Open Access Article
Open Access ArticleDynamic changes in DNA populations revealed by split–combine selection†
Olga
Plückthun
a,
Julia
Siegl
a,
Laura Lledo
Bryant
a and
Günter
Mayer
 *ab
*ab
aChemical Biology and Chemical Genetics, Life and Medical Sciences (LIMES) Institute, University of Bonn, Gerhard-Domagk-Str. 1, 53121 Bonn, Germany. E-mail: gmayer@uni-bonn.de
bCenter of Aptamer Research and Development (CARD), University of Bonn, Gerhard-Domagk-Str. 1, 53121 Bonn, Germany
First published on 14th August 2020
Abstract
Clickmers are chemically modified aptamers representing an innovative reagent class for developing binders for biomolecules with great impact on therapeutic and diagnostic applications. To establish a novel layer for screening various chemical entities, we developed a split–combine selection strategy simultaneously enriching for clickmers having different modifications. Due to the inherent design of this strategy, dynamic changes of DNA populations are traceable at an individual sequence level. Besides off-rate guided enrichment, the process makes the survival of the sequences most adapted to the applied selection condition observable. The underlying strategy provides unprecedented molecular insight into the selection process, based on which more sophisticated procedures will become pliable in the future.
Introduction
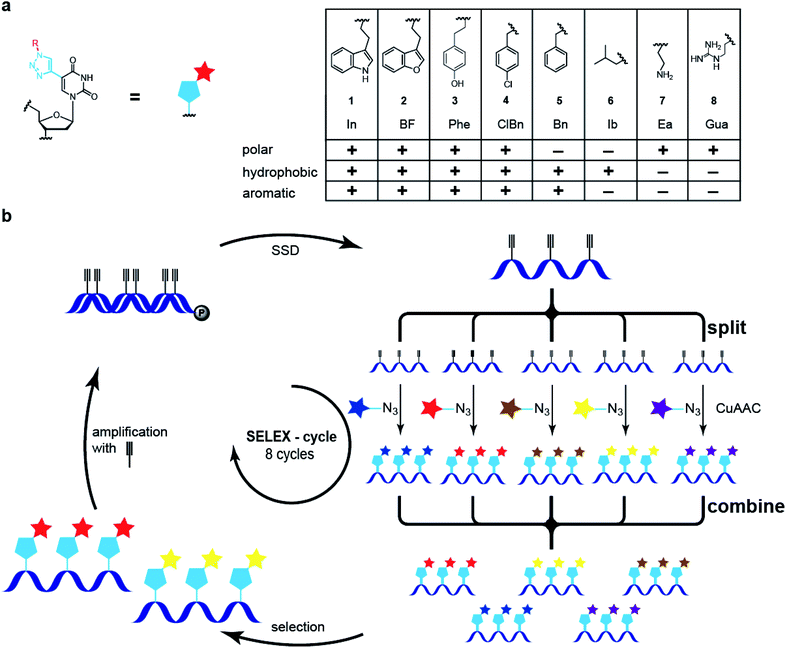
The selection of nucleic acid ligands for defined target molecules using modified nucleobases, novel backbone modifications or an extended nucleotide alphabet has emerged in the past decade.1–8 For example, so-called SOMAmers bearing hydrophobic amino acid side-chain like modifications have been identified for numerous plasma proteins and used in proteomic approaches to study differences in disease vs. normal organismic states. SOMAmers reveal novel structural features, e.g., so-called benzyl-zipper motifs that form hydrophobic interaction networks.3 Likewise, the Hirao and Benner groups established two additional base pairs and subjected DNA libraries that have these base pairs to in vitro selection schemes, yielding aptamers for specific protein targets.4,5 These extended alphabet aptamers have been shown to have high affinities compared to previously reported non-modified DNA aptamers for the same protein targets. Overall, modified DNA aptamers reveal superior characteristics compared to those solely built from canonical nucleotides, e.g., slower off-rates and higher affinity. Most previous examples of nucleobase-modifications make use of one or two6 for selection experiments or three different functionalities, exemplified by the selection of deoxyribozymes.7,8 Recently, we and others established a so-called click-SELEX procedure, in which Cu(I)-catalyzed azide alkyne-coupling (CuAAC) is used to introduce chemical modifications into DNA libraries at the C5-position of uridine or the 2′-position of the deoxyribose position.4–6,9,10 Aptamers obtained by click-SELEX are termed clickmers. Here, we extended this approach towards a split–combine procedure enabling the simultaneous screening of five or even more different modifications, permitting the rapid identification of chemical modifications that best mediate binding to a target molecule. We applied