Open Access Article
Open Access ArticleA photo zipper locked DNA nanomachine with an internal standard for precise miRNA imaging in living cells†
Yue
Zhang
a,
Yue
Zhang
a,
Xiaobo
Zhang
a,
Yuyi
Li
a,
Yuling
He
a,
Ying
Liu
 *ab and
Huangxian
Ju
*ab and
Huangxian
Ju
 a
a
aState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China
bChemistry and Biomedicine Innovation Center, Nanjing University, Nanjing 210023, China. E-mail: yingliu@nju.edu.cn
First published on 26th May 2020
Abstract
DNA nanomachines are capable of converting tiny triggers into autonomous accelerated cascade hybridization reactions and they have been used as a signal amplification strategy for intracellular imaging. However, the “always active” property of most DNA nanomachines with an “absolute intensity-dependent” signal acquisition mode results in “false positive signal amplification” by extracellular analytes and impairs detection accuracy. Here we design a photo zipper locked miRNA responsive DNA nanomachine (PZ-DNA nanomachine) based on upconversion nanoparticles (UCNPs) with a photo-cleavable DNA strand to block the miRNA recognition region, which provided sufficient protection to the DNA nanomachine against nonspecific extracellular activation and allowed satisfactory signal amplification for sensitive miRNA imaging after intracellular photoactivation. Multiple emissions from the UCNPs were also utilized as an internal standard to self-calibrate the intracellular miRNA responsive fluorescence signal. The presented PZ-DNA nanomachine demonstrated the sensitive imaging of intracellular miRNA from different cell lines, which resulted in good accordance with qRT-PCR measurements, providing a universal platform for precise imaging in living cells with high spatial–temporal specificity.
Introduction
DNA nanomachines are artificially designed DNA self-assembly structures based on sequence specific interactions.1–4 With DNA reaction strands compacted in the synthesized nano-structures,5–8 DNA nanomachines accelerate cascade hybridization reactions by converting tiny triggers like nucleic acids,9–12 proteins5,13 and pH14–16 into autonomous mechanical motions and resulting outputs, such as the conformational change of DNA assemblies. Self-quenched DNA nanomachines have been constructed by conjugating both fluorescent molecules and quenchers to DNA strands with close distance in between,17–19 and the machine motion triggered by a specific target resulted in the continuous configuration change of the DNA strands with amplified fluorescence recovery in a short time, which efficiently enhanced the target signal and was applied as a signal amplification strategy for bioanalysis and imaging.20–23Considering the low expression level of miRNA per cell18 and the complex intracellular environment,3,24–26 DNA nanomachines have been used as valuable fluorescence signal amplification tools for intracellular miRNA imaging.17,27–30 Conjugating with a DNAzyme, a DNA walking machine was activated by target miRNA and automatically cleaved substrate DNA strands in a continuous fashion for sensitive intracellular imaging.31–33 Though being capable of amplifying the existence of a low amount miRNA to an appreciable fluorescence signal in a short time, the further application of DNA nanomachines for precise intracellular miRNA imaging still faces the challenges of: (1) the “always active” design of most DNA nanomachines makes them susceptible to extracellular target miRNA in the tumor microenvironment or serum, which would cause nonspecific fluorescence signal amplification before intracellular delivery and result in a false positive signal;18,33–38 (2) the uptake efficiency variation for different cells would also contribute to intracellular signal differences due to the “absolute intensity-dependent” mode with a single luminance channel for signal acquisition.39–41 Both of these factors impair the detection accuracy. DNA nanomachines with controllable activation and an internal standard are needed for precise miRNA intracellular imaging.
Taking advantage of multiple luminescence emissions from upconversion nanoparticles (UCNPs) under near infrared (NIR) light excitation,42–44 here we present a photo zipper locked DNA nanomachine (PZ-DNA nanomachine) with an internal standard for precise intracellular miRNA imaging. The PZ-DNA nanomachine was composed of UCNPs with surface functionalization of a target miRNA responsive DNA walker and its corresponding substrate DNA strands labelled with quencher BHQ2. Dye Cy3 was also co-immobilized on the UCNP surface to facilitate energy transfer from the interior UCNP emission at 540 nm under NIR irradiation to BHQ2 labelled at the terminus of the substrate DNA strands. To protect the DNA nanomachine against unwanted activation before it is located inside tumor cells, a UV cleavable DNA strand was used to block the recognition of the DNA nanomachine by the extracellular target miRNA. After internalization and subsequent photo-activation, the PZ-DNA nanomachine was operated by intracellular miRNA and continuously cleaved the BHQ2 labelled substrate DNA strands, with corresponding Cy3 fluorescence recovery for intracellular miRNA imaging. The concentration of the imaging dye Cy3 on the UCNP surface benefited intracellular imaging. It not only contributed to high signal output compared with cytoplasm dispersed imaging dyes, but also realized fluorescence emission upon NIR irradiation. The UCNP emission at 658 nm under NIR light excitation remained stable during the PZ-DNA nanomachine operation process, which served as the internal standard for self-correction of the Cy3 fluorescence to improve the detection accuracy. This PZ-DNA nanomachine will provide a versatile strategy for precise intracellular imaging of low amounts of biomarkers.
