Open Access Article
Open Access ArticleSDS-induced multi-stage unfolding of a small globular protein through different denatured states revealed by single-molecule fluorescence†
Georg
Krainer‡§
 *a,
Andreas
Hartmann‡
a,
Vadim
Bogatyr‡¶
*a,
Andreas
Hartmann‡
a,
Vadim
Bogatyr‡¶
 a,
Janni
Nielsen
c,
Michael
Schlierf
a,
Janni
Nielsen
c,
Michael
Schlierf
 *ab and
Daniel E.
Otzen
*ab and
Daniel E.
Otzen
 *c
*c
aB CUBE – Center for Molecular Bioengineering, TU Dresden, Tatzberg 41, 01307 Dresden, Germany. E-mail: georg.krainer@tu-dresden.de; michael.schlierf@tu-dresden.de
bCluster of Excellence Physics of Life, TU Dresden, 01062 Dresden, Germany
cInterdisciplinary Nanoscience Center (iNANO), Aarhus University, Gustav Wieds Vej 14, 8000 Aarhus, Denmark. E-mail: dao@inano.au.dk
First published on 10th August 2020
Abstract
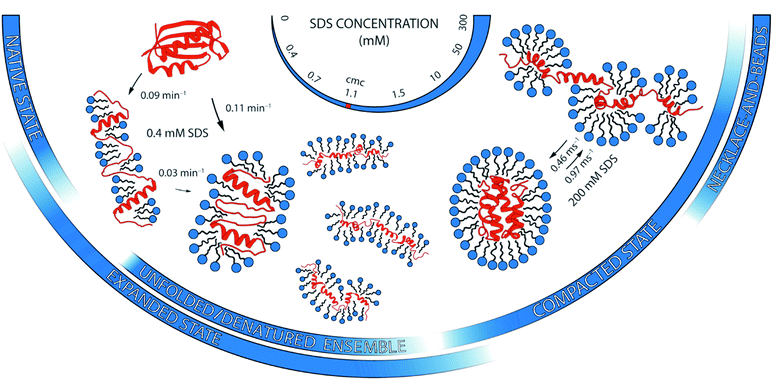
Ionic surfactants such as sodium dodecyl sulfate (SDS) unfold proteins in a much more diverse yet effective way than chemical denaturants such as guanidium chloride (GdmCl). But how these unfolding processes compare on a molecular level is poorly understood. Here, we address this question by scrutinising the unfolding pathway of the globular protein S6 in SDS and GdmCl with single-molecule Förster resonance energy transfer (smFRET) spectroscopy. We show that the unfolding mechanism in SDS is strikingly different and convoluted in comparison to denaturation in GdmCl. In contrast to the reversible two-state unfolding behaviour in GdmCl characterised by kinetics on the timescale of seconds, SDS demonstrated not one, but four distinct regimes of interactions with S6, dependent on the surfactant concentration. At ≤1 mM SDS, S6 and surfactant molecules form quasi-micelles on a minute timescale; at millimolar [SDS], the protein denatures through an unfolded/denatured ensemble of highly heterogeneous states on a multi-second timescale; at tens of millimolar of SDS, the protein unfolds into a micelle-packed conformation on the second timescale; and >50 mM SDS, the protein unfolds with millisecond timescale dynamics. We propose a detailed model for multi-stage unfolding of S6 in SDS, which involves at least three different types of denatured states with different level of compactness and dynamics and a continually changing landscape of interactions between protein and surfactant. Our results highlight the great potential of single-molecule fluorescence as a direct probe of nanoscale protein structure and dynamics in chemically complex surfactant environments.
Introduction
Interactions of proteins with surfactants have been the focus of studies for many years, not only because of the remarkable variety of conformations that surfactants induce in proteins, but also due to the importance of protein–surfactant interactions in many applications such as detergency, food, and cosmetics—not to mention SDS-PAGE, which is probably the most wide-spread protein analysis method worldwide.1–3 Surfactants differ fundamentally from chaotropic denaturants (e.g., urea and guanidinium chloride (GdmCl)) in their impact on protein conformations. Chemical denaturants are highly polar/ionic small molecules that interact weakly with protein backbone/sidechain groups and stabilise preferentially unfolded states due to their greater solvent-accessible surface area.4,5 Through this effect they destabilise compact states such as the native state, yet molar concentration levels are required to significantly displace the equilibrium away from the native state. Surfactants, by contrast, are relatively large amphiphilic molecules with both hydrophobic and hydrophilic moieties and are active at much lower concentrations (i.e., typically in the millimolar range).2 While non- and zwitter-ionic surfactants prefer to self-assemble into micelles at their critical micellar concentration (CMC) rather than interacting with proteins, ionic surfactants—particularly the anionic kind—interact strongly with proteins through a combination of hydrophobic and complementary electrostatic interactions and thus are able to effectively disrupt native protein conformations already below the CMC. The best-studied example is sodium dodecyl sulfate (SDS).Below the CMC (which is around 1–5 mM for SDS in most buffer systems), where it predominantly exists in its monomeric state, SDS can bind to proteins almost like a ligand stabilising the native state, rather than denaturing the protein as a chemical denaturant.6,7 With increasing numbers of bound SDS molecules, surfactant clusters form typically around the protein, leading to local or global disruption of protein structure. For example, the formation of shared micelles involving several protein molecules around a common micelle core has been reported.8 Above the CMC, proteins form complexes with SDS, which are generally thought to be built up of already intact SDS micelles decorated with partially unfolded protein.9–13 However, the picture is more complex as there is not just one generic protein–SDS complex present in solution: the shapes and sizes of protein–surfactant complexes depend on the protein and surfactant in question as well as the solvent conditions such as surfactant concentration, ionic strength, pH, and temperature.14 Also, micelles have been shown to rearrange around the protein during the initial stages of unfolding according to both computational15 and synchrotron-SAXS16 studies. Another important difference compared to the chemically denatured state is that SDS-denatured proteins are not random coils but typically retain significant, albeit non-native, secondary structure elements, which are often rich in α-helices.
The globular protein S6 has been used as a model system for protein unfolding studies in both chemical denaturants17,18 and SDS,19–21 hence making it particularly useful in efforts to understand the action mechanism of chaotropes and surfactants on protein unfolding. In GdmCl, S6 unfolds reversibly in a two-state manner involving only the native and the denatured state;17 only high concentrations of stabilising inorganic salts such as sodium sulfate lead to a transient accumulation of a metastable off-pathway collapsed state at very low GdmCl concentrations.22 The situation is significantly more complex in SDS. S6 denatures below the CMC of SDS, indicating that monomeric SDS is a potent denaturing agent.21 The denaturation takes place in several phases according to multiple different spectroscopic and calorimetric techniques. Under these conditions the CMC is around 3 mM SDS. The early phase (up to 1 mM SDS in the absence of salt) involves binding of <10 SDS molecules and likely only involves local rearrangements or simple binding of individual SDS molecules to the native state. Between 1 and 3 mM SDS, up to 20 more SDS molecules bind, leading to small clusters on the protein surface, at which the protein undergoes a conformational change, according to both Trp fluorescence and circular dichroism (CD) spectroscopy, which can be monitored kinetically and leads to an exponential increase in the rate constant with [SDS].21 At 3 mM SDS, bulk micelles start to form and the rate of denaturation stays constant right up to 150 mM SDS, after which it starts to rise again, probably due to the formation of more cylindrical or “sausage-shaped” micelles, which alter the kinetics of SDS–protein interactions.20 Higher ionic strength promotes formation of these cylindrical micelles, probably by screening the electrostatic repulsion between the sulfate head groups and reducing the CMC, and this also leads to a more steady increase in the rate of unfolding with [SDS], abolishing the flat plateau seen in the absence of salt.14 Thus, conditions that modulate the strength of protein–surfactant interactions clearly affect the mechanism of denaturation.