next-generation sequencing (NGS) to analyze the DNA populations along with bioinformatic analysis.11–13 Correlating these data with interaction analysis, the enrichment process was found to be not only driven by specific modifications but also by off-rate kinetic terms. Clickmers having slow off-rates outcompete fast off-rate binders in the population. Notwithstanding, the latter species can be rescued by changing the selection criteria, e.g., limiting availability of modifications. Individual sequence-modification pairs can be traced back in the DNA libraries shedding light on dynamic changes of DNA populations during in vitro selection experiments, correlated with selection conditions.We synthesized a series of azides with distinct properties, e.g., polar, hydrophobic, charged, and/or aromatic (Scheme 1a). These azides were used to separately functionalize a C5-ethynyl-2′-deoxy uridine modified DNA library.12,13 The products of the click reaction were analyzed by nuclease digest followed by liquid chromatography and mass spectrometry.14–17 These analyses reveal a near quantitative modification of the DNA (see ESI†). Subsequently, the five libraries were mixed at equimolar concentrations and subjected to a split–combine click-SELEX procedure (Scheme 1b).9,15 After incubation, separation and the recovery of bound species, the single-stranded DNA of the amplified library was produced and divided into five samples (Scheme 1b, split). Each of them was modified with one of the five azides, separately, and after pooling (Scheme 1b, combine), this library was subjected to the next selection cycle.
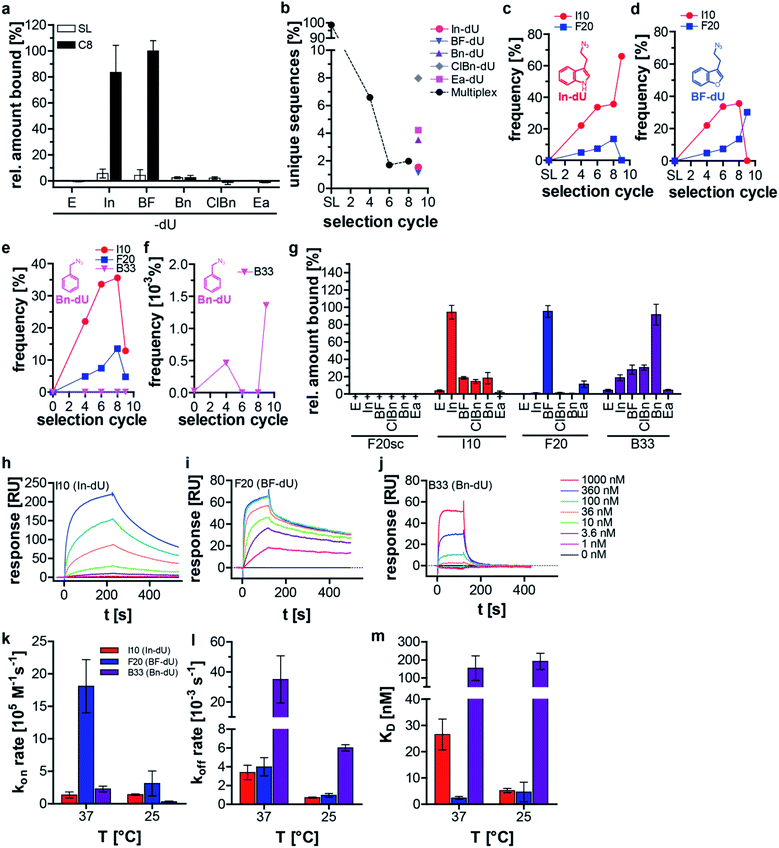
We applied this split–combine SELEX procedure to two different target proteins and started using C3-GFP.15 Besides indole (In), we used benzofuran (BF), benzyl (Bn), 4-chloro-benzyl (ClBn), and ethylamine (Ea), as modifications (ESI Table 1, ESI Fig. 1a and b†). An increase in C3-GFP binding was observed after eight selection cycles only when the DNA was modified with In or BF, whereas neither of the other three modifications (Bn, ClBn, Ea) nor the unmodified library (E) showed C3-GFP binding (Fig. 1a). An additional ninth selection cycle was performed, dubbed deconvolution cycle, in which the ‘combine’-step of the libraries (Scheme 1b) was omitted. Instead, each modified library was subjected separately to a click-SELEX cycle. The purpose of introducing this deconvolution cycle was to increase the copy numbers of sequences that bind to C3-GFP when modified with a specific residue. This procedure would simplify the identification of individual clickmers and assigning them to the specific azide. All obtained libraries were analyzed by next-generation sequencing (NGS), which revealed a reduction of unique sequences in the DNA libraries obtained from cycles 4, 6, and 8 (Fig. 1b). The five DNA libraries obtained from the deconvolution cycle (i.e., selection cycle 9) reveal either a further decrease (when using In or BF) or an increase of the number of unique sequences (when using Bn, ClBn, or Ea) (Fig. 1b). Likewise, the distribution of nucleotides in the 44-nucleotide random region changed strongly from the starting to the enriched libraries of selection cycles 4, 6, and 8 (ESI Fig. 2†). Compared to the library