Results and discussion
Principle of the PZ-DNA nanomachine with an internal standard for miRNA imaging
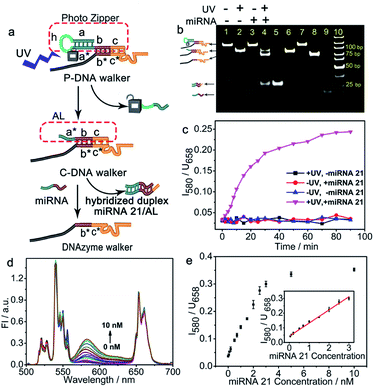
To prepare the PZ-DNA nanomachine, core–shell structured UCNPs NaYF4:Yb,Er,Gd@NaYF4 were synthesized, ligand exchanged with alendronic acid (ADA) to functionalize the surface with amine groups, and covalently coupled with a photo zipper locked DNAzyme walker (P-DNA walker) as well as its substrate DNA strand with the quencher BHQ2 labelled at the terminus (S-DNA-BHQ2). To prepare the P-DNA walker, the DNAzyme walker was hybridized with the “photo zipper” DNA strand to block the catalytic reaction of the DNAzyme towards S-DNA-BHQ2, and equipped with a 40 nt polyT spacer strand for walking. The photo zipper was composed of three regions: a 6 nt DNAzyme block region (c) that hybridized with the DNAzyme anchor position; a 22 nt miRNA responsive region (a* + b) with complementary sequence to the target miRNA; and a 15 nt miRNA block region (a + h) that partially hybridized with the miRNA responsive region. Pc-linker, a UV light cleavable molecule as the photocleavable linker, was embedded between h and a*, which locked the recognition of the miRNA responsive region (a* + b) to miRNA in the absence of light to protect the P-DNA walker from extracellular nonspecific activation during its delivery process. Dye Cy3 was also co-immobilized on the UCNP surface, which bridged the two continuous luminance resonance energy transfer (LRET) processes of concentrating the UCNP 540 nm luminance under NIR light irradiation from the interior of the nanoparticle to the UCNP surface (intra-LRET) and the subsequent quenching of the Cy3 luminance by BHQ2 at the DNA strand S-DNA-BHQ2 terminus (external LRET) (Scheme 1a). After internalization of the PZ-DNA nanomachine, the miRNA block region of the photo zipper (a + h) was cleaved off via UV irradiation to activate the miRNA responsive region (a* + b), and the corresponding hybridized intracellular target miRNA zippered off the photo zipper to activate the DNAzyme walker for S-DNA-BHQ2 cleavage in the presence of Mn2+. After the cleavage of S-DNA-BHQ2, the DNAzyme walker was liberated because of insufficient binding base pairs and moved to the next S-DNA-BHQ2. The successive cleavage of S-DNA-BHQ2 by the autonomous walking of the DNA nanomachine resulted in continuous release of BHQ2-containing DNA fragments from the UCNP surface with Cy3 fluorescence recovery at 580 nm under NIR light irradiation for precisely amplified intracellular miRNA imaging. The operation of the PZ-DNA nanomachine only utilized UCNP emission at 540 nm under NIR light irradiation, while another UCNP emission at 658 nm wasn't affected, therefore serving as an internal standard, and the intensity of Cy3 luminance at 580 nm was ratioed over the UCNP luminance at 658 nm for miRNA imaging self-correction (Scheme 1b).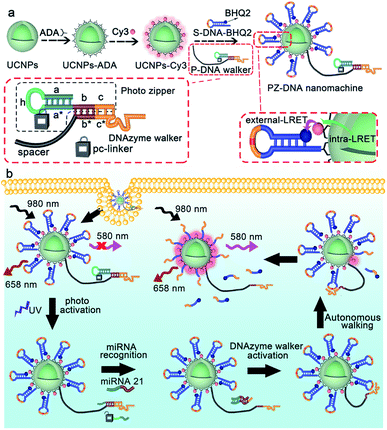 | ||
| Scheme 1 Schematic illustrations of (a) the preparation and (b) activation of the photo zipper locked DNA nanomachine, as well as its operation in response to miRNA in living cells. | ||
Preparation and characterization of the PZ-DNA nanomachine
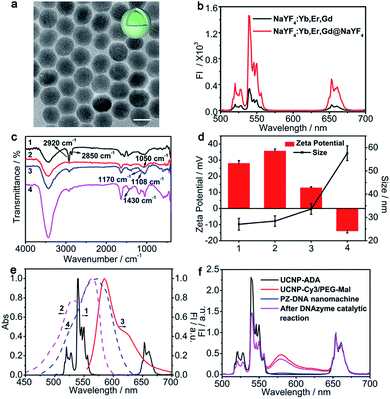
β-NaYF4 crystalline structures co-doped with Er3+ (2%), Yb3+ (18%) and Gd3+ (10%) were synthesized according to a previously reported solvothermal method45 and coated with NaYF4 to prevent surface quenching and enhance the upconversion luminance. Gd3+ was doped to control the size and shape of the UCNPs.46 Compared with UCNPs NaYF4:Er,Yb,Gd, the as-prepared core–shell structured UCNPs NaYF4:Er,Yb,Gd@NaYF4 exhibited increased particle size from 20.3 ± 0.7 nm (Fig. S1a†) to 22.6 ± 1.1 nm (Fig. 1a) and showed much enhanced luminance intensities for the emission peaks at 520 nm, 540 nm, and 658 nm corresponding to the 2H11/2 → 4I15/2, 4S3/2 → 4I15/2, and 4F9/2 → 4I15/2 transitions of Er3+ (ref. 47) (Fig. 1b). Oleic acid (OA), the surface ligand of the UCNPs, was replaced with ADA, a bisphosphine ligand with amine terminal groups due to the high binding affinity between bidentate phosphates and lanthanide ions, which improved the dispersion of the UCNPs in aqueous solution and facilitated the subsequent functionalization. The successful surface ligand exchange process was confirmed by FTIR, which showed the IR absorption of OA at 2920 cm−1 and 2850 cm−1 for the OA functionalized UCNPs (UCNPs-OA) due to the asymmetric and symmetric stretching vibrations of the –CH2– group, respectively (line 1, Fig. 1c). After ligand exchange with ADA, the characteristic peaks for the –CH2– group were strongly suppressed, corresponding with the appearance of an absorption peak at 1050 cm−1 attributed to the stretching vibration of P–O for ADA (line 2, Fig. 1c). The as-prepared ADA stabilized UCNPs (UCNPs-ADA) exhibited an increased zeta potential of 35.7 ± 1.2 mV compared with the bare UCNPs and a hydrodynamic size of 28.4 ± 2.3 nm similar to that of the bare UCNPs (Fig. 1d).Cy3, with its maximum absorption well overlapped with the UCNP emission at 540 nm (Fig. 1e), was chosen as the bridge dye connecting two continuous LRET processes and covalently immobilized on the UCNP surface via amidation to facilitate luminance energy transfer from the interior of the UCNPs to the surface extended DNA strands that were subsequently modified. NHS-PEG-Mal was co-immobilized on the UCNP surface with Cy3 to facilitate subsequent DNA strand immobilization, and the molar ratio of Cy3 over NHS-PEG-Mal was optimized to maximize the UCNP energy transfer efficiency to Cy3, which achieved the highest Cy3 luminance intensity at 580 nm with Cy3/NHS-PEG-Mal of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 (Fig. S1b and c†). To quantify the amount of Cy3 immobilized on each UCNP, Cy3 fluorescence from UCNPs-Cy3/PEG-Mal was measured at 545 nm and compared with the Cy3 standard calibration curve to get 170 ± 16 Cy3 per UCNP (Fig. S1d and e†). The efficient energy transfer from the interior of the UCNPs to Cy3 was confirmed by the obvious fluorescence intensity decrease at 520 nm and 540 nm and increase at 580 nm (Fig. S1b† and 1f). The successful synthesis of UCNPs-Cy3/PEG-Mal was also confirmed by the characteristic IR absorption peaks at 1108 cm−1 and 1170 cm−1 attributed to the stretching and asymmetric stretching vibrations of C–O–C groups, respectively (line3, Fig. 1c), and the decrease of the zeta potential to 12.9 ± 0.6 mV due to PEG conjugation with an increase in hydrodynamic size to 33.6 ± 2.3 nm (Fig. 1d).