One major drawback with measurements performed at the ensemble level is that they only describe average conformational changes and fail to provide insights into the heterogeneity of these transitions. This is particularly distressing when studying protein–surfactant interactions where one expects a complex mixture of different interconverting species due to the many types of interactions that occur between the protein and the surfactant. Here we address this gap of knowledge by the use of single-molecule Förster resonance energy transfer (smFRET) spectroscopy, which in conjunction with ensemble spectroscopic techniques allows us to uniquely assess the structural heterogeneity and conformational dynamics of S6 upon SDS denaturation. smFRET,23,24 owing to its remarkable sensitivity and temporal resolution, is a powerful technique for disentangling intricate folding scenarios and extracting folding kinetics in surfactant micelles from equilibrium measurements on timescales from hours down to nanoseconds,25,26 without the need of synchronisation or complications arising from the superposition of micellar reorganization.27,28 Because the signal in smFRET measurements is recorded on single molecules in the absence of ensemble averaging, subpopulation-specific conformational heterogeneity can be resolved and nanoscale dynamics can be observed, thus providing a new route for obtaining insights into the denaturation mechanism of S6 by chaotropes and surfactants. Our data obtained herein reveal that the unfolding mechanism in SDS is strikingly different and structurally and temporally convoluted in comparison to the denaturation in GdmCl. In contrast to the reversible two-state unfolding behaviour in GdmCl characterised by kinetics on the timescale of seconds, SDS demonstrated not one, but four distinct regimes of interactions with S6, dependent on the surfactant concentration. These lead to a multitude of different species of unfolded states with different levels of compactness, caused by a complex interplay with SDS monomers and micelles. The region just below and above the CMC is particularly complex and full of coexisting species but additional species continue to form and disappear at significantly higher concentrations, indicating a continually changing and complex landscape of interactions between protein and surfactant.
Results and discussion
Ensemble stopped-flow kinetic analysis indicates complex unfolding behaviour of S6 in SDS
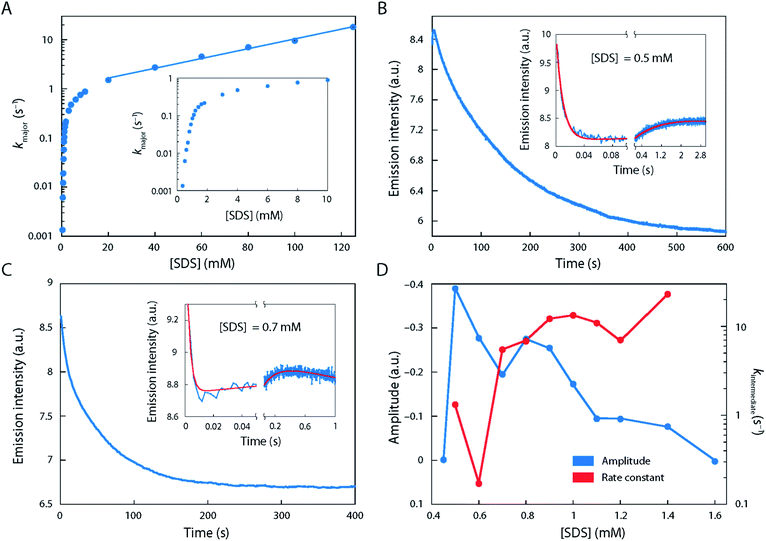
The starting point of our investigations, which prompted us to study the unfolding behaviour of S6 by single-molecule methodology, was the remarkable complexity observed in ensemble stopped-flow kinetic measurements. Fig. 1 shows the kinetic time profiles of S6 as a function of [SDS], recorded from around 0.35 to 125 mM SDS with a large number of data points recorded around the CMC of SDS (under our buffer conditions, i.e. 50 mM TRIS, 150 mM NaCl, pH 7.4, CMCSDS = 1.1 mM, see Fig. S1†). At all [SDS], we observed a major unfolding relaxation phase, seen as a reduction in fluorescence over time (i.e., a positive amplitude Amajor), whose associated rate constant kmajor increases monotonically with [SDS]. kmajor showed an almost 100-fold increase from 0.35 mM SDS (kmajor ∼ 0.001 s−1) up to 1 mM SDS (kmajor ∼ 0.1 s−1), after which there was a decreasingly steep rise up to ∼10 mM SDS (kmajor ∼ 1 s−1) (Fig. 1A). Between 10 and 125 mM SDS, kmajor increased much more slowly, eventually reaching ∼20 s−1.In addition to this major phase, three additional phenomena were observed at low [SDS]:
(i) Between 0.35 and 0.7 mM SDS, there was a second 20-fold faster relaxation phase, also with a positive amplitude Aminor, with a magnitude ∼30–40% of Amajor (data not shown). Between 0.8 and 1.0 mM SDS, the two relaxation phases came close to each other in rate constants (∼2–3-fold difference in magnitude), making it difficult to distinguish them. Above 1.0 mM SDS, there was (along with the major phase) consistently a slow phase with rate constant <0.02 s−1 with a small and positive amplitude.
(ii) A very fast phase with rate constants between 60 and ∼300 s−1 and positive amplitude was observed just above 0.35 mM SDS (cfr. Fig. 1B and C). It showed the greatest magnitude around 0.5–0.6 mM SDS, after which it gradually declined and disappeared by 1.1 mM SDS.
(iii) Between 0.5 and 1.4 mM SDS, there was also an intermediate phase with a negative amplitude over the first few seconds (rate constant kintermediate 0.1–22 s−1) before the major phase took over (examples in Fig. 1B and C; summarised in Fig. 1D). The length of this burst phase was around 5–10 s at 0.6 mM SDS, but it declined to 1.0 s at 1.0 mM SDS and disappears above 1.4 mM SDS.
Thus, we conclude that there is a complex series of kinetic developments at these [SDS], indicating possible heterogeneity which is hard if not impossible to access in ensemble methods. We will now examine to what extent smFRET can shed light on these phenomena.