from selection cycle 8, the ninth deconvolution cycle led to either a largely unchanged nucleotide distribution (Ea) or changes that almost reflect single sequences (In, BF), or more distinct variations (Bn, ClBn) (ESI Fig. 2†). The dominating sequences in the population of selection cycle 8 were I10 (33%) and F20 (14%) (Fig. 1c–e, ESI Table 2†). The deconvolution cycle either increased or decreased their copy numbers when using the matching azides (Fig. 1c and d). For example, using In in the deconvolution cycle led to an increase of I10's frequency to 64%, whereas F20's decreased to 0.05% (Fig. 1c, ESI Table 2†). The contrary behavior was observed when using BF. Here, I10's frequency decreased to 0.08% whereas F20's increased to 25% (Fig. 1d, ESI Table 2†). When all other modifications were used, the copy numbers of I10 and F20 decreased, although to different extents (Fig. 1e, ESI Table 2†). The frequency of sequence C1 is 0.9% in selection cycle 8 and was enriched 10-fold when using ClBn or Bn as modifications in the deconvolution cycle (to 9.7% and 9.6%, respectively). When using In, BF or Ea the occurrence of C1 decreased to 0.007% (In) and 0.24 (BF) or slightly increased to 1.9% (Ea) (ESI Table 2†). Interaction studies revealed that C1 preferentially binds to C3-GFP when modified with BF and to a lesser extent modified with ClBn, whereas no interaction was detected when using Bn (ESI Fig. 3†). These data indicate that C1 is preferentially a BF-dependent binder. During the course of the selection, however it was impossible for C1 to compete with F20 as it is the predominant BF-dependent sequence. Only when the selection pressure changed, e.g., by using ClBn C1 gained evolutionary advantages as it also interacts with C3-GFP when modified with this entity whereas F20 and I10, the sequences dominating selection cycle 8, do not. When using Bn in the deconvolution cycle, the most dominant enriched sequences are either relatives of C1, I10 or F20. However, B33 was not detected in selection cycle 8 within the sequencing depth, whereas its copy number increased (0.0014%) strongly in the ninth selection cycle using Bn (Fig. 1f, ESI Table 2†). Interaction analysis reveals binding to C3-GFP of I10, F20 and B33, while F20 revealed the most pronounced click-in dependency (Fig. 1g). When using the aromatic click-in residues (In, BF, and ClBn), the interaction of I10 and B33 with C3-GFP was less intense, but above background. However, interaction intensities were much stronger when using the correctly assigned modifications (Fig. 1g). All sequences did not interact with C3-GFP at the ethynyl-state (E) and a scrambled control sequence, based on F20, also did not reveal any C3-GFP binding properties (Fig. 1g). We next performed kinetic studies using surface plasmon resonance (Fig. 1h–j, Table 1, ESI Fig. 4–9†). These studies demonstrate that I10 and F20 have superior kinetic properties with regard to on-rates (Fig. 1k) and off-rates (Fig. 1l) as well as with regard to the dissociation constant compared to B33 (Fig. 1m). These characteristics underline why B33 is not strongly enriched in the split–combine selection cycles 1–8, and only had a chance to level up, although to low copy numbers, when selected using Bn in the deconvolution cycle.