7 (Fig. S1b and c†). To quantify the amount of Cy3 immobilized on each UCNP, Cy3 fluorescence from UCNPs-Cy3/PEG-Mal was measured at 545 nm and compared with the Cy3 standard calibration curve to get 170 ± 16 Cy3 per UCNP (Fig. S1d and e†). The efficient energy transfer from the interior of the UCNPs to Cy3 was confirmed by the obvious fluorescence intensity decrease at 520 nm and 540 nm and increase at 580 nm (Fig. S1b† and 1f). The successful synthesis of UCNPs-Cy3/PEG-Mal was also confirmed by the characteristic IR absorption peaks at 1108 cm−1 and 1170 cm−1 attributed to the stretching and asymmetric stretching vibrations of C–O–C groups, respectively (line3, Fig. 1c), and the decrease of the zeta potential to 12.9 ± 0.6 mV due to PEG conjugation with an increase in hydrodynamic size to 33.6 ± 2.3 nm (Fig. 1d).
Before modification of DNA strands on the UCNP surface, the performance of the catalytic reaction for the DNAzyme walker in solution was verified via gel electrophoresis with a polyT linked DNAzyme walker and substrate DNA (S-DNA). In the presence of Mn2+, S-DNA was cleaved into two fragments (F1, F2) by the DNAzyme walker (Fig. S2a†), and gave two distinct bands at their individual corresponding positions in the gel electrophoresis images (lane 5, Fig. S2b†). A thiol terminus P-DNA walker was then modified on UCNPs-Cy3/PEG-Mal with S-DNA-BHQ2 to complete the PZ-DNA nanomachine, which showed DNA characteristic absorption peaks at 260 nm in the UV-Vis absorption spectrum (Fig. S1f†) and 1430 cm−1 in the FTIR spectra with a series of peaks below 900 cm−1 attributed to the bending vibrations of the heterocycle in DNA48 (line 4, Fig. 1c). The resulting PZ-DNA nanomachine demonstrated decreased zeta potential of −14.0 ± 1.2 mV due to the negative charges of the DNA strands with a highly increased hydrodynamic size of 57.6 ± 3.1 nm (Fig. 1d).
The maximum absorption of BHQ2 was located at 572 nm, overlapped with the Cy3 emission peak at 585 nm (Fig. 1e) and the PZ-DNA nanomachine with S-DNA-BHQ2 functionalization showed a complete suppression of Cy3 luminance compared with UCNPs-Cy3/PEG-Mal, while the UCNP luminance at 658 nm remained unchanged (Fig. 1f), indicating the efficient energy transfer from UCNP luminance at 540 nm to the S-DNA-BHQ2 terminus BHQ2 via bridge dye Cy3, which facilitated two continuous LRET processes of intra-LRET from the interior of the UCNPs to the UCNP surface and external LRET from the UCNP surface to BHQ2. In comparison, considering the inefficient long distance energy transfer from the interior of the UCNPs to the surface extended DNA strand, there was little decrease of the UCNP emission peak at 540 nm for the S-DNA-BHQ1 functionalized UCNPs (UCNPs-DNA-BHQ1) in the absence of the bridge dye (Fig. S2c†) despite the good matching of the UCNP emission peak at 540 nm and the BHQ1 absorption peak at 534 nm (Fig. S2d†). The operation of the DNAzyme walker on the UCNP surface was verified with the DNAzyme walker and S-DNA-BHQ2 functionalized UCNPs upon the addition of Mn2+, which activated the DNAzyme catalytic reaction to cleave off the F1 fragment (with the BHQ2 labeled terminus) of S-DNA-BHQ2 from the UCNP surface, and the corresponding termination of external LRET from Cy3 to BHQ2 resulted in the recovery of the Cy3 luminance. The cleavage of S-DNA-BHQ2 liberated the DNAzyme walker to continuously react with the next S-DNA-BHQ2 on the UCNP surface, and the catalytic reaction of the DNAzyme walker with numerous S-DNA-BHQ2 resulted in amplified luminance recovery of Cy3 (Fig. 1f) and guaranteed high operational efficiency of the DNAzyme-based nanomachine. During the operation process of the DNAzyme catalytic reaction, the UCNP luminance at 658 nm remained the same, indicating its capability as an internal standard for intracellular miRNA imaging (Fig. 1f). To enhance the quenching efficiency and improve the activated fluorescence recovery for the DNA nanomachine, the molar ratios of the S-DNA-BHQ2/DNAzyme walker and the length of the DNAzyme linker polyT were optimized. The results showed low Cy3 background (Fig. S3a,† quenching) with maximized Cy3 recovery at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (Fig. S3a,† recovery) and length of 40T for the polyT spacer linker (Fig. S3b†). The surface immobilization density of the DNA substrate strands was further calculated by measuring the FAM fluorescence for the FAM labelled DNA nanomachine UCNPs-Cy3/S-DNA-FAM/DNAzyme walker and comparing it with the standard calibration curve of S-DNA-FAM, which resulted in 133 ± 11 DNA substrate strands per UCNP (Fig. S3c and d†).