smFRET unveils structural heterogeneity and complex unfolding behaviour of S6 in SDS
For our smFRET analysis, a fluorescently labelled S6 variant (Fig. 2A) was diluted to 10 pM in a buffer containing 0–300 mM SDS. Fluorescence bursts from a large number of individual S6 molecules diffusing through the confocal detection volume were analysed to generate FRET efficiency (E) histograms (Fig. 2B; see also Fig. S2†). Such histograms report on the diversity of conformations with different levels of compactness (i.e., distance between the two dyes). For example, a protein populating two states with significantly different levels of compactness (e.g., a folded and an unfolded state) would lead to two well-defined peaks in such a histogram. However, a complex non-two state unfolding behaviour of S6 was observed upon addition of SDS. The FRET efficiency histograms exhibited a large degree of conformational heterogeneity when going from 0 mM SDS to very high [SDS]. Various subpopulations classified by a change in mean FRET efficiency positions (〈E〉) and widths (σ) were observed, corroborating the observations from kinetic stopped-flow measurements that S6 unfolding in SDS follows a complex unfolding scheme.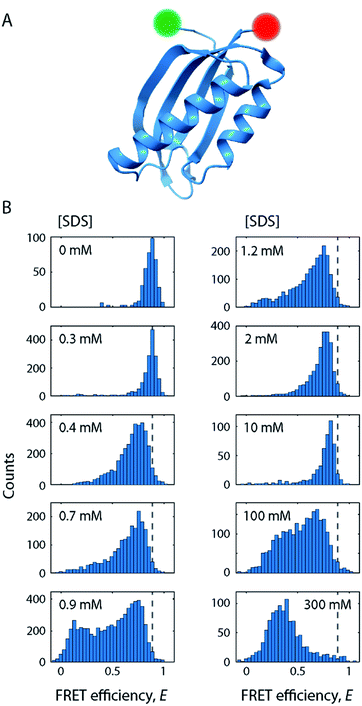 | ||
| Fig. 2 SDS-mediated unfolding of S6 monitored by smFRET. (A) Schematic of S6 labelled at positions 1 and 97 with donor (green) and acceptor (red) fluorophores. (B) FRET efficiency histograms of S6 at increasing [SDS] as indicated in the subpanels. Dashed lines indicate the position of the native state. The full series of histograms is given in Fig. S2.† | ||
In the following, we provide structural and dynamic insights into this heterogeneity by performing in-depth analysis of smFRET data along the unfolding pathway of S6.
The native state of S6 remains well-folded and unperturbed up to 0.3 mM SDS
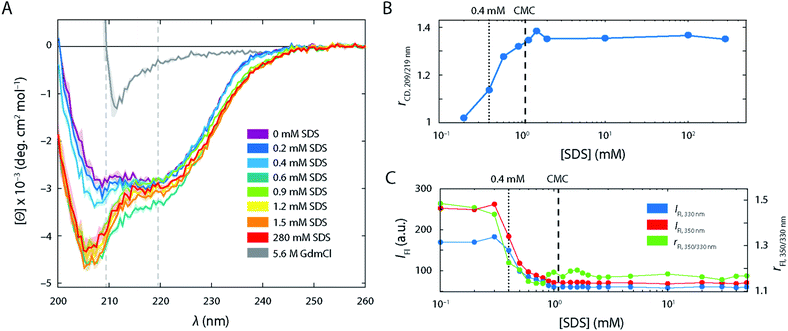
In the absence of SDS, the FRET efficiency histogram (Fig. 2B, top left histogram) exhibited a single narrow peak centred at 〈E〉 = 0.86 (σ = 0.08) as expected for a well-folded globular protein. Increasing [SDS] up to approximately 0.3 mM (Fig. 2B, left column) did not induce any noticeable changes to the shape of the FRET efficiency histograms, suggesting that SDS in this range does not perturb the structure of S6.CD and steady-state intrinsic Trp protein fluorescence experiments support this observation (Fig. 3). CD spectra (Fig. 3A) in the range up to 0.3 mM SDS exhibited minima in the mean molar residual ellipticity [Θ] at 209 and 219 nm, in agreement with the secondary structure content seen in the crystal structure, which comprises a mixture of α-helices and β-sheets. Also, the ratio rCD,209/219 nm of [Θ]-values at 209 and 219 nm did not change until 0.4 mM SDS (Fig. 3B). Furthermore, Trp fluorescence emission spectra of the single Trp residue at position 62 of S6 showed only minimal shifts in this region and the ratio rFl 330/350 nm of fluorescence emission intensities IFl at 350 and 330 nm remained constant (Fig. 3C), indicating no alteration of the chromophores' surroundings (i.e., global protein structure) up to 0.3 mM SDS.
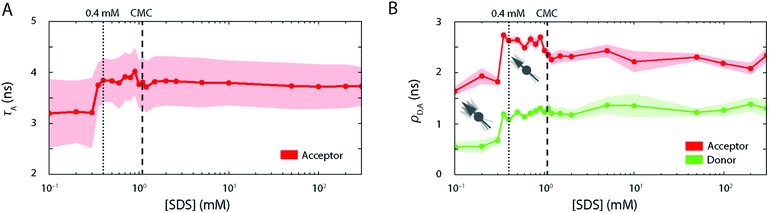
Further, the fluorescence lifetime τA of the attached FRET acceptor fluorophore, which serves as a sensitive reporter of local quenching effects (Fig. 4A), remained constant in the region below 0.3 mM (τA = 3.2 ± 0.6 ns), indicating that the acceptors' molecular environment remains largely unchanged. We also probed the steady-state rotational correlation times ρ of both the acceptor and the donor fluorophores (Fig. 4B) and found that increasing [SDS] from 0 to 0.3 mM led only to a minor slowdown of the rotational correlation time of both the acceptor (from ρA = 1.64 ± 0.08 ns to 1.82 ± 0.08 ns) and donor (from ρD = 0.55 ± 0.08 ns to 0.67 ± 0.09 ns) labels, possibly arising from a gradual association of detergent monomers close to the fluorophores.
In conclusion, at up to 0.3 mM SDS, the protein remains in a well-folded, native state, here denoted as N, that is largely unperturbed by the addition of SDS.
S6 undergoes a structural expansion through an intermediate state with slow kinetics as the CMC of SDS (0.3–1.5 mM SDS) is approached and passed
As we increased the [SDS] within the sub-millimolar range, we observed between 0.3 and 0.4 mM SDS a marked broadening and shift of the high FRET efficiency peak (〈E〉 = 0.86, σ = 0.08) to lower FRET efficiencies (〈E〉 = 0.72, σ = 0.17) (Fig. 2B, see also Fig. S2†). The peak was two-fold broader than the corresponding peak at and below 0.3 mM SDS (i.e., native conditions). In order to exclude that the broadening originates from changes in quantum yield and a restricted rotational freedom of the dyes, we confirmed an unaltered fluorescence lifetime and sufficient rotational averaging at 0.2, 0.6, and 10 mM SDS (Table S1†).This indicated that S6 underwent a structural change to an expanded, more loosely folded state, in the following denoted the expanded state Ex. This state was discernible up to about 1.5 mM SDS, a concentration slightly above the CMC. The range 0.35–1.5 mM SDS corresponded in stopped-flow experiments to the region of the onset of equilibrium unfolding and greatest kinetic complexity with multiple relaxation phases and rate constants ranging from 0.001 s−1 to >100 s−1. In addition to the shift and broadening of the high FRET efficiency peak we observed an increasing population of low and mid FRET efficiencies in the range between 0.7–1.5 mM SDS, as shown for 0.9 mM SDS in Fig. 5A. This region coincides with the intermediate “overshoot” phase observed in stopped-flow experiments with kintermediate = 0.1–22 s−1 (Fig. 1).