| T | I10 (In) | F20 (BF) | B33 (Bn) | |
|---|---|---|---|---|
| k on rate [105 M−1 s−1] | 25 °C | 14.0 ± 1.1 | 31.2 ± 19.2 | 3.3 ± 0.8 |
| 37 °C | 13.5 ± 4.8 | 181.0 ± 41.1 | 22.8 ± 4.4 | |
| k off rate [10−4 s−1] | 25 °C | 7.2 ± 0.5 | 9.6 ± 2.0 | 60.1 ± 3.4 |
| 37 °C | 33.9 ± 7.6 | 39.9 ± 9.8 | 350.3 ± 156.9 | |
| K D [nM] | 25 °C | 5.2 ± 0.8 | 4.7 ± 3.7 | 191.2 ± 45.1 |
| 37 °C | 26.5 ± 5.9 | 2.3 ± 0.6 | 153.8 ± 68.4 |
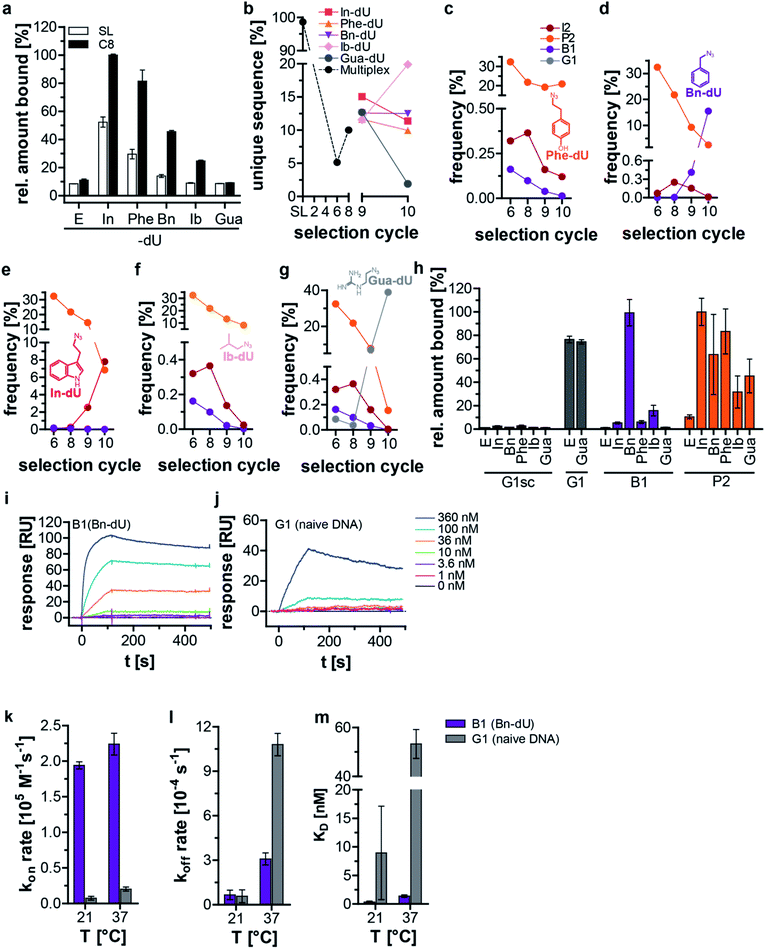
We used streptavidin (SA) as a second target protein in the split–combine SELEX procedure. While DNA aptamers have been described for SA,18 chemically modified aptamers and clickmers are yet not known. As for C3-GFP we used the indole (In) and benzyl (Bn), but instead of BF, ClBn and Ea we employed phenol (Phe), isobutyl (Ib) and guanidine (Gua) as modifications (Scheme 1a, ESI Table 3, Fig. 1c and d†). These residues exemplify pleiotropic characteristics, e.g., aromatic, polar, and non-polar allowing various interaction modalities of the DNA with SA. After eight split–combine selection cycles, the enriched library revealed an increase in SA binding compared to the respective starting library when using the clicked-in modifications In, Phe, Bn, or Ib (Fig. 2a). No improvement of interaction properties was observed when the library was modified with Gua (Fig. 2a). Of note, the interaction intensity of the starting library strictly depends on the nature of the click-in residue (Fig. 2a). While aromatic and polar residues (In, Phe) promote a pronounced interaction of the starting library with SA, the non-polar aromatic residue Bn showed much lower interaction capabilities, which were further reduced when the non-polar and non-aromatic but hydrophobic residue Ib or charged Gua were used (Fig. 2a). The latter two residues revealed similar interaction intensities to the non-modified ethynyl (E) starting library (Fig. 2a). These findings are in line with previous data, in which varying interaction properties of starting libraries towards the small molecule Δ9-tetrahydrocannabinol were determined when different click-in residues with distinct aromatic and hydrophobic characteristics were employed.16,19 After conducting eight split–combine SELEX cycles, we did two consecutive deconvolution cycles (selection cycles 9 and 10) followed by NGS analysis. NGS revealed a reduction of unique sequences in the DNA libraries obtained from cycles 6 and 8 (Fig. 2b). The five DNA libraries obtained from the deconvolution cycles (i.e., selection cycle 9 and 10) revealed either a further decrease (when using Gua), only a slight variation (when using In, Phe, Bn), or an increase of the number of unique sequences (when using Ib) (Fig. 2b). Likewise, the distribution of nucleotides within the 42-nucleotide random region changed strongly from the starting (ESI Fig. 10a†) to the enriched libraries of selection cycles 6 and 8 (ESI Fig. 10b and c†). Compared to the library from selection cycle 8, the deconvolution cycles 9 and 10 led to similar nucleotide distribution profiles (when using In, Phe, Bn, Ib, ESI Fig. 10d–k†), whereupon Gua resulted in significant changes of the nucleotide distribution (ESI Fig. 10l and