1 molar ratio (Fig. S3a,† recovery) and length of 40T for the polyT spacer linker (Fig. S3b†). The surface immobilization density of the DNA substrate strands was further calculated by measuring the FAM fluorescence for the FAM labelled DNA nanomachine UCNPs-Cy3/S-DNA-FAM/DNAzyme walker and comparing it with the standard calibration curve of S-DNA-FAM, which resulted in 133 ± 11 DNA substrate strands per UCNP (Fig. S3c and d†).
Activation of the P-DNA walker and in vitro response of the PZ-DNA nanomachine to miRNA 21
miRNAs play important roles in various cellular processes and the progression of many diseases, therefore, it is significant to determine their expression levels in living cells. As a potential diagnostic biomarker,49 miRNA 21 is critical to a lot of biological functions and disease progressions, and was chosen here as the model target to trigger the operation of the PZ-DNA nanomachine.28 To prepare the P-DNA walker, a photo-cleavable hairpin structured photo zipper was hybridized with the DNAzyme walker to block its anchoring region with S-DNA-BHQ2. The feasibility of photo-responsive activation of the P-DNA walker was first verified in homogeneous solution. 5 min UV (7 mW cm−2) exposure cleaved off the miRNA block region a + h, and generated a cleaved DNA walker (C-DNA walker) with partially exposed target miRNA 21 recognition region a* + b (Fig. 2a). The efficient photo cleavage of the miRNA block region was confirmed by the complete disappearance of the P-DNA walker band, accompanied by the clear appearance of the C-DNA walker band with higher mobility when the P-DNA walker was exposed to UV light (lane 2, Fig. 2b). The dropped fragment (a + h) with short strand length did not appear in the gel electrophoresis images. The subsequent miRNA 21 recognition completely unzipped the C-DNA walker and liberated the DNAzyme for the enzymatic catalytic reaction (Fig. 2a). Incubation of the P-DNA walker with miRNA 21 after photo activation resulted in two new bands with much higher mobility, which were at the same positions of the DNAzyme walker band and hybridized duplex miRNA 21/AL band, respectively (lane 4, Fig. 2b), indicating the successful activation of the P-DNA walker in the presence of miRNA 21 after photo irradiation. On the contrary, miRNA 21 didn't react with the P-DNA walker in the absence of photo irradiation, and individual miRNA 21 and P-DNA walker bands were observed in gel electrophoresis (lane 3, Fig. 2b).S-DNA-BHQ2 and the P-DNA walker were then conjugated to UCNPs-Cy3/PEG-Mal with a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to prepare the PZ-DNA nanomachine, and the intensity ratio of Cy3 fluorescence recovery over UCNP luminance at 658 nm (I580/U658) was measured according to time to confirm the photo activation of the PZ-DNA nanomachine and its response to miRNA 21. With UV exposure, the mixture of 2 nM miRNA 21 with the PZ-DNA nanomachine resulted in an extensive increase of I580/U658 according to time and the signal increase quickly saturated at 80 min, indicating the high reaction efficiency of the PZ-DNA nanomachine, while little increase of I580/U658 was observed either in the absence of light irradiation or miRNA 21 (Fig. 2c). After photo activation, the PZ-DNA nanomachine was challenged with miRNA 21 of different concentrations from 0 to 10 nM, which resulted in a gradual increase of the Cy3 fluorescence peak at 580 nm, while the UCNP luminance at 658 nm remained unchanged (Fig. 2d). A linear correlation between I580/U658 and miRNA 21 concentration was obtained in the range from 0.05 to 3.00 nM with a limit of detection (LOD) of 3.71 pM (Fig. 2e), which is much improved compared with many previous reported UCNP-based miRNA detection probes.50 The introduction of the DNA nanomachine on the UCNP surface contributed to impressive enhancement of the detection sensitivity.23,51,52 To further demonstrate the amplification effect of the PZ-DNA nanomachine, a FAM labelled P-DNA walker was prepared by conjugating the dye FAM to the photo zipper, and immobilized on the UCNP surface with S-DNA at a molar ratio of 1
1 to prepare the PZ-DNA nanomachine, and the intensity ratio of Cy3 fluorescence recovery over UCNP luminance at 658 nm (I580/U658) was measured according to time to confirm the photo activation of the PZ-DNA nanomachine and its response to miRNA 21. With UV exposure, the mixture of 2 nM miRNA 21 with the PZ-DNA nanomachine resulted in an extensive increase of I580/U658 according to time and the signal increase quickly saturated at 80 min, indicating the high reaction efficiency of the PZ-DNA nanomachine, while little increase of I580/U658 was observed either in the absence of light irradiation or miRNA 21 (Fig. 2c). After photo activation, the PZ-DNA nanomachine was challenged with miRNA 21 of different concentrations from 0 to 10 nM, which resulted in a gradual increase of the Cy3 fluorescence peak at 580 nm, while the UCNP luminance at 658 nm remained unchanged (Fig. 2d). A linear correlation between I580/U658 and miRNA 21 concentration was obtained in the range from 0.05 to 3.00 nM with a limit of detection (LOD) of 3.71 pM (Fig. 2e), which is much improved compared with many previous reported UCNP-based miRNA detection probes.50 The introduction of the DNA nanomachine on the UCNP surface contributed to impressive enhancement of the detection sensitivity.23,51,52 To further demonstrate the amplification effect of the PZ-DNA nanomachine, a FAM labelled P-DNA walker was prepared by conjugating the dye FAM to the photo zipper, and immobilized on the UCNP surface with S-DNA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to prepare a PF-DNA nanomachine (Fig. S4a†). After photo exposure, the operation of the PF-DNA nanomachine in response to 2 nM miRNA 21 released hybridized duplex miRNA 21/AL-FAM, and the FAM fluorescence intensity from the supernatant was measured. Since FAM dye was labelled on the P-DNA walker instead of S-DNA, the operation of the PF-DNA nanomachine with continuous cleavage of S-DNA did not contribute to fluorescence amplification, and only resulted in very limited FAM fluorescence intensity after the reaction with miRNA 21 (Fig. S4b†). In comparison, the SF-DNA nanomachine with FAM labelled S-DNA (Fig. S4a†) was also prepared with S-DNA-FAM and the P-DNA walker immobilized on the UCNPs at a molar ratio of 10
10 to prepare a PF-DNA nanomachine (Fig. S4a†). After photo exposure, the operation of the PF-DNA nanomachine in response to 2 nM miRNA 21 released hybridized duplex miRNA 21/AL-FAM, and the FAM fluorescence intensity from the supernatant was measured. Since FAM dye was labelled on the P-DNA walker instead of S-DNA, the operation of the PF-DNA nanomachine with continuous cleavage of S-DNA did not contribute to fluorescence amplification, and only resulted in very limited FAM fluorescence intensity after the reaction with miRNA 21 (Fig. S4b†). In comparison, the SF-DNA nanomachine with FAM labelled S-DNA (Fig. S4a†) was also prepared with S-DNA-FAM and the P-DNA walker immobilized on the UCNPs at a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In response to photo exposure and 2 nM miRNA 21, the SF-DNA walker continuously cleaved the FAM labelled F1 segment (F1-FAM) off the UCNP surface, and resulted in a substantial increase in the FAM fluorescence recovery compared with the PF-DNA nanomachine due to the catalytic amplification reaction of the DNAzyme walker (Fig. S4c†).