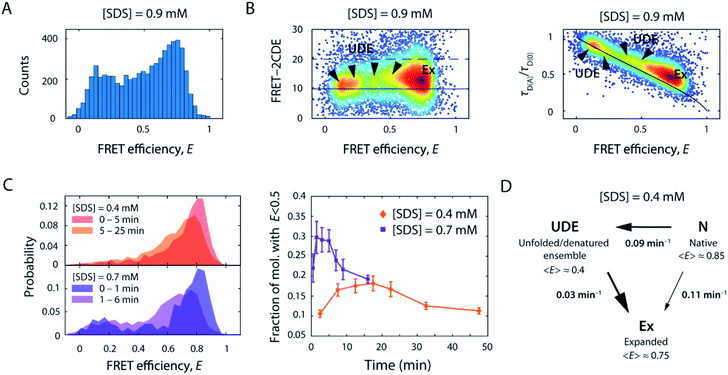 | ||
| Fig. 5 Structural heterogeneity and slow kinetics of S6 below and around the CMC of SDS. (A) FRET efficiency histogram of S6 in the presence of 0.9 mM SDS. (B) 2D scatter plots of FRET-2CDE score versus FRET efficiency (left) and relative donor lifetime (τD(A)/τD(0)) versus FRET efficiency (right), respectively. The normalised density of single molecules is colour coded (scaled from blue to red). In the FRET-2CDE plot, the black lines represent the static FRET line at FRET-2CDE scores of 10 (solid) and the threshold for dynamic events with FRET-2CDE scores bigger than 20 (dashed), respectively. In the relative donor lifetime plot, the solid black line denotes the static FRET line including linker motion.45 Ex, expanded state; UDE, unfolded/denatured ensemble. (C) Temporal change of the FRET efficiency histograms (left) and the time evolution of the fraction of molecules below E = 0.5 for 0.4 mM and 0.7 mM SDS (right). (D) Kinetic reaction scheme as obtained from slow dynamic PDA, employing a two-way three-state reaction scheme to extract unfolding rates, k, between the native N, intermediate unfolded/denatured ensemble UDE, and the expanded state Ex, and their respective mean FRET efficiency values (〈E〉). | ||
The emergence of additional populations was also clearly visible when plotting FRET-2CDE versus FRET efficiency (Fig. 5B, left panel) as well as relative donor lifetime (τD(A)/τD(0)) versus FRET efficiency (Fig. 5B, right panel; exemplarily showing data for 0.9 mM SDS). These plots report on sample dynamics at the observational millisecond to (sub)-microsecond timescale, based on photon density fluctuations in the donor and acceptor channels or fluorescence lifetime fluctuations in the donor channel, respectively. At least three areas of higher density (arrows) were spread between the FRET efficiency range from E = 0.1–0.6, in addition to the most populated state Ex (〈E〉 = 0.72). As seen for the FRET histogram-based analyses in Fig. 2B and S2,† these low and mid FRET states reached maximum occupancy at around 0.9 mM SDS and then decreased as the [SDS] approached 1.5 mM. Notably, the expanded state Ex, and to some extent the populations at mid FRET efficiencies, are displaced from the static FRET line in the relative donor lifetime (τD(A)/τD(0)) versus FRET efficiency plot (Fig. 5B, right panel). This displacement indicates ultrafast dynamics of these states, on microsecond timescales, even faster than the rapid rate constants of 60–300 s−1 seen between 0.35 and 0.7 mM SDS in stopped-flow unfolding experiments. Conversely, populations at low FRET efficiency were found on the static FRET line, indicating that these are static on the observation timescale. Overall, the states at mid and low FRET efficiencies represent a heterogeneous ensemble of unfolded/denatured states. These intermediate states are denoted as the unfolded/denatured ensemble UDE in the following.
To evaluate the kinetic evolution below and around the CMC of SDS between the native state N, the expanded state Ex, and the unfolded/denatured ensemble UDE, time-resolved smFRET unfolding experiments were performed by rapid mixing of fluorescently-labelled S6 into solutions containing 0.4–1.5 mM SDS, after which FRET efficiency histograms were acquired in time intervals of several minutes. In Fig. 5C representative histograms of the timeseries at 0.4 and 0.7 mM SDS are shown. At 0.4 mM SDS, the FRET efficiency distribution gradually shifted from the native state in the first minutes to an expanded peak. Strikingly, in the course of the reaction, the number of bursts in the range below E = 0.5 first increased and then decreased over 30 min, suggesting the presence of a transient state in this region. Comparison with the time course at 0.7 mM SDS (Fig. 5C) reaffirmed this observation, as the normalised distribution of bursts in the low FRET region during the first 10 min corresponded to an additional, extended state that ceased to exist afterwards. These observations are in line with ensemble stopped-flow unfolding kinetics measurements, which show complex double exponential decays, indicating the existence of at least two sets of unfolding transitions at comparable yet distinct timescales.
To assess the timescale of this conformational rearrangement, time series histograms at 0.4 and 0.7 mM SDS were fitted with an adapted probability distribution analysis (PDA) method for quantifying slow kinetics (see ESI†). We employed a three-state model to extract state fractions and interconversion rates between the three states N, UDE, and Ex. Three out of six resulting kinetic rates were negligible (<0.001 min−1), thus the scheme of the reaction transformed into unidirectional transitions between N, UDE, and Ex (Scheme S2†). The extracted rates of the slow dynamic PDA fit at 0.4 mM SDS were kN→UDE = (0.09 ± 0.006) min−1, kUDE→Ex = (0.030 ± 0.002) min−1 and kN→Ex = (0.108 ± 0.007) min−1 for the N-to-UDE, UDE-to-Ex and N-to-Ex transitions, respectively. At 0.7 mM SDS the rates were found to be kN→UDE = (0.22 ± 0.07) min−1, kUDE→Ex = (0.15 ± 0.04) min−1 and kN→Ex = (0.39 ± 0.07) min−1. These values were close to the average weighted rate constant of 0.112 min−1 measured by stopped-flow. Accordingly, at 0.4 mM SDS the unfolding occurred with similar transition probability either directly from the native state N to the expanded state Ex or via the intermediate unfolded/denatured ensemble UDE. In the latter scenario, the transition was significantly slower as the molecules spent an average dwell time of ∼33 min in the intermediate state UDE. On the contrary, at 0.7 mM SDS the direct transition to the expanded state Ex appeared to be more favourable. In addition, the unfolding transition via UDE was accelerated with a decreased dwell time of ∼7 min. Both, N-to-UDE and UDE-to-Ex, transition rates increased 2.4–5 times with 0.4 to 0.7 mM [SDS], respectively. This agrees well with ensemble measurements, which indicated a ∼5-fold acceleration of unfolding (increase in kmajor from 0.112 to 0.57 min−1) for the same change in SDS.
The large changes in S6 tertiary structure and dynamics between 0.3 and 1.0 mM SDS as observed by smFRET were accompanied by secondary structure alterations (Fig. 3A). When going from 0.2 mM SDS to 0.4 mM SDS and above, a strong minimum at 207 nm in CD spectra emerged while the minimum at 220 nm becomes less pronounced. This is in line with the ability of SDS to promote the formation of α-helices in the regions which used to be β-sheets in the native conformation29,30 and the ability of SDS to denature proteins into an α-helical rich state with random coil elements.31 The alteration of the secondary structure was also reflected in the rCD-ratio (Fig. 3B). The steep increase in rCD at 0.4 mM matched the emergence of the extended state Ex in the FRET efficiency histograms. The ratio increased with [SDS] up to 1.0–1.5 mM (i.e., slightly beyond the CMC of SDS (1.1 mM)). Above these concentrations the ratio values levelled off. This indicated that the major secondary structure loss and/or rearrangements into α-helical and random coil structures occurred at sub-CMC conditions, and no further changes were observed after the onset of micellization.