m†). The number of unique sequences in the DNA libraries from selection cycles 6 and 8 decreased to 5% and 10%, respectively (Fig. 2b). In the deconvolution cycles a further decrease of the diversity of DNA libraries was observed when using Gua, whereas the use of Phe, Bn and In did not reveal a significant variation of the number of unique sequences compared to the DNA population from selection cycle 8 (Fig. 2b). In turn, when using Ib, the number of unique sequences increased (Fig. 2b). The dominating sequence after 6 and 8 selection cycles was P2 (32% and 22%, respectively, ESI Table 4†). The deconvolution cycles 9 and 10 kept P2's frequency stable when using Phe as modification (Fig. 2c, ESI Table 4†). In all other cases, the copy numbers of P2 decreased, most pronounced when using Gua (0.15%, Fig. 2d–g, ESI Table 4†). The sequence B1 has low copy numbers after 6 and 8 selection cycles (0.0001% and 0.0058%) and only when using Bn in the deconvolution cycles its frequency increased significantly by a factor of 2583 (15.5%, Fig. 2d, ESI Table 4†). Using all other modifications led to a further decrease of B1's frequency (ESI Table 4†). The frequency of sequence I2 only increased when using In in the deconvolution step (Fig. 2e). G1 revealed a 973-fold increase of copy numbers, with a frequency of 0.04% in cycle 8 and 38.9% in the corresponding deconvolution cycle 10 (ESI Table 4†), but only when using Gua as modification (Fig. 2g). Interaction experiments revealed binding of the sequences P2, B1, and G1 to SA, whereas P2 showed tolerance regarding the click-in residues (Fig. 2h), although with preference for the click-ins In and Phe. B1 preferentially binds to SA when modified with Bn and with much lower intensity when using Ib. Of note, P2 and B1 did not interact with SA at the ethynyl-stage (Fig. 2h). In contrast, G1 interacts with SA independent of the Gua modification (Fig. 2h). A scrambled variant of G1 was used as a non-binding control sequence (G1sc), which did not bind to SA independent of any modification (Fig. 2h). We next performed kinetic analysis by SPR using B1 and G1 as both sequences revealed a clear interaction pattern depending on either Bn or no modification, respectively (Fig. 2i, j, Table 2, ESI Fig. 11–14†). B1 has a faster on-rate (Fig. 2k) and a slower off-rate (Fig. 2l) compared to G1 at 37 °C and the dissociation constant by which B1 interacts with SA is significantly lower than the one obtained by G1 (Fig. 2m). The off-rate at 21 °C of both sequences is similar (Fig. 2l). Due to these kinetic properties, G1 significantly increases in copy number compared to B1 and P2 only when Gua is used in the deconvolution cycle, because B1 does not interact and P2 only weakly interacts with SA when modified with Gua. G1 resembles structural motifs of common DNA-based SA binders (ESI Fig. 15†),18,20 indicating that it belongs to a similar sequence family and it binds to SA with similar affinities to previously reported DNA aptamers (Table 2).18 B1, in turn, does not share these structural motifs and binds with a 30-fold higher affinity to SA compared to G1. Therefore, B1 can be considered as a new class of ligands interacting with SA.