1. In response to photo exposure and 2 nM miRNA 21, the SF-DNA walker continuously cleaved the FAM labelled F1 segment (F1-FAM) off the UCNP surface, and resulted in a substantial increase in the FAM fluorescence recovery compared with the PF-DNA nanomachine due to the catalytic amplification reaction of the DNAzyme walker (Fig. S4c†).
The reaction specificity of the PZ-DNA nanomachine to miRNA 21 was also investigated with nonspecific miRNAs of three base mismatch, single base mismatch, miRNA 141 and miRNA 199-a (Fig. S4d†). The three base mismatched miRNA and nonspecific miRNAs all showed very low ratio of luminance intensities for Cy3 at 580 nm over UCNPs at 658 nm (I580/U658) close to the blank control, indicating impressive reaction specificity of the PZ-DNA nanomachine in response to miRNA 21. The I580/U658 for single base mismatched miRNA 21 was slightly higher due to approximate thermodynamic energy with miRNA 21.11
Live cell imaging of miRNA 21 with the PZ-DNA nanomachine
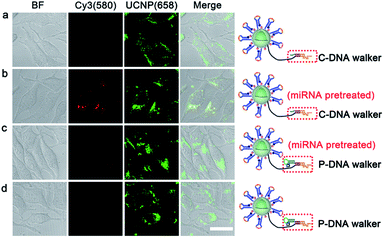
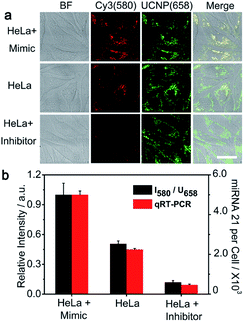
HeLa cells, with high miRNA 21 expression profile, were chosen as the model cells to demonstrate the feasibility of the PZ-DNA nanomachine for in situ visualization of intracellular miRNA. After internalization, the PZ-DNA nanomachine accumulated inside the endosomes at first and then escaped from the endosomes. The PZ-DNA nanomachine in the absence of BHQ2 was incubated with HeLa cells to demonstrate the lysosome escape process, which demonstrated progressive separation of Cy3 fluorescence (red) and Lysotracker fluorescence (green) in the cytoplasm (Fig. S5†). Considering the steric hindrance from the nanomaterial to prevent the access of nucleases and protect the nanomaterial-assembled nucleic acids from degradation,53–55 the PZ-DNA nanomachine retained its integrity in the lysosome, which was indicated by the complete overlap of the DNA conjugated Cy5 fluorescence (blue) and UCNP fluorescence (red) (Fig. S6†). After the lysosome escape process, the HeLa cells were then exposed to UV irradiation (7 mW cm−2) for 5 min to activate the photo zipper, and the de-protected PZ-DNA nanomachine was subsequently activated by intracellular miRNA 21. Considering the low level of Mn2+ in native cells, the required Mn2+ concentration for the intracellular operation of the PZ-DNA nanomachine was achieved by incubating the cells with Mn2+ solution.33,56 Intracellular Cy3 fluorescence recovery began show up in the TCS SP5 confocal laser scanning microscope (CLSM) at 0.5 h, and the intensity of Cy3 fluorescence recovery gradually increased with incubation time and saturated at 2 h (Fig. S7†). Instead of dropping from the nanocarrier and diffusing in the cytoplasm after the DNAzyme catalytic reaction, the imaging dyes were concentrated on the UCNP surface after reacting with the target miRNA. This improved the signal output and allowed efficient emission upon NIR irradiation. To confirm the complete protection of the PZ-DNA nanomachine with the photo zipper, as well as its efficient unzipping intracellularly, HeLa cells were incubated with the PZ-DNA nanomachine (functionalized with P-DNA walker) and an unprotected DNA nanomachine (functionalized with C-DNA walker) respectively, and there was little Cy3 fluorescence recovery from the PZ-DNA nanomachine incubated HeLa cells in the absence of UV irradiation, indicating good protection from the photo zipper (Fig. S8a†). After 5 min of UV activation, the activated PZ-DNA nanomachine incubated HeLa cells demonstrated similar intensity of intracellular Cy3 fluorescence recovery compared with that of the unprotected DNA nanomachine incubated HeLa cells (Fig. S8b and c†). To demonstrate photo zipper protection of the PZ-DNA nanomachine against unwanted extracellular activation, a miRNA 21 inhibitor, a synthetic 22 nt RNA strand with complementary sequence to miRNA 21, was used to treat HeLa cells to suppress intracellular miRNA 21 expression. The unprotected DNA nanomachine (functionalized with C-DNA walker) and PZ-DNA nanomachine (functionalized with P-DNA walker) were pre-treated with 2 nM miRNA 21 for 9 h in vitro to simulate the nonspecific activation during the systemic circulation process, and were subsequently incubated with inhibitor treated HeLa cells. Strong UCNP luminance at 658 nm was observed from all the cells, indicating the efficient intracellular delivery of the PZ-DNA nanomachine and unprotected DNA nanomachine. Cy3 fluorescence recovery was barely observed from both the unprotected DNA nanomachine (Fig. 3a) and PZ-DNA nanomachine (Fig. 3d) incubated HeLa cells due to the suppressed expression of miRNA intracellularly, while obvious Cy3 fluorescence was observed from the HeLa cells incubated with a miRNA 21 pretreated unprotected DNA nanomachine (Fig. 3b); the “false positive” fluorescence signal came from the activation of the unprotected DNA nanomachine before endocytosis. In contrast, the miRNA 21 pre-treated PZ-DNA nanomachine demonstrated little fluorescence recovery intracellularly (Fig. 3c), indicating the effective protection of the DNA nanomachine from the photo zipper against nonspecific activation and the impressive suppression of the background fluorescence signal.The expression levels of identical miRNA change at different periods of tumorigenesis,57 therefore the accurate imaging of the miRNA expression level in living cells is of great significance in disease diagnosis and evaluation of therapeutic effects. The potential of the PZ-DNA nanomachine for quantitative evaluation of the relative expression levels of miRNA in living cells was further confirmed. miRNA 21 mimic, a synthetic RNA mimicking miRNA 21, and