorakis, J. Am. Chem. Soc., 2006, 128, 398–399 CrossRef.
- Z. G. Yang, Y. X. He, J. H. Lee, N. Y. Park, M. Suh, X. J. Peng, H. G. Jung, C. H. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 9181–9185 CrossRef CAS.
- T. Chen, Z. K. Chen, R. Y. Liu and S. B. Zheng, Org. Biomol. Chem., 2019, 17, 6398–6403 RSC.
- H. B. Cheng, Y. Y. Li, B. Z. Tang and J. Y. Yoon, Chem. Soc. Rev., 2020, 49, 21–31 RSC.
- M. K. Kuimova, S. W. Botchway, A. W. Parker, M. Balaz, H. A. Collins, H. L. Anderson, K. Suhling and P. R. Ogilby, Nat. Chem., 2009, 1, 69–73 CrossRef CAS.
- D. E. Fischer, A. Theodorakis and M. A. Haidekker, Nat. Protoc., 2007, 2, 227–236 CrossRef CAS.
- Z. Zou, Q. Yan, S. X. Ai, P. Qi, H. Yang, Y. F. Zhang, Z. H. Qing, L. H. Zhang, F. Feng and R. H. Yang, Anal. Chem., 2019, 91, 8574–8581 CrossRef CAS.
- X. J. Peng, Z. G. Yang, J. Y. Wang, J. L. Fan, Y. X. He, F. L. Song, B. S. Wang, S. G. Sun, J. L. Qu, J. Qi and M. Yan, J. Am. Chem. Soc., 2011, 133, 6626–6635 CrossRef CAS.
- H. Xu, H. Xu, S. N. Ma, X. N. Chen, L. X. Huang, J. W. Chen, F. Gao, R. Wang, K. Y. Lou and W. Wang, J. Am. Chem. Soc., 2018, 140, 16408–16412 CrossRef CAS.
- M. Z. Ye, X. H. Wang, J. B. Tang, Z. Q. Guo, Y. Q. Shen, H. Tian and W. H. Zhu, Chem. Sci., 2016, 7, 4958–4965 RSC.
- Y. Z. Zhao, Q. X. Hu, F. X. Cheng, N. Su, A. X. Wang, Y. J. Zou, H. Y. Hu, X. J. Chen, H. M. Zhou, X. Z. Huang, K. Yang, Q. Zhu, X. Wang, J. Yi, L. Y. Zhu, X. H. Qian, L. X. Chen, Y. Tang, J. Loscalzo and Y. Yang, Cell Metab., 2015, 21, 777–789 CrossRef CAS.
- R. J. Wanders, S. Ferdinandusse, P. Brites and S. Kemp, Biochim. Biophys. Acta, 2010, 1801, 272–280 CrossRef CAS.
- P. B. Lazarow, J. Biol. Chem., 1978, 253, 1522–1528 CAS.
- C. Wang, Z. L. LV, Y. J. Kang, T. X. Xing, P. L. Wang and Z. Jiang, Int. J. Mol. Med., 2013, 32, 1159–1165 CrossRef CAS.
- L. A. DelRio, Peroxisomes and their Key Role in Cellular Singaling and Metabolism, 2013 Search PubMed.
- F. Q. Yu, X. Y. Jing and W. Y. Lin, Sens. Actuators, B, 2020, 302, 127207 CrossRef CAS.
- M. C. Heffern, H. M. Park, H. A. Yeung, C. V. de Bittner, C. M. Ackerman, A. Stahlb and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 14219–14224 CrossRef CAS.
- G. Kanuri and I. Bergheim, Int. J. Mol. Sci., 2013, 14, 11963–11980 CrossRef.
- M. Kohjima, M. Enjoji, N. Highuchl, M. Kato, K. Koth, T. Yoshimoto, T. Fujino, M. Yada, R. Yada, N. Harada, R. I. Takayanag and M. Nakamut, Int. J. Mol. Med., 2007, 20, 351–358 CAS.
- J. Y. Kim, R. G. Carbonell, S. Yamachika, P. Zhao, D. Dhar, R. Loomba, R. J. Kaufman, A. R. Saltiel and M. Karin, Cell, 2018, 175, 133–145 CrossRef CAS.
- A. Sadowska, Ther. Adv. Respir. Dis., 2012, 6, 127–135 CrossRef CAS.
- K. F. Schoenberg, M. Wiedmann, K. J. Georgieff and M. D. Applikation, Anaesthesist, 1995, 44, 651–658 CrossRef.
- G. A. Coque, M. Hernandez, V. Camanas and C. M. Fernandez, Analyst, 1989, 84, 975–977 RSC.
- M. Jaworska, G. Szulinska, M. Wilk and J. Taut, J. Chromatogr. A, 1999, 853, 479–485 CrossRef CAS.
- H. Kitade, G. L. Chen, Y. H. Ni and T. Ota, Nutrients, 2017, 9, 387 CrossRef.
- J. E. Lavine, J. Pediatr., 2000, 5, 16–22 Search PubMed.
- T. Hardy, Q. M. Anstee and C. P. Day, Curr. Opin. Gastroenterol., 2015, 31, 175–183 CrossRef CAS.
- V. G. Athyros, T. K. Alexandrides, H. Bilianou, E. Cholongitas, M. Doumas, E. S. Ganotakis and J. Goudevenos, Metabolism, 2017, 71, 17–32 CrossRef CAS.
- T. Schreuder, B. J. Verwer, C. V. Nieuwkerk and C. J. Mulder, World J. Gastroenterol., 2008, 28(14), 2474–2486 CrossRef.
- K. M. Comar and R. K. Sterling, Aliment. Pharmacol. Ther., 2005, 23, 207–215 CrossRef.
- A. Federico, C. Zulli, I. D. Sio, A. D. Prete, M. Dallio, M. Masarone and C. Loguercio, World J. Gastroenterol., 2014, 20, 16841–16857 CrossRef CAS.
- Y. Chang, S. Ryu, E. j. Sung and Y. Jang, Clin. Chem., 2007, 53, 686–692 CrossRef CAS.
- H. J. Oh, T. H. Kim, Y. W. Sohn, Y. S. Kim, Y. R. Oh, E. Y. Cho, S. Y. Shim, S. R. Shin, A. L. Han, S. J. Yoon and H. C. Kim, J. Hepatol., 2011, 17, 27–36 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, cells/mice culture, additional figures. See DOI: 10.1039/d0sc02922j |
| This journal is © The Royal Society of Chemistry 2020 |