| T | B1 (Bn) | G1 | |
|---|---|---|---|
| k on rate [104 M−1 s−1] | 37 °C | 22.42 ± 1.53 | 2.04 ± 0.28 |
| 21 °C | 19.42 ± 0.50 | 0.75 ± 0.26 | |
| k off rate [10−4 s−1] | 37 °C | 3.09 ± 0.40 | 10.75 ± 0.79 |
| 21 °C | 0.66 ± 0.32 | 0.57 ± 0.43 | |
| K D [nM] | 37 °C | 1.4 ± 0.20 | 53.28 ± 5.89 |
| 21 °C | 0.34 ± 0.16 | 8.92 ± 8.20 |
In conclusion, we present a split–combine selection procedure enabling the simultaneous enrichment of clickmers binding to a target protein but having different chemical modifications. In this way, various clickmers are accessible and many modifications can be screened within a shorter time, allowing the identification of the best possible combinations. We exemplarily chose three sequences of each selection and analyzed them further regarding click-in dependency and kinetic properties. We found sequences with clearly assigned modifications (F20, B1), those that are tolerant to more than one (P2) and a sequence that is independent of the modification bound to the target protein (G1). Due to the large sequence space we anticipate that even more binders might be identified in the enriched DNA populations. Our approach provides molecular insight into population dynamics during selection experiments, which is hardly achieved by SELEX experiments using canonical nucleotides. The combination of introducing chemical modifications by CuAAC and splitting–combining the libraries in each selection cycle establishes a strong competitive environment. In this environment we detect sequences that are enriched and due to the deconvolution cycles we are able to assign the binding properties of each sequence to a distinct modification. We can trace back their frequency, thereby obtaining evolutionary profiles of individual sequence-modification pairs. Our data underline that the described enrichment process is mainly guided by off-rate kinetics, observing the slowest for the most enriched sequences. Nevertheless, unfavorable sequences, e.g., those with faster off-rates are not entirely lost but kept at very low copy numbers in the respective DNA library. Once the selection pressure changes, i.e. by removing all but one modification, these sequences thrive and gain copy numbers. At the population level, this behavior was not monitored before in selection experiments. During the deconvolution cycles, we clearly observe enrichment or de-enrichment of sequences that either do or no longer interact when their cognate modification is present or missing. Of note, our selection approach also gives rise to sequences that do bind independently of the modification or in the absence of any, e.g., as naïve DNA. The former is reasonable, as due to the design of our selection scheme, sequences that bind with various modifications have a stronger selective advantage. Their copy numbers are in sum greater than that of any of the others that bind only in the presence of one modification, but are still present and equipped with the other (here four) modifications. It would be interesting to analyze further sequences of the enriched libraries, which we were not able to do yet. Especially, those sequences with low copy numbers even after one or two deconvolution cycles, similar to B33, are of interest. It may be worthwhile to add more deconvolution cycles to enrich those sequences to a greater extent. Notwithstanding, bioinformatic tools are able to identify them even when less abundant. We anticipate that our findings might be informative for the design of new selection schemes and for analyzing the boundaries of the split–combine approach with regard to the number of modifications that can be screened simultaneously.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been made possible by funding from the German Research Council (DFG) to G.M. (MA3442/4-2 and SFB1089 P1) and the ZIM Project (ZF4613701CR8). We thank Jörg Kutscher and Gerhard Brändle for the synthesis of some of the azides used in this study. We thank Joachim Schultze and members of the PRECISE platform for support in NGS analysis of the enriched libraries.References
- K. H. Lee, K. Hamashima, M. Kimoto and I. Hirao, Genetic alphabet expansion biotechnology by creating unnatural base pairs, Curr. Opin. Biotechnol., 2018, 51, 8–15 CrossRef CAS PubMed.