miRNA 21 inhibitor were incubated with HeLa cells separately to adjust the intracellular miRNA 21 expression to mimic the natural change of miRNA expression upon biological stimulus. After being incubated with the PZ-DNA nanomachine and irradiated with UV light (7 mW cm−2) for 5 min, the intracellular Cy3 fluorescence recovery was imaged via CLSM and the fluorescence intensity was compared with the UCNP luminance at 658 nm to evaluate the intracellular miRNA 21 expression level. Both the miRNA 21 mimic and inhibitor treated HeLa cells demonstrated similar fluorescence intensity for the UCNP luminance at 658 nm, indicating the efficient endocytosis process of the PZ-DNA nanomachine (Fig. 4a). Compared with the Cy3 fluorescence recovery from untreated HeLa cells, the miRNA 21 mimic treated HeLa cells demonstrated obviously enhanced Cy3 fluorescence, while Cy3 fluorescence recovery was barely observed from the miRNA 21 inhibitor treated HeLa cells due to the decrease of intracellular miRNA 21 expression, providing the specific and quantitative response of the PZ-DNA nanomachine to intracellular miRNA 21. The luminance intensities for Cy3 at 580 nm and UCNPs at 658 nm as an internal standard were calculated from CLSM images, and I580/U658 for the miRNA 21 mimic and inhibitor treated HeLa cells were 197.6% and 23.6%, respectively, compared with that of the untreated HeLa cells (Fig. 4b). Meanwhile, real time PCR (qRT-PCR) was performed to verify the accuracy of intracellular imaging by means of a miRNA 21 calibration curve with a linear range of 1 pM to 10 nM (Fig. S9†), and the miRNA 21 copy number for the miRNA mimic-treated HeLa cells was 2.2 fold that of the untreated HeLa cells, while the miRNA 21 copy number for the miRNA inhibitor treated HeLa cells was 20.5% that of the untreated HeLa cells, which demonstrated a similar tendency to the result from CLSM imaging, confirming the accuracy of the PZ-DNA nanomachine for intracellular miRNA 21 imaging.
The cytotoxicity of the PZ-DNA nanomachine was evaluated with a standard 3-(4,5-dimethylthiazol-2-yl)-2-diphenyltetrazolium bromide (MTT) assay at a series of PZ-DNA nanomachine concentrations (Fig. S10a†). Even at a very high concentration of PZ-DNA nanomachine, the HeLa cells still kept 91.4% viability. In addition, 5 min of UV irradiation (7 mW cm−2) didn't influence the viability for both the HeLa cells and PZ-DNA nanomachine loaded HeLa cells (Fig. S10b†). The good biocompatibility of the PZ-DNA nanomachine and the duration of cells over short time UV irradiation guarantee its application in intracellular cell imaging. Though little influence on cell viability and satisfactory photo control for precise cellular imaging have been demonstrated, the involvement of UV irradiation might limit the application of the PZ-DNA nanomachine in the living body considering its low penetration depth. Our on-going follow up project will utilize NIR irradiation for activation of the DNA nanomachine, which hopefully will extend its in vivo applications.
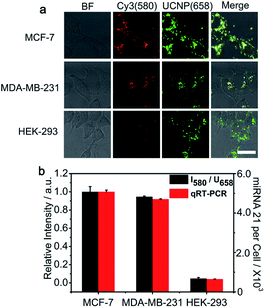
The PZ-DNA nanomachine was further applied for miRNA 21 imaging of different kinds of cell lines, including MCF-7 cells and MDA-MB-231 cells with high miRNA 21 expression and HEK-293 cells with low miRNA 21 expression. After incubation with the PZ-DNA nanomachine and 5 min light irradiation, strong Cy3 fluorescence recovery was observed from the MCF-7 cells and MDA-MB-231 cells, while weak Cy3 fluorescence recovery was observed from HEK-293 cells with similar UCNP luminance intensity at 658 nm for all three cells lines in the CLSM images (Fig. 5a). The relative fluorescence signals obtained after normalization with internal standard corresponded with the copy numbers calculated from qRT-PCR (Fig. 5b) and previously reported results,36 indicating the applicability of the PZ-DNA nanomachine for precise intracellular miRNA imaging.
Conclusions
A PZ-DNA nanomachine with an internal standard based on UCNPs was presented in this work for accurate intracellular miRNA imaging. A miRNA responsive DNAzyme walker was immobilized on the UCNP surface with its corresponding substrate DNA strands, and the miRNA responsive region of the DNAzyme was blocked by a photo-cleavable DNA strand, which well protected the unwanted activation of the DNA nanomachine before intracellular delivery and effectively avoided “false positive” signals for miRNA imaging in living cells. After photoactivation, the PZ-DNA nanomachine was operated by intracellular miRNA, and gave impressive luminance signal amplification for miRNA imaging in living cells. The concentration of the imaging dye on the UCNP surface improved the signal output and achieved intracellular imaging upon NIR irradiation. Multiple emissions from the UCNPs were used as an internal standard to enhance the detection accuracy. Meanwhile, the design allowed the presented PZ-DNA nanomachine to demonstrate high detection sensitivity for miRNA imaging from different cell lines, and the results were in good accordance with qRT-PCR measurements, providing a universal platform for the precise imaging of biomarkers in living cells.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21974064 and 21635005), the State Key Laboratory of Analytical Chemistry for Life Sciences (5431ZZXM2003), Fundamental Research Funds for the Central Universities (14380472), the National Research Foundation for Thousand Youth Talents Plan of China, the Specially-appointed Professor Foundation of Jiangsu Province, and the Program for innovative Talents and Entrepreneurs of Jiangsu Province.Note and references
- D. S. Liu and S. Balasubramanian, Angew. Chem., Int. Ed., 2003, 42, 5734–5736 CrossRef CAS PubMed.