Intrinsic fluorescence parameters, which rely on the single Trp in position 62, also dramatically changed in the range between 0.35 and 1.0 mM SDS (Fig. 3C). Steady-state intrinsic Trp protein fluorescence experiments showed a strong drop of both absolute emission at 350 and 330 nm, as well as the ratio between the two emission wavelengths rFL which decreases from 1.5 down to 1.15, in line with the reported blue shift of the Trp fluorescence spectrum at these conditions.20 This suggests that the microenvironment of Trp turns less polar upon expansion of S6, i.e. less accessible to water molecules,32 which implies an increasing association of Trp with the hydrophobic tails of the surfactants. This situation did not change upon further titration with SDS, indicating that Trp is embedded within the micelle core above the CMC. There were minor fluctuations of rFl in the range between 0.9–1.5 mM SDS (Fig. 3C), which may recapitulate the structural heterogeneity observed in smFRET experiments in this range, seen as a set of low and mid FRET states at this set of conditions.
Further, the lifetime τA of the attached FRET acceptor fluorophore increased to 3.8 ± 0.5 ns (Fig. 4A), hinting at a larger spatial separation of the acceptor fluorophore and any nearby quencher (e.g., Trp62), likely caused by the overall expansion of the protein as seen in FRET histograms. τA values in this range are associated with fluctuations in the acceptor lifetime, which likely underline the heterogeneity of S6 states in this SDS range. Similarly, ρ increased strongly for both dyes in the range of 0.35–1.1 mM SDS (Fig. 4B) and saturated around ρA = 2.6 ± 0.2 ns and ρD = 1.2 ± 0.15 ns for the acceptor and donor fluorophores, respectively. This suggests a more extensive binding of SDS molecules to the local environment of the fluorophores, and, by extension, to the protein. Beyond the CMC of SDS, ρA dropped to 2.2 ± 0.2 ns, while ρD is not affected by a further increase in [SDS].
S6 is compact and densely packed in SDS micelles above the CMC and up to 10 mM SDS
Strikingly, above 1.5 mM SDS, FRET efficiency histograms morph into more and more compact, unimodal distributions (Fig. 2B, right column). They appear very similar to the ones seen in the native state, but differed by exhibiting slightly lower mean FRET efficiency values. This culminated at 10 mM SDS, where the peak with 〈E〉 = 0.80 (σ = 0.06) was most compressed. The peak shape was narrower than the one of the expanded state Ex (σ = 0.06 vs. 0.17 at 0.4 mM SDS) and had a lower mean FRET efficiency than the one corresponding to the native fold N (〈E〉 = 0.80 vs. 〈E〉 = 0.86 at 0 mM SDS), suggesting a new distinct conformation of unfolded S6 at 10 mM SDS. SDS micelles were previously shown to denature S6,20 therefore a plausible structural explanation for the narrow high FRET peak is that the denatured protein is densely packed into micelles. We shall refer to this state in the following as the compact state C. The transitions observed in this regime have parallels to ensemble data. In the same range up to 10 mM SDS, the stopped-flow ensemble unfolding kinetics became dominated by a single exponential decay, indicating a streamlining of the unfolding process to one dominating pathway. At 10 mM, we also observed a transition from a steep, hyperbolic rise in kmajor with [SDS] (<10 mM SDS) to a more linear increase (>10 mM SDS, Fig. 1A).The secondary structure of state C at ≥1 mM SDS contains a large amount of α-helical and some random coil elements (Fig. 3A). This suggests that S6 undergoes “reconstructive denaturation”33 within the micelle around and above the CMC. Interestingly, intrinsic Trp fluorescence supported a micellar embedment as rFl remained largely unchanged and similar to values around and above the CMC (Fig. 3C). Further, the steady-state rotational correlation times ρA,D and τA remained largely unchanged above 1 mM SDS (Fig. 4A and B), while FRET efficiencies showed a strong compaction of S6 in this range, particularly close to 10 mM SDS. This indicates that SDS monomers in the compact state C efficiently shield direct chromophore interactions and thus prevent acceptor quenching, as we observed it in the native state.
S6 expands to a dynamic state with millisecond kinetics at high micellar density
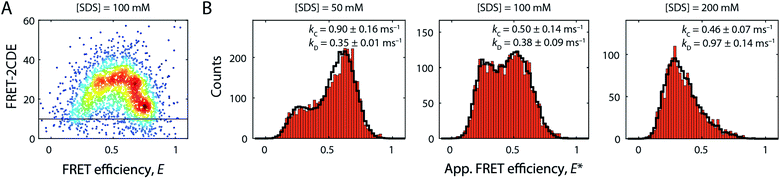
Beyond 50 mM SDS, another remarkable characteristic of S6 became apparent (Fig. 2B, right column). FRET efficiency histograms in this high [SDS] regime exhibited bimodal distributions, where the high FRET efficiency peak gradually converted to a low FRET efficiency state, indicating a two-state interconversion behaviour. Notably, the two states in the transition region were not well-separated, but connected by a pronounced bridge-like population.28 FRET-2CDE analysis revealed that this interconnecting population arises from dynamics on the millisecond timescale (∼1 ms−1) as implied by the arc-shaped distributions in the FRET-2CDE plots (e.g., at 100 mM SDS in Fig. 6A). FRET-2CDE allowed us to separate dynamic and static molecules by a cut-off at FRET-2CDE = 12 (Fig. S4†). This filtering revealed the two states of S6, one at high FRET efficiencies, identical to the compact state C centred around 〈E〉 = 0.80 with σ = 0.06, and a low FRET efficiency peak centred around 〈E〉 = 0.23 with σ = 0.10, denoted in the following as the denatured state D, given the high degree of secondary structure even at very high [SDS] (Fig. 3A).To quantify the kinetic rate constants between these two states, histograms at 50, 100, and 200 mM SDS were subjected to dynamic two-state probability distribution analysis (dPDA) (see ESI†). This approach uses a Monte Carlo simulation-based method to estimate kinetic rates.25 Resulting dPDA histogram fits and extracted collapse/denaturation rates are shown in Fig. 6B. By increasing the [SDS] from 50 to 200 mM, the collapsing rates kC decreased two-fold from 0.9 ± 0.16 to 0.46 ± 0.07 ms−1. Conversely, the denaturation rates kD exhibit an increase from 0.35 ± 0.01 to 0.97 ± 0.14 ms−1 in the same regime. Thus, both rates show a strong dependency on [SDS]. Note that these interconversions are significantly more rapid than the unfolding rates we observe in ensemble kinetic experiments in this range (rate constants between 2 and 20 s−1, i.e. half-lives of 50–500 ms, Fig. 1A). This is likely a consequence of smFRET and stopped-flow experiments probing different structural transitions. During smFRET experiments, we probe steady-state equilibrium kinetics between two conformations (i.e., between states C and D); a conformational change that is inaccessible to stopped-flow experiments. Stopped-flow experiments, conversely, probe the entire unfolding process from the native state to the ensemble of states at the final [SDS]; hence, structural processes that occur transiently during bulk unfolding may not necessarily be populated to significant levels under steady-state conditions. Nonetheless, overall, S6 shows a significant, almost 6 × 105-fold speed-up in unfolding in smFRET experiments and a ∼2 × 104 increase in ensemble experiments. This disparity in rate constants likely reflects the difference between steady-state dynamics obtained from smFRET and the gross shifts in ensemble populations caused by addition of SDS in stopped-flow measurements.
Comparison to unfolding in the chemical denaturant GdmCl
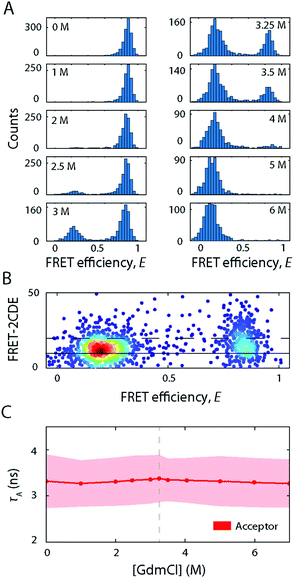
Having explored the structural dynamics and conformational heterogeneity involved in the SDS-induced unfolding of S6, we sought to characterise how unfolding of S6 by a chaotropic chemical denaturant compares to unfolding by SDS. To this end, we exposed labelled S6 to increasing [GdmCl] and performed smFRET experiments. Representative FRET efficiency histograms are depicted in Fig. 7A. S6 exhibited a two-state folding behaviour in GdmCl as manifested by the coexistence of two FRET efficiency peaks representing the folded (〈E〉 = 0.85) and unfolded states (〈E〉 = 0.15). There is no evidence for a compact off-pathway intermediate, which can only be observed indirectly during refolding experiments19,22,34 and is not observed under equilibrium conditions. With increasing GdmCl concentration, the occupancy of the folded state with high FRET efficiency gradually decreased, while the low-FRET peak representing the unfolded state increased. The mid-point at which unfolding occurs was around 3.2 M GdmCl. The two populations were well separated along the GdmCl concentration series. There is a slight decrease in the average FRET efficiency of the unfolded state between 4 and 7 M GdmCl (〈E〉 = 0.22 at 4 M, 〈E〉 = 0.15 at 7 M), suggesting an expansion of the unfolded state with increasing GdmCl concentration, as observed for many globular proteins.35 The absence of a bridge-like population between the folded and unfolded states indicated that the kinetic rates were slower than the millisecond observation timescale of confocal smFRET measurements. This was confirmed by FRET-2CDE scoring, where no arc-like shaped population connecting the two states was observed (Fig. 7B). These observations are in agreement with stopped-flow kinetic measurements where second-to-minute timescale rate constants of 0.032 s−1 were observed at mid-point conditions (3.25 M GdmCl).17In CD experiments, the protein exhibited a significant decrease in secondary structure when exposing the protein to high [GdmCl] where only the unfolded state peak appears in smFRET experiments (Fig. 3A). The spectrum at 5.6 M GdmCl showed a single drop between 220 and 210 nm, which suggests a fully unfolded random coil polypeptide structure. Further, τA remained largely constant (Fig. 7C), indicating that the acceptor's molecular environment remains largely unchanged at increasing [GdmCl] (τA = 3.3 ± 0.5 ns).
Taken together, S6 unfolding in GdmCl follows a simple two-state behaviour characterised by kinetics on the timescale of seconds. The folded and unfolded states are the only states present at the equilibrium unfolding conditions studied here with the unfolded state being an ensemble of unfolded protein molecules that behave like fully unfolded random coils.
Conclusion
Multiple pathways of S6 unfolding in SDS
A complementary suite of smFRET and ensemble-based spectroscopic methods allowed us to draw a picture of unprecedented detail of the structural heterogeneity and conformational dynamics of S6 unfolding upon SDS denaturation. In contrast to a distinct two-state unfolding with GdmCl, SDS induced complex multi-pathway unfolding of S6, which can be described in four stages. Each of them is described below and summarised in Fig. 8.The large heterogeneity of states in S6 with multiple, even parallel transitions revealed by smFRET are mirrored by the complex kinetic developments in stopped-flow kinetic measurements. However, the exact values of the rate constants observed from ensemble measurements are not matched by the smFRET values. This emphasises that smFRET and ensemble stopped-flow measurements may not probe the exact same reaction coordinate. Species, which occur transiently during bulk unfolding in [SDS], will not necessarily be populated to significant levels under steady-state conditions. Similarly, there is no obvious relationship between a protein's hydrogen exchange at equilibrium and its folding pathway.36 In support of this, a compact state known to accumulate transiently during refolding of S6 from the GdmCl-denatured state22 is not observed in equilibrium FRET distributions of S6 at 0 M GdmCl. It is also important to note that rapid stopped-flow mixing leads to dynamic rearrangements of both protein and micellar structures. These processes can occur on similar timescales and are thus difficult if not impossible to separate. Nonetheless, the overall correspondence between smFRET and ensemble measurements in levels of high and low complexity at different [SDS] indicates that they both reflect existence of multiple types of denatured states.
Taken together, the rich conformational heterogeneity and timescales involved in the SDS-induced unfolding of S6 contrast with the simple two-state unfolding in GdmCl. This showcases the differences in the denaturation mode of these two denaturants and the diversity of mechanisms by which surfactants interact with proteins.2 The basis for this complexity resides of course in the self-assembling abilities of surfactants, which are completely lacking in chemical denaturants. This means that surfactants can bind, for example, in clusters of different sizes on protein surfaces well below the CMC,8 and the sizes of these clusters and their impact on protein structure can change markedly with [SDS].21 Furthermore, proteins can rearrange around micelles as part of the unfolding process, as shown by recent synchrotron SAXS studies.16
The intricacy of SDS-mediated unfolding makes it imperative to study this process by multiple, complementary approaches. It is here that single-molecule measurements add a unique perspective on the nanoscale structural and dynamic complexity of proteins in surfactant environments to ensemble measurements of secondary and tertiary rearrangements. Single-molecule methods provide direct access to molecular heterogeneities and can be used as precise measurement tools for structural characterizations.40 In this study, we monitored with smFRET one reaction coordinate—the distance between residue 1 and 97—and found a large number of unfolded states that show significant differences in their structural expansion by several nanometres. Yet this can be expanded to more residues allowing a three-dimensional trilateration of protein conformations41 or disordered states of proteins42 that are hard to characterise with classical structural tools that often require high sample homogeneities.
Experimental
Protein design and production
A double-Cys variant of the 101-amino-acid-residue protein S6 from Thermus thermophilus was constructed in which Met1 and Phe97 were replaced with Cys. The DNA sequence of the S6 gene (constructed using Escherichia coli-optimised codons) was custom synthesised (GenScript) and the fragment cloned into a pET28a expression vector using NcoI/XhoI restriction sites. This cloning resulted in an open reading frame without any affinity tag and the sequence Met-Gly placed immediately before Cys1. The protein was expressed in E. coli BL21 (DE3) and purified by anion-exchange chromatography followed by size-exclusion chromatography to >95% purity as described.17 For smFRET experiments, the protein was site-specifically labelled via thiol–maleimide chemistry with maleimide-functionalised FRET donor (ATTO 532, Atto-Tec) and acceptor (Abberior STAR 635P, Abberior) fluorophores, following standard procedures. The labelled protein was separated from unbound dyes by size-exclusion chromatography.Stopped-flow fluorescence spectroscopy
1.8 μM (ca. 20 μg ml−1) unlabelled double-Cys S6 in 50 mM TRIS pH 7.4, 150 mM NaCl and 2 mM tris(2-carboxyethyl)phosphine (TCEP) was mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with SDS in the same buffer at 25 °C using a Chirascan spectrophotometer with stopped-flow accessory (Applied Photophysics) equipped with a Xenon lamp. Trp fluorescence was monitored through excitation at 283 nm with a 2 nm bandwidth and emission measured using a 320 nm cut-off filter. Data were collected in logarithmic time mode over 5–1000 s (depending on the [SDS]) and fitted using single or double exponential decays. Double decays were used when a single exponential decay was unable to describe the data sufficiently well. This was the case when (i) there were phases with amplitudes of opposing signs (e.g., an initial rapid decrease in signal followed by an increase in signal) or (ii) when oscillating residuals from a single exponential fit indicated the need for an extra phase. In the latter case the two rate constants had to differ by at least a factor 10 in order to be considered an acceptably robust double exponential fit.