- F. Lipi, S. Chen, M. Chakravarthy, S. Rakesh and R. N. Veedu, In vitro evolution of chemically-modified nucleic acid aptamers: pros and cons, and comprehensive selection strategies, RNA Biol., 2016, 13, 1232–1245 CrossRef PubMed.
- A. D. Gelinas, D. R. Davies and N. Janjic, Embracing proteins: structural themes in aptamer–protein complexes, Curr. Opin. Struct. Biol., 2016, 36, 122–132 CrossRef CAS PubMed.
- M. Kimoto, R. Yamashige, K.-I. Matsunaga, S. Yokoyama and I. Hirao, Generation of high-affinity DNA aptamers using an expanded genetic alphabet, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS PubMed.
- K. Sefah, et al., In vitro selection with artificial expanded genetic information systems, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1449–1454 CrossRef CAS PubMed.
- B. N. Gawande, et al., Selection of DNA aptamers with two modified bases, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2898–2903 CrossRef CAS PubMed.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+), Nucleic Acids Res., 2009, 37, 1638–1649 CrossRef CAS PubMed.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates, ACS Comb. Sci., 2013, 15, 174–182 CrossRef CAS PubMed.
- C. K. L. Gordon, et al., Click-Particle Display for Base-Modified Aptamer Discovery, ACS Chem. Biol., 2019, 14(12), 2652–2662 CrossRef CAS PubMed.
- Q. Shao, et al., Selection of Aptamers with Large Hydrophobic 2'-Substituents, J. Am. Chem. Soc., 2020, 142, 2125–2128 CrossRef CAS PubMed.
- T. Schütze, et al., Probing the SELEX process with next-generation sequencing, PLoS One, 2011, 6, e29604 CrossRef PubMed.
- W. H. Thiel, et al., Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection, PLoS One, 2012, 7, e43836 CrossRef CAS PubMed.
- J. Hoinka, et al., Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery, Nucleic Acids Res., 2015, 43, 5699–5707 CrossRef CAS PubMed.
- M. Slavíčková, et al., Turning Off Transcription with Bacterial RNA Polymerase through CuAAC Click Reactions of DNA Containing 5-Ethynyluracil, Chemistry, 2018, 24, 8311–8314 CrossRef PubMed.
- F. Tolle, G. M. Brändle, D. Matzner and G. Mayer, A Versatile Approach Towards Nucleobase-Modified Aptamers, Angew. Chem., Int. Ed. Engl., 2015, 54, 10971–10974 CrossRef CAS PubMed.
- F. Pfeiffer, et al., Identification and characterization of nucleobase-modified aptamers by click-SELEX, Nat. Protoc., 2018, 13, 1153–1180 CrossRef CAS PubMed.
- A. Panattoni, R. Pohl and M. Hocek, Flexible Alkyne-Linked Thymidine Phosphoramidites and Triphosphates for Chemical or Polymerase Synthesis and Fast Postsynthetic DNA Functionalization through Copper-Catalyzed Alkyne-Azide 1,3-Dipolar Cycloaddition, Org. Lett., 2018, 20, 3962–3965 CrossRef CAS PubMed.
- T. Bing, X. Yang, H. Mei, Z. Cao and D. Shangguan, Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories, Bioorg. Med. Chem., 2010, 18, 1798–1805 CrossRef CAS.
- M. Rosenthal, F. Pfeiffer and G. Mayer, A Receptor-Guided Design Strategy for Ligand Identification, Angew. Chem., Int. Ed. Engl., 2019, 58, 10752–10755 CrossRef CAS PubMed.
- F. Opazo, et al., Modular Assembly of Cell-targeting Devices Based on an Uncommon G-quadruplex Aptamer, Mol. Ther.--Nucleic Acids, 2015, 4, e251 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc01952f |
| This journal is © The Royal Society of Chemistry 2020 |