- S. Modi, M. G. Swetha, D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- K. Leung, K. Chakraborty, A. Saminathan and Y. Krishnan, Nat. Nanotechnol., 2019, 14, 176–183 CrossRef CAS PubMed.
- S. Ranallo, C. Prevost-Tremblay, A. Idili, A. Vallee-Belisle and F. Ricci, Nat. Commun., 2017, 8, 15150 CrossRef CAS PubMed.
- H. Q. Zhang, M. D. Lai, A. Zuehlke, H. Y. Peng, X. F. Li and X. C. Le, Angew. Chem., Int. Ed., 2015, 54, 14326–14330 CrossRef CAS PubMed.
- X. M. Qu, D. Zhu, G. B. Yao, S. Su, J. Chao, H. J. Liu, X. L. Zuo, L. H. Wang, J. Y. Shi, L. H. Wang, W. Huang, H. Pei and C. H. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- J. Zheng, A. L. Jiao, R. H. Yang, H. M. Li, J. S. Li, M. L. Shi, C. Ma, Y. Jiang, L. Deng and W. H. Tan, J. Am. Chem. Soc., 2012, 134, 19957–19960 CrossRef CAS PubMed.
- J. Elbaz, A. Cecconello, Z. Y. Fan, A. O. Govorov and I. Willner, Nat. Commun., 2013, 4, 2000 CrossRef PubMed.
- R. Z. Peng, X. F. Zheng, Y. F. Lyu, L. J. Xu, X. B. Zhang, G. L. Ke, Q. L. Liu, C. J. You, S. Y. Huan and W. H. Tan, J. Am. Chem. Soc., 2018, 140, 9793–9796 CrossRef CAS PubMed.
- P. Zhang, J. Jiang, R. Yuan, Y. Zhuo and Y. Q. Chai, J. Am. Chem. Soc., 2018, 140, 9361–9364 CrossRef CAS PubMed.
- Y. Y. Li, G. A. Wang, S. D. Mason, X. L. Yang, Z. C. Yu, Y. N. Tang and F. Li, Chem. Sci., 2018, 9, 6434–6439 RSC.
- X. L. Yang, Y. N. Tang, S. D. Mason, J. B. Chen and F. Li, ACS Nano, 2016, 10, 2324–2330 CrossRef CAS PubMed.
- F. Li, Y. W. Lin, A. Lau, Y. N. Tang, J. B. Chen and X. C. Le, Anal. Chem., 2018, 90, 8651–8657 CrossRef CAS PubMed.
- S. Surana, J. M. Bhat, S. P. Koushika and Y. Krishnan, Nat. Commun., 2011, 2, 340 CrossRef PubMed.
- A. Porchetta, A. Idili, A. Vallee-Belisle and F. Ricci, Nano Lett., 2015, 15, 4467–4471 CrossRef CAS PubMed.
- W. H. Fu, L. L. Tang, G. H. Wei, L. Fang, J. Zeng, R. J. Zhan, X. M. Liu, H. Zuo, C. Z. Huang and C. D. Mao, Angew. Chem., Int. Ed., 2019, 58, 16405–16410 CrossRef CAS PubMed.
- S. Z. Yue, X. Y. Song, W. L. Song and S. Bi, Chem. Sci., 2019, 10, 1651–1658 RSC.
- C. P. Liang, P. Q. Ma, H. Liu, X. G. Guo, B. C. Yin and B. C. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed.
- F. Yang, Y. R. Cheng, Y. Cao, H. F. Dong, H. T. Lu, K. Zhang, X. D. Meng, C. H. Liu and X. J. Zhang, Chem. Sci., 2019, 10, 1709–1715 RSC.
- Q. M. Wei, J. Huang, J. Li, J. L. Wang, X. H. Yang, J. B. Liu and K. M. Wang, Chem. Sci., 2018, 9, 7802–7808 RSC.
- Z. Wu, G. Q. Liu, X. L. Yang and J. H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS PubMed.
- C. C. Wu, S. Cansiz, L. Q. Zhang, I. T. Teng, L. P. Qiu, J. Li, Y. Liu, C. S. Zhou, R. Hu, T. Zhang, C. Cui, L. Cui and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 4900–4903 CrossRef CAS PubMed.
- K. W. Ren, Y. F. Xu, Y. Liu, M. Yang and H. X. Ju, ACS Nano, 2018, 12, 263–271 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2018, 173, 20–51 CrossRef CAS PubMed.
- A. Ventura and T. Jacks, Cell, 2009, 136, 586–591 CrossRef CAS PubMed.
- M. Ghandi, F. W. Huang, J. Jane-Valbuena, G. V. Kryukov, C. C. Lo, E. R. McDonald 3rd, J. Barretina, E. T. Gelfand, C. M. Bielski, H. X. Li, K. Hu, A. Y. Andreev-Drakhlin, J. Kim, J. M. Hess, B. J. Haas, F. Aguet, B. A. Weir, M. V. Rothberg, B. R. Paolella, M. S. Lawrence, R. Akbani, Y. L. Lu, H. L. Tiv, P. C. Gokhale, A. de Weck, A. A. Mansour, C. Oh, J. Shih, K. Hadi, Y. Rosen, J. Bistline, K. Venkatesan, A. Reddy, D. Sonkin, M. Liu, J. Lehar, J. M. Korn, D. A. Porter, M. D. Jones, J. Golji, G. Caponigro, J. E. Taylor, C. M. Dunning, A. L. Creech, A. C. Warren, J. M. McFarland, M. Zamanighomi, A. Kauffmann, N. Stransky, M. Imielinski, Y. E. Maruvka, A. D. Cherniack, A. Tsherniak, F. Vazquez, J. D. Jaffe, A. A. Lane, D. M. Weinstock, C. M. Johannessen, M. P. Morrissey, F. Stegmeier, R. Schlegel, W. C. Hahn, G. Getz, G. B. Mills, J. S. Boehm, T. R. Golub, L. A. Garraway and W. R. Sellers, Nature, 2019, 569, 503–508 CrossRef CAS PubMed.
