Open Access Article
Open Access ArticleModular bioengineered kinase sensors via scaffold protein-mediated split-luciferase complementation†
Xiaolu
Xu‡
,
Lenne J. M.
Lemmens‡
 ,
Anniek
den Hamer
,
Anniek
den Hamer
 ,
Maarten
Merkx
,
Maarten
Merkx
 ,
Christian
Ottmann
,
Christian
Ottmann
 and
Luc
Brunsveld
and
Luc
Brunsveld
 *
*
Laboratory of Chemical Biology, Department of Biomedical Engineering, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Den Dolech 2, 5612AZ, Eindhoven, the Netherlands. E-mail: l.brunsveld@tue.nl
First published on 12th May 2020
Abstract
Phosphorylation is a key regulation event in cellular signaling. Sensing the underlying kinase activity is of crucial importance for its fundamental understanding and for drug development. For this, modular kinase activity sensing concepts are urgently needed. We engineered modular serine kinase sensors based on complementation of split NanoBiT luciferase on protein assembly platforms generated from the scaffold protein 14-3-3. The bioengineered platforms are modular and easy adaptable as exemplary shown using novel sensors for the kinases PKA, PKB, and CHK1. Two designs were conceptualized, both relying on binding of defined mono- or bivalent kinase recognition motifs to the 14-3-3 platform upon phosphorylation, resulting in reconstitution of active split-luciferase. Especially the design based on double phosphorylation and bivalent 14-3-3 binding exhibits high efficiency for signal amplification (>1000-fold) and sensitivity to specific kinases, including in cellular lysates.
Introduction
Cells respond to a diversity of internal and external signals with the appropriate physiological output through tightly controlled networks predominantly comprising signalling proteins.1 Regulation of proteins in signal transduction is coordinated by distinct mechanisms,2 with posttranslational modifications (PTMs) as important, fast, and reversible central mode-of-action.3–8 Phosphorylation is the most prevalent PTM and more than 500 kinases catalyse the phosphorylation of specific substrates. Misregulation of kinases has been connected to diverse diseases, such as various cancers and developmental-, metabolic-, and neurological disorders.9 Sensors for protein phosphorylation and kinase activity have therefore been developed and have seen applications ranging from screening in drug discovery to cellular activity studies. Notwithstanding these great achievements current sensors typically feature certain limitations such as lack of modularity10 or specificity,11 restrain to only one kinase,12 need for purified kinases,13 small dynamic ranges,14,15 or the need for genetic modification of the kinase of interest.16,17 As such, there is a need for conceptually novel sensors that can measure activities of various kinases, show a large dynamic range, and ideally are modular18 to allow the same sensor design to be applied to different kinases. Bioluminescence based sensors serve great potential over the use of fluorescence based sensors,19 as their dynamic range and sensitivity is typically higher.14-3-3 proteins are scaffold proteins that act as the main reader proteins of serine phosphorylation events and bind as dimers to a large variety of both single and double phosphorylated protein motifs.20,21 Previous work has shown the amenability of 14-3-3 proteins to protein engineering approaches towards protein assembly platforms based on the binding of phosphorylated peptide motifs.22–24 Here, we designed and engineered a set of modular serine kinase sensors comprising the 14-3-3 protein as general assembly platform for kinase phosphorylation motifs, complemented with the NanoBiT split-luciferase system.25 The modular kinase sensor design was exemplary evaluated using sensors for three clinically relevant kinases PKA, PKB/Akt, and CHK1. With these three kinases as input, split-luciferase complementation-based sensors were made with diverse peptide recognition motifs that feature large dynamic ranges combined with promising sensing selectivity.
Results and discussion
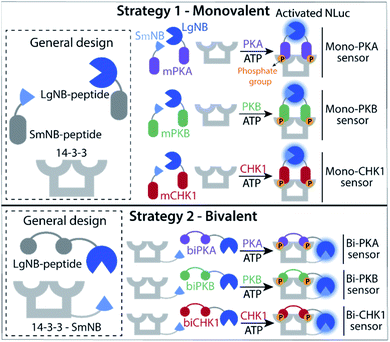
Two kinase sensing concepts were designed, based on binding of kinase-specific single or double phosphorylated peptide motifs to the 14-3-3 protein platform. The monovalent strategy (Fig. 1, strategy 1) relies on the binding and functional luciferase activation of two peptides each built up with the same kinase motifs but with either a small or large half of the NanoBiT (NB) split luciferase (SmNB-peptide and LgNB-peptide) to 14-3-3 upon phosphorylation. The intrinsic affinity for dimerization of the LgNB and SmNB elements was chosen to be relatively weak (KD = 190 μM)25 to suppress background complementation. Specific peptide motifs for the three kinases defined the Mono-PKA, Mono-PKB, and Mono-CHK1 sensors. The bivalent strategy (Fig. 1, strategy 2) is based on a 14-3-3 construct engineered to be connected to the small fragment of the split NanoBiT luciferase (14-3-3-SmNB) and constructs based on the large fragment of NanoBiT together with a peptide motif that features two phosphorylation sites for the specific kinase at hand (LgNB-peptide), resulting in the Bi-PKA, Bi-PKB, and Bi-CHK1 sensors. Phosphorylation of the LgNB-peptide would lead to a bivalent binding to 14-3-3-SmNB, enabling luciferase complementation and signal amplification. We expected the monovalent sensor to selectively form the hetero split-luciferase complex on the 14-3-3 platform because of the additional cooperative interaction between the two different luciferase parts23,26 and the bivalent sensors to assemble with a potentially higher affinity because of the underlying multivalency principle as also used by 14-3-3 in natural protein recognition events.24,27 Detailed design considerations, the selection and origin of the specific peptide sequences within the sensors, linker lengths used for the different sensors and connected references are provided in the ESI (SI1.1),† as well as protein expression protocols and protein characterization results (SI1.2, 2.1†).Monovalent sensing strategy
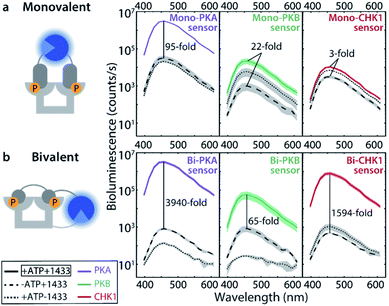
The monovalent sensing strategy was first evaluated. Mass spectrometric analyses showed that the SmNB-peptide and LgNB-peptide elements of the Mono-PKA, Mono-PKB, and Mono-CHK1 sensors could all be phosphorylated by their corresponding kinases (Fig. S3†). When studying the effect of this phosphorylation on the luciferase signal amplification, the Mono-PKA sensor featured an impressive 95-fold bioluminescence increase upon phosphorylation, with all sensor components (SmNB-peptide, LgNB-peptide, 14-3-3 dimer) present at 50 nM (Fig. 2a). As anticipated, also the presence of the 14-3-3 assembly platform was crucially important to facilitate the luciferase complementation of the phosphorylated components. The Mono-PKB and Mono-CHK1 were similarly evaluated and showed, respectively, a 22-fold and 3-fold increase in luciferase signal intensity upon phosphorylation (Fig. 2a). The strong signal amplification, as observed for the Mono-PKA sensor, probably relates to the high 14-3-3 binding affinity of the connected phosphorylated peptide motif used for this sensor.23Bivalent sensing strategy
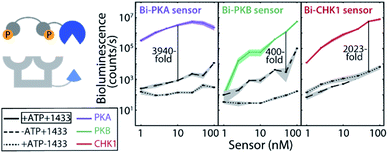
Effective two-fold phosphorylation of the three LgNB-peptide constructs from the bivalent sensors, by their respective kinases, could also be shown using mass spectrometry analyses (Fig. S4†). Functionally, the phosphorylation of the bivalent sensors led to even more impressive levels of signal amplification as compared to the monovalent sensors (Fig. 2b). Phosphorylation of the Bi-PKA sensor induced an almost 4000-fold increase in luciferase activity. Interestingly, the Bi-PKA sensor achieved a similar luciferase signal intensity as for the Mono-PKA sensor, at a 5-fold lower concentration of the sensor elements and simultaneously a 4-fold lower substrate concentration (furimazine). These features also result in a lowering of the background signals generated by the large NanoBiT element of the unphosphorylated sensor, or in the absence of the 14-3-3-SmNB assembly platform. The Bi-PKB and Bi-CHK1 sensors similarly showed attractive levels of signal amplification upon phosphorylation of around 65- and 1600-fold respectively. For these two kinases the bivalent binding of the phosphorylation motifs to the 14-3-3 platform thus crucially enhances the affinity for complex formation, as compared to the monovalent sensors. This enhanced binding affinity for the bivalent constructs20,27 allows usage of lower sensor concentrations with concomitantly lowered background activity, but still with enhanced sensor reconstitution. Notwithstanding the promising data for the monovalent sensors, we thus focused the subsequent in depth evaluation of the sensors on the bivalent platform.The dynamic range of the bivalent sensors was evaluated by analysing the concentration dependences of the sensors. All three bivalent sensors showed impressive signal to background values over a broad concentration window (Fig. 3 and S5–7†). Each sensor featured its own ideal concentration for optimal signal intensity vs. background signal, typically at sensor concentrations between 10 and 100 nM and most likely relating to a combination and interplay of intermolecular events, including the binding affinities of the specific phospho-epitopes to the 14-3-3 platform and the background affinities of the peptide-protein interaction and of the NanoBiT (NB) split luciferase elements. The data also showed that even at low sensor concentration, down to 1 nM for the Bi-PKA and the Bi-CHK1 sensors, impressive signal intensity and amplification is still achieved.
An alternative for modulating and understanding the stability of the sensor responses is by modulating the ratio of the two sensor elements. Titration of the 14-3-3-SmNB platform (itself showing no background luciferase activity) to a constant concentration of the phosphorylated kinase peptide epitopes connected to the LgNB element revealed a remarkable high and constant signal output (Fig. S8–S10†). The 14-3-3-SmNB platform could be added in up to 100-fold excess without affecting the signal intensity, nor signal-to-background values, strongly. All these features together hint to a sensor design which is applicable and signal-stable over broad concentrations windows.
The bivalent sensors were subsequently evaluated regarding their real-time luciferase activity upon kinase addition (Fig. S11a and S12†). Incubation of each sensor with its respective kinase and ATP led to a clearly detectable time-dependent increase of the bioluminescence of several orders of magnitude. The responsiveness of the sensors is almost immediate, on the time-scale of the measurement, as adding the kinases directly resulted in detectable signal enhancements. This result testifies to the rapid phosphorylation by the kinases and subsequent rapid sensor assembly on the 14-3-3 platform and split-luciferase reconstitution. When performing these analyses over a range of kinase concentrations (Fig. S11a and S12†) a similar fast response was observed. Even though the Bi-PKB sensor has a ten-fold lower signal-to-background ratio in comparison to the Bi-PKA sensor (3940-fold), also this sensor shows an excellent time-responsiveness even for low PKB concentrations (Fig. S12a†). These results demonstrate the applicability of the sensors for real-time activity sensing and their relevant sensitivity over a range of different kinase concentrations; all at low sensor concentrations of 10 nM only.
The time-responsiveness of the Bi-PKA sensor (10 nM) upon varying PKA concentrations was compared to the response of the commercial PKA activity kit AssayQuant PhosphoSens (10 μM sensor concentration). Linear regression was applied to the initial linear part of the time traces to determine the velocity of signal growth for the various PKA concentrations (Fig. 4b and S11†). Both assays show a clear PKA concentration dependent increase in velocity. The Bi-PKA 0.02 U μL−1 datapoint approaches the limit of detection (LOD) slightly closer as compared to the AssayQuant assay, suggesting higher sensitivity for the commercial kit. In contrast, the Bi-PKA sensor shows a bigger spread in velocity values, suggesting higher precision compared to the AssayQuant assay.
The kinase selectivity of the three bivalent sensors was tested within the panel of the three kinases studied at set kinase concentrations (Fig. 4c–e). All three bivalent sensors performed with gratifying selectivity towards the kinase they were specifically designed for. Differences in bioluminescence signal intensity could already be observed within minutes after addition of the kinases to the sensors. While such selectivity effects are not of relevance in, for example, biochemical kinase inhibitor screens, they do provide an exciting entry for application of the sensors in complex biological media.
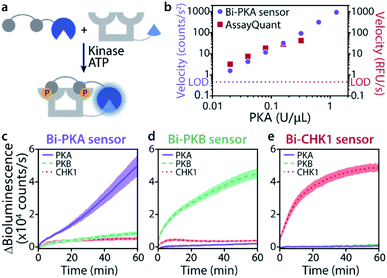 | ||
Fig. 4 Time-responsiveness (b) and kinase selectivity of bivalent sensors (c–e). (a) Schematic representation of experiment. (b) Velocity upon phosphorylation of Bi-PKA (10 nM) or AssayQuant with varying PKA concentrations (0.02–1.28 and 0.02–0.32 U μL−1, respectively). Signal intensities were normalized to background, the velocity was derived from the linear part of the curves. Error bars represent the standard deviation of triplicates, smaller than datapoint symbols. The limit of detection (LOD) is defined as three times the standard deviation of the background. Time-response curves and regression can be found in SI2.3.† (c–e) Bi-PKA, Bi-PKB, and Bi-CHK1 (10 nM) were each incubated with PKA (0.1 U μL−1), PKB (0.008 U μL−1) and CHK1 (0.1 U μL−1). Furimazine was used at a final dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 (to avoid its depletion) and background (without kinase) was subtracted. Error clouds represent the standard error of triplicates. 2000 (to avoid its depletion) and background (without kinase) was subtracted. Error clouds represent the standard error of triplicates. | ||
Kinase activity sensing in complex media
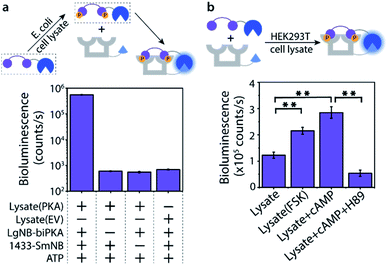
The Bi-PKA sensor was used to test the applicability of the novel sensing concept to detect kinase activity in complex biological media like cell lysates. First the activity of a PKA overexpressed in E. coli cells was investigated. The LgNB-biPKA component of the sensor, containing the two PKA phosphorylation sites, was incubated with the E. coli lysate and additional ATP for 3 hours after which the 14-3-3-SmNB platform was added to reconstitute the functional sensor. A strong bioluminescent signal was observed only in the presence of all sensor components (Fig. 5a, S13 and S14†), testifying to the large dynamic range of the sensor system also in complex media. The around 1000-fold lower luminescence signal generated by the E. coli lysate without PKA (EV; empty vector) or when either of the two protein elements of the Bi-PKA sensor is absent pin-point the selective activation of the sensor by the PKA, the effectiveness of the sensor in the lysate, and the need for full sensor complementation.The activity of native PKA, as present in eukaryotic cells, was evaluated using the exemplary human 293T cell line. Incubation of the Bi-PKA sensor with 293T lysate resulted in a substantial bioluminescence signal, not present in absence of lysate (Fig. 5b and S15†). The signal output of the Bi-PKA sensor was sensitive, and increased, to stimulation of the PKA signalling pathway by culturing the cells in the presence of forskolin (FSK), a small molecule activator of the adenylate cyclase pathway. Direct addition of cyclic AMP (cAMP), an activator of PKA, to the 293T lysate could also be sensed, and resulted in a more than 3-fold increase in signal output by the Bi-PKA sensor. Furthermore, the Bi-PKA sensor was also capable of sensing PKA inhibition in the cellular lysate, as revealed by the inhibition of the bioluminescence upon addition of the PKA inhibitor H89.
Conclusions
The new protein designs presented in this manuscript address the need for modular kinase sensors with large dynamic ranges and facile applicability, as illustrated by sensors for three exemplary serine kinases. The modular sensors are based on the 14-3-3 scaffold protein as integrator platform of protein serine phosphorylation combining binding and NanoBiT split-luciferase reconstitution. Of the two conceptual designs, the monovalent sensors showed attractive and selective signal amplification, which was even surpassed by the bivalent sensors with an additional broad concentration efficiency and kinase selectivity. As an example, the Bi-PKA sensor was shown also to be effective in diverse cellular lysates and capable of sensing PKA presence and activity in complex media. The flexibility of the sensor design allows development of sensors for different kinases by implementing one or two kinase recognition motifs in the bioengineered protein constructs. While the need for a synthetic luciferase substrate might limit easy applications in cells, we envision these kind of sensors to lead to broad applications for in vitro small molecule screenings and diagnostic kinase activity profiling.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the Netherlands Organization for Scientific Research (NWO) (Gravity Program 024.001.035 and Vici grant 016.150.366).Notes and references
- W. A. Lim, Nat. Rev. Mol. Cell Biol., 2010, 11, 393–403 CrossRef CAS PubMed.
- M. J. Lee and M. B. Yaffe, Cold Spring Harbor Perspect. Biol., 2016, 8, a005918 CrossRef PubMed.
- C. T. Walsh, S. Garneau-Tsodikova and G. J. Gatto, Angew. Chem., Int. Ed., 2005, 44, 7342–7372 CrossRef CAS PubMed.
- R. M. Gordley, R. E. Williams, C. J. Bashor, J. E. Toettcher, S. Yan and W. A. Lim, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 13528–13533 CrossRef CAS PubMed.
- X. Li, M. Wilmanns, J. Thornton and M. Kohn, Sci. Signaling, 2013, 6, rs10 CrossRef.
- Y. Chen, Y. Liu, T. Lan, W. Qin, Y. Zhu, K. Qin, J. Gao, H. Wang, X. Hou, N. Chen, J. P. Friedmann Angeli, M. Conrad and C. Wang, J. Am. Chem. Soc., 2018, 140, 4712–4720 CrossRef CAS PubMed.
- N. M. Riley, A. S. Hebert, M. S. Westphall and J. J. Coon, Nat. Commun., 2019, 10, 1311 CrossRef PubMed.
- S. Liokatis, A. Dose, D. Schwarzer and P. Selenko, J. Am. Chem. Soc., 2010, 132, 14704–14705 CrossRef CAS PubMed.
- P. Lahiry, A. Torkamani, N. J. Schork and R. A. Hegele, Nat. Rev. Genet., 2010, 11, 60–74 CrossRef CAS PubMed.
- J. G. Albeck, G. B. Mills and J. S. Brugge, Mol. Cell, 2013, 49, 249–261 CrossRef CAS PubMed.
- Y. Liu, J. Lee, L. Perez, A. D. Gill, R. J. Hooley and W. Zhong, J. Am. Chem. Soc., 2018, 140, 13869–13877 CrossRef CAS PubMed.
- Y. Chen, J. L. Saulnier, G. Yellen and B. Sabatini, Front. Pharmacol., 2014, 5, 56 Search PubMed.
- H. Zegzouti, M. Zdanovskaia, K. Hsiao and S. A. Goueli, Assay Drug Dev. Technol., 2009, 7, 560–572 CrossRef CAS PubMed.
- G. C. H. Mo, B. Ross, F. Hertel, P. Manna, X. Yang, E. Greenwald, C. Booth, A. M. Plummer, B. Tenner, Z. Chen, Y. Wang, E. J. Kennedy, P. A. Cole, K. G. Fleming, A. Palmer, R. Jimenez, J. Xiao, P. Dedecker and J. Zhang, Nat. Methods, 2017, 14, 427–434 CrossRef CAS PubMed.
- S. Regot, J. J. Hughey, B. T. Bajar, S. Carrasco and M. W. Covert, Cell, 2014, 157, 1724–1734 CrossRef CAS PubMed.
- J. D. Vasta, C. R. Corona, J. Wilkinson, C. A. Zimprich, J. R. Hartnett, M. R. Ingold, K. Zimmerman, T. Machleidt, T. A. Kirkland, K. G. Huwiler, R. F. Ohana, M. Slater, P. Otto, M. Cong, C. I. Wells, B.-T. Berger, T. Hanke, C. Glas, K. Ding, D. H. Drewry, K. V. M. Huber, T. M. Willson, S. Knapp, S. Müller, P. L. Meisenheimer, F. Fan, K. V. Wood and M. B. Robers, Cell Chem. Biol., 2018, 25, 206–214 CrossRef CAS PubMed.
- H. Eishingdrelo, J. Cai, P. Weissensee, P. Sharma, M. J. Tocci and P. S. Wright, Curr. Chem. Genomics, 2011, 5, 122–128 CrossRef CAS PubMed.
- Z. Guo, W. A. Johnston, J. Whitfield, P. Walden, Z. Cui, E. Wijker, S. Edwardraja, I. Retamal Lantadilla, F. Ely, C. Vickers, J. P. J. Ungerer and K. Alexandrov, J. Am. Chem. Soc., 2019, 141, 8128–8135 CAS.
- L. B. Peterson, M. B. Yaffe and B. Imperiali, Biochemistry, 2014, 53, 5771–5778 CrossRef CAS PubMed.
- M. B. Yaffe, K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon and L. C. Cantley, Cell, 1997, 91, 961–971 CrossRef CAS PubMed.
- C. Johnson, M. Tinti, N. T. Wood, D. G. Campbell, R. Toth, F. Dubois, K. M. Geraghty, B. H. C. Wong, L. J. Brown, J. Tyler, A. Gernez, S. Chen, S. Synowsky and C. MacKintosh, Mol. Cell. Proteomics, 2011, 10, M110.005751 CrossRef PubMed.
- M. Skwarczynska, M. Molzan and C. Ottmann, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, E377–E386 CrossRef CAS PubMed.
- A. den Hamer, L. J. M. Lemmens, M. A. D. Nijenhuis, C. Ottmann, M. Merkx, T. F. A. de Greef and L. Brunsveld, Chembiochem, 2017, 18, 331–335 CrossRef CAS PubMed.
- S. J. A. Aper, A. den Hamer, S. F. A. Wouters, L. J. M. Lemmens, C. Ottmann, L. Brunsveld and M. Merkx, ACS Synth. Biol., 2018, 7, 2216–2225 CrossRef CAS PubMed.
- A. S. Dixon, M. K. Schwinn, M. P. Hall, K. Zimmerman, P. Otto, T. H. Lubben, B. L. Butler, B. F. Binkowski, T. Machleidt, T. A. Kirkland, M. G. Wood, C. T. Eggers, L. P. Encell and K. V. Wood, ACS Chem. Biol., 2016, 11, 400–408 CrossRef CAS PubMed.
- R. P. G. Bosmans, J. M. Briels, L.-G. Milroy, T. F. A. de Greef, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed. Engl., 2016, 55, 8899–8903 CrossRef CAS PubMed.