1 with SDS in the same buffer at 25 °C using a Chirascan spectrophotometer with stopped-flow accessory (Applied Photophysics) equipped with a Xenon lamp. Trp fluorescence was monitored through excitation at 283 nm with a 2 nm bandwidth and emission measured using a 320 nm cut-off filter. Data were collected in logarithmic time mode over 5–1000 s (depending on the [SDS]) and fitted using single or double exponential decays. Double decays were used when a single exponential decay was unable to describe the data sufficiently well. This was the case when (i) there were phases with amplitudes of opposing signs (e.g., an initial rapid decrease in signal followed by an increase in signal) or (ii) when oscillating residuals from a single exponential fit indicated the need for an extra phase. In the latter case the two rate constants had to differ by at least a factor 10 in order to be considered an acceptably robust double exponential fit.
Circular dichroism spectroscopy
S6 samples were prepared as for stopped-flow fluorescence experiments at a final protein concentration of 0.2 mg ml−1 (ca. 20 μM) at different [SDS] in the range of 0–280 mM and at 5.6 M GdmCl. CD spectra were recorded on a Chirascan-plus spectrometer (Applied Photophysics) at room temperature in the wavelength range 200–260 nm with a step size of 0.5 nm, a spectral bandwidth of 1 nm, a scanning sped of 30 nm per min, a digital integration time of 1 s, and a 1 mm path length quartz glass cuvette. Each sample was scanned 5 times and averaged. Spectra were blank corrected. Raw ellipticity signal was converted to mean residue ellipticity [Θ] using standard formulae.Dynamic light scattering
The CMC of SDS at 150 mM NaCl and pH 7.4 was measured on a Zetasizer Nano S (Malvern) in a low-volume quartz cuvette. Each of the [SDS] in the 0–5 mM range was measured three times and the Z-average was taken as the value of the particle size.Single-molecule FRET
Experiments were carried out using a custom-built dual-colour, dual-polarization single-molecule confocal fluorescence microscope as described previously.28,42 Measurements were performed at room temperature on freely diffusing molecules in a custom-built sample chamber by placing the confocal volume into solution at an axial position 60 μm above the surface of the cover slide. Labelled S6 was diluted prior to measurements from stock solutions into buffer (150 mM NaCl, 50 mM TRIS pH 7.4, 2 mM TCEP) containing the respective denaturing agent (i.e., SDS or GdmCl) to a final protein concentration of ∼75 pM to obtain appropriate burst rates under single-molecule conditions. The illumination power was 110 μW for excitation of the donor dye and 90 μW for excitation of the acceptor dye, both measured before the objective.Analysis of single-molecule FRET experiments
Data analysis was performed with custom-written Matlab scripts (Mathworks) following procedures described previously.25,28 Briefly, after identification of bursts with a search algorithm (using a maximum interphoton time of 50 μs, a minimum total number of 70 photons, and a Lee filter of 4), FRET efficiency histograms with bin widths of 0.033 were constructed from bursts exhibiting a stoichiometry ratio of S = 0.2–0.75 and an alternating laser excitation-two-channel kernel-based density distribution estimator (ALEX-2CDE) score <10.44 Additionally, an asymmetric burst filter was applied using a channel-asymmetry time of <50 μs.45 Individual E-values were corrected for background, direct acceptor excitation (α = 0.075), donor cross-talk in the acceptor channel (β = 0.029), as well as differences in detector efficiencies and quantum yields between the dyes (γ = 0.535), as described previously.28 Apparent FRET efficiencies (E*) used in probability-distribution analysis (PDA) were calculated without applying the aforementioned corrections. Calculation of fluorescence lifetimes (τD,A) and anisotropies (rD,A) as well as steady-state rotational correlation times (ρD,A) were performed as described previously;26 also plots of relative donor fluorescence lifetime (τD(A)/τD(0)) versus E were created as described.43 Analysis of single-molecule bursts for millisecond dynamics was done by FRET-2CDE analysis.44 Quantification of millisecond interconversion dynamics from FRET efficiency histograms was performed by dPDA as described in the ESI† and detailed elsewhere.25,28 Details about PDA kinetic analysis procedures used to fit the time-resolved kinetic unfolding experiments are also provided in the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank all members of the Otzen and Schlierf laboratories for discussions. We thank Dr Anja Buttstedt (TU Dresden) for her support with CD measurements. This research was supported by the Deutsche Forschungsgemeinschaft with grant SCHL 1896/3-1 to M. S. and the Independent Research Foundation Denmark | Technology and Production (grant 9041-00123B) to D. E. O.References
- O. Kirk, T. V. Borchert and C. C. Fuglsang, Curr. Opin. Biotechnol., 2002, 13, 345–351 CrossRef CAS PubMed.
- D. Otzen, Biochim. Biophys. Acta, Proteins Proteomics, 2011, 1814, 562–591 CrossRef CAS PubMed.
- D. E. Otzen, Curr. Opin. Colloid Interface Sci., 2015, 20, 161–169 CrossRef CAS.
- J. L. England and G. Haran, Annu. Rev. Phys. Chem., 2011, 62, 257–277 CrossRef CAS PubMed.
- D. R. Canchi and A. E. García, Annu. Rev. Phys. Chem., 2013, 64, 273–293 CrossRef CAS PubMed.
- A. Yonath, A. Podjarny, B. Honig, A. Sielecki and W. Traub, Biochemistry, 1977, 16, 1418–1424 CrossRef CAS PubMed.
- C. Tanford, The Hydrophobic Effect: Formation of Micelles and Biological Membranes, Wiley, New York, 2nd edn, 1980 Search PubMed.
- K. K. Andersen, C. L. Oliveira, K. L. Larsen, F. M. Poulsen, T. H. Callisen, P. Westh, J. S. Pedersen and D. Otzen, J. Mol. Biol., 2009, 391, 207–226 CrossRef CAS PubMed.
- H. G. Mortensen, J. K. Madsen, K. K. Andersen, T. Vosegaard, G. R. Deen, D. E. Otzen and J. S. Pedersen, Biophys. J., 2017, 113, 2621–2633 CrossRef CAS PubMed.
- Y. Sun, P. L. O. Filho, J. C. Bozelli, J. Carvalho, S. Schreier and C. L. P. Oliveira, Soft Matter, 2015, 11, 7769–7777 RSC.
- J. Narayanan, A. S. Abdul Rasheed and J. R. Bellare, J. Colloid Interface Sci., 2008, 328, 67–72 CrossRef CAS PubMed.
- L. Giehm, C. L. P. Oliveira, G. Christiansen, J. S. Pedersen and D. E. Otzen, J. Mol. Biol., 2010, 401, 115–133 CrossRef CAS PubMed.
- J. D. Kaspersen, A. Søndergaard, D. J. Madsen, D. E. Otzen and J. S. Pedersen, Biophys. J., 2017, 112, 1609–1620 CrossRef CAS PubMed.
- D. E. Otzen, Biophys. J., 2002, 83, 2219–2230 CrossRef CAS PubMed.
- A. H. Poghosyan, N. P. Schafer, J. Lyngsø, A. A. Shahinyan, J. S. Pedersen and D. E. Otzen, Protein Eng., Des. Sel., 2019, 32, 175–190 CrossRef CAS PubMed.
- J. N. Pedersen, J. Lyngsø, T. Zinn, D. E. Otzen and J. S. Pedersen, Chem. Sci., 2020, 11, 699–712 RSC.
- D. E. Otzen, O. Kristensen, M. Proctor and M. Oliveberg, Biochemistry, 1999, 2, 6499–6511 CrossRef PubMed.
- D. E. Otzen and M. Oliveberg, J. Mol. Biol., 2002, 317, 613–627 CrossRef CAS PubMed.
- D. E. Otzen and M. Oliveberg, J. Mol. Biol., 2001, 313, 479–483 CrossRef CAS PubMed.
- D. E. Otzen and M. Oliveberg, J. Mol. Biol., 2002, 315, 1231–1240 CrossRef CAS PubMed.
- D. E. Otzen, L. W. Nesgaard, K. K. Andersen, J. Høeg, G. Christiansen, H. Doe and P. Sehgal, Biochim. Biophys. Acta, 2008, 1784, 400–414 CrossRef CAS PubMed.
- D. E. Otzen and M. Oliveberg, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11746–11751 CrossRef CAS PubMed.
- B. Schuler and H. Hofmann, Curr. Opin. Struct. Biol., 2013, 23, 36–47 CrossRef CAS PubMed.
- G. Krainer, S. Keller and M. Schlierf, Curr. Opin. Struct. Biol., 2019, 58, 124–137 CrossRef CAS PubMed.
- G. Krainer, A. Hartmann, A. Anandamurugan, P. Gracia, S. Keller and M. Schlierf, J. Mol. Biol., 2018, 430, 554–564 CrossRef CAS PubMed.
- G. Krainer, P. Gracia, E. Frotscher, A. Hartmann, P. Gröger, S. Keller and M. Schlierf, Biophys. J., 2017, 113, 1280–1289 CrossRef CAS PubMed.
- E. Frotscher, G. Krainer, A. Hartmann, M. Schlierf and S. Keller, ACS Omega, 2018, 3, 12026–12032 CrossRef CAS PubMed.
- A. Hartmann, G. Krainer, S. Keller and M. Schlierf, Anal. Chem., 2015, 87, 11224–11232 CrossRef CAS PubMed.
- C. Michaux, N. C. Pomroy and G. G. Privé, J. Mol. Biol., 2008, 375, 1477–1488 CrossRef CAS PubMed.
- W. L. Mattice, J. M. Riser and D. S. Clark, Biochemistry, 1976, 15, 4264–4272 CrossRef CAS PubMed.
- K. Takeda, H. Sasaoka, K. Sasa, H. Hirai, K. Hachiya and Y. Moriyama, J. Colloid Interface Sci., 1992, 154, 385–392 CrossRef CAS.
- J. T. Vivian and P. R. Callis, Biophys. J., 2001, 80, 2093–2109 CrossRef CAS PubMed.
- B. Jirgensons, Die Makromol. Chemie, Rapid Commun., 1981, 2, 213–217 CrossRef CAS.
- D. E. Otzen, Biochim. Biophys. Acta, Proteins Proteomics, 2005, 1750, 146–153 CrossRef CAS PubMed.
- A. Borgia, W. Zheng, K. Buholzer, M. B. Borgia, A. Schüler, H. Hofmann, A. Soranno, D. Nettels, K. Gast, A. Grishaev, R. B. Best and B. Schuler, J. Am. Chem. Soc., 2016, 138, 11714–11726 CrossRef CAS PubMed.
- J. Clarke, L. S. Itzhaki and A. R. Fersht, Trends Biochem. Sci., 1997, 22, 284–287 CrossRef CAS PubMed.
- X. H. Guo, N. M. Zhao, S. H. Chen and J. Teixeira, Biopolymers, 1990, 29, 335–346 CrossRef CAS PubMed.
- N. J. Turro, X.-G. Lei, K. P. Ananthapadmanabhan and M. Aronson, Langmuir, 1995, 11, 2525–2533 CrossRef CAS.
- M. Samsó, J. R. Daban, S. Hansen and G. R. Jones, Eur. J. Biochem., 1995, 232, 818–824 CrossRef.
- B. Hellenkamp, S. Schmid, O. Doroshenko, O. Opanasyuk, R. Kühnemuth, S. Rezaei Adariani, B. Ambrose, M. Aznauryan, A. Barth, V. Birkedal, M. E. Bowen, H. Chen, T. Cordes, T. Eilert, C. Fijen, C. Gebhardt, M. Götz, G. Gouridis, E. Gratton, T. Ha, P. Hao, C. A. Hanke, A. Hartmann, J. Hendrix, L. L. Hildebrandt, V. Hirschfeld, J. Hohlbein, B. Hua, C. G. Hübner, E. Kallis, A. N. Kapanidis, J.-Y. Kim, G. Krainer, D. C. Lamb, N. K. Lee, E. A. Lemke, B. Levesque, M. Levitus, J. J. McCann, N. Naredi-Rainer, D. Nettels, T. Ngo, R. Qiu, N. C. Robb, C. Röcker, H. Sanabria, M. Schlierf, T. Schröder, B. Schuler, H. Seidel, L. Streit, J. Thurn, P. Tinnefeld, S. Tyagi, N. Vandenberk, A. M. Vera, K. R. Weninger, B. Wünsch, I. S. Yanez-Orozco, J. Michaelis, C. A. M. Seidel, T. D. Craggs and T. Hugel, Nat. Methods, 2018, 15, 669–676 CrossRef CAS PubMed.
- H. Sanabria, D. Rodnin, K. Hemmen, T.-O. Peulen, S. Felekyan, M. R. Fleissner, M. Dimura, F. Koberling, R. Kühnemuth, W. Hubbell, H. Gohlke and C. A. M. Seidel, Nat. Commun., 2020, 11, 1231 CrossRef CAS PubMed.
- A. Borgia, M. B. Borgia, K. Bugge, V. M. Kissling, P. O. Heidarsson, C. B. Fernandes, A. Sottini, A. Soranno, K. J. Buholzer, D. Nettels, B. B. Kragelund, R. B. Best and B. Schuler, Nature, 2018, 555, 61–66 CrossRef CAS PubMed.
- G. Krainer, A. Treff, A. Hartmann, T. A. Stone, M. Schenkel, S. Keller, C. M. Deber and M. Schlierf, Commun. Biol., 2018, 1, 154 CrossRef PubMed.
- T. E. Tomov, R. Tsukanov, R. Masoud, M. Liber, N. Plavner and E. Nir, Biophys. J., 2012, 102, 1163–1173 CrossRef CAS PubMed.
- V. Kudryavtsev, M. Sikor, S. Kalinin, D. Mokranjac, C. A. M. Seidel and D. C. Lamb, ChemPhysChem, 2012, 13, 1060–1078 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary figures and table (Fig. S1–S4 and Table S1) and supplementary methods (Schemes S1 and S2). See DOI: 10.1039/d0sc02100h |
| ‡ These authors contributed equally to this work. |
| § Present address: Centre for Misfolding Diseases, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK. E-mail: E-mail: gk422@cam.ac.uk. |
| ¶ Present address: Department of Physics and Astronomy, LaserLaB Amsterdam, Vrije Universiteit Amsterdam, De Boelelaan 1081, 1081 HV Amsterdam, Netherlands. |
| This journal is © The Royal Society of Chemistry 2020 |