- J. G. Xu, Z. S. Wu, Z. M. Wang, J. Q. Le, T. T. Zheng and L. Jia, Biomaterials, 2017, 120, 57–65 CrossRef CAS PubMed.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef CAS PubMed.
- X. D. Meng, K. Zhang, W. H. Dai, Y. Cao, F. Yang, H. F. Dong and X. J. Zhang, Chem. Sci., 2018, 9, 7419–7425 RSC.
- L. Li, J. Feng, H. Y. Liu, Q. L. Li, L. L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC.
- Y. J. Yang, J. Huang, X. H. Yang, K. Quan, H. Wang, L. Ying, N. L. Xie, M. Ou and K. M. Wang, Anal. Chem., 2016, 88, 5981–5987 CrossRef CAS PubMed.
- Y. J. Yang, J. Huang, X. H. Yang, X. X. He, K. Quan, N. L. Xie, M. Ou and K. M. Wang, Anal. Chem., 2017, 89, 5850–5856 CrossRef CAS PubMed.
- H. Y. Peng, X. F. Li, H. Q. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- H. Q. Zhang, F. Li, B. Dever, X. F. Li and X. C. Le, Chem. Rev., 2013, 113, 2812–2841 CrossRef CAS PubMed.
- P. Q. Ma, C. P. Liang, H. H. Zhang, B. C. Yin and B. C. Ye, Chem. Sci., 2018, 9, 3299–3304 RSC.
- J. Zhao, H. Q. Chu, Y. Zhao, Y. Lu and L. L. Li, J. Am. Chem. Soc., 2019, 141, 7056–7062 CrossRef CAS PubMed.
- L. P. Qiu, C. C. Wu, M. X. You, D. Han, T. Chen, G. Z. Zhu, J. H. Jiang, R. Q. Yu and W. H. Tan, J. Am. Chem. Soc., 2013, 135, 12952–12955 CrossRef CAS PubMed.
- C. Xue, S. X. Zhang, C. H. Ouyang, D. R. Chang, B. J. Salena, Y. F. Li and Z. S. Wu, Angew. Chem., Int. Ed., 2018, 57, 9739–9743 CrossRef CAS PubMed.
- K. Zhang, X. D. Meng, Z. Yang, Y. Cao, Y. R. Cheng, D. D. Wang, H. T. Lu, Z. J. Shi, H. F. Dong and X. J. Zhang, Adv. Mater., 2019, 31, e1807888 CrossRef PubMed.
- Z. M. Ying, Z. Wu, B. Tu, W. H. Tan and J. H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779–9782 CrossRef CAS PubMed.
- X. L. Huang, J. B. Song, B. C. Yung, X. H. Huang, Y. H. Xiong and X. Y. Chen, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC.
- C. Drees, A. N. Raj, R. Kurre, K. B. Busch, M. Haase and J. Piehler, Angew. Chem., Int. Ed., 2016, 55, 11668–11672 CrossRef CAS PubMed.
- C. Homann, L. Krukewitt, F. Frenzel, B. Grauel, C. Wurth, U. Resch-Genger and M. Haase, Angew. Chem., Int. Ed., 2018, 57, 8765–8769 CrossRef CAS PubMed.
- C. Wurth, S. Fischer, B. Grauel, A. P. Alivisatos and U. Resch-Genger, J. Am. Chem. Soc., 2018, 140, 4922–4928 CrossRef PubMed.
- L. L. Li, R. B. Zhang, L. L. Yin, K. Z. Zheng, W. P. Qin, P. R. Selvin and Y. Lu, Angew. Chem., Int. Ed., 2012, 51, 6121–6125 CrossRef CAS PubMed.
- V. Muhr, C. Wurth, M. Kraft, M. Buchner, A. J. Baeumner, U. Resch-Genger and T. Hirsch, Anal. Chem., 2017, 89, 4868–4874 CrossRef CAS PubMed.
- D. Lu, S. K. Cho, S. Ahn, S. Brun, C. J. Summers and W. Park, ACS Nano, 2014, 8, 7780–7792 CrossRef CAS PubMed.
- H. Ren, Z. Long, X. T. Shen, Y. Zhang, J. H. Sun, J. Ouyang and N. Na, ACS Appl. Mater. Interfaces, 2018, 10, 25621–25628 CrossRef CAS PubMed.
- L. He, J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon and S. M. Hammond, Nature, 2005, 435, 828–833 CrossRef CAS PubMed.
- M. Huo, S. Q. Li, P. W. Zhang, Y. M. Feng, Y. R. Liu, N. Wu, H. X. Ju and L. Ding, Anal. Chem., 2019, 91, 3374–3381 CrossRef CAS PubMed.
- P. Liu, X. H. Yang, S. Sun, Q. Wang, K. M. Wang, J. Huang, J. B. Liu and L. L. He, Anal. Chem., 2013, 85, 7689–7695 CrossRef CAS PubMed.
- F. Yang, Y. R. Cheng, Y. Cao, Y. Y. Zhang, H. F. Dong, H. T. Lu and X. J. Zhang, Anal. Chem., 2019, 91, 9828–9835 CrossRef CAS PubMed.
- D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258–3261 CrossRef CAS PubMed.
- Y. X. Xu, Q. Wu, Y. Q. Sun, H. Bai and G. Q. Shi, ACS Nano, 2010, 4, 7358–7362 CrossRef CAS PubMed.
- J. Wang, D. X. Wang, A. N. Tang and D. M. Kang, Anal. Chem., 2019, 91, 5244–5251 CrossRef CAS PubMed.
- J. T. Nofiele, G. J. Czarnota and H. L. M. Cheng, Mol. Imaging, 2014, 13, 1–8 Search PubMed.
- Y. Zhao, E. Samal and D. Srivastava, Nature, 2005, 436, 214–220 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc00394h |
| This journal is © The Royal Society of Chemistry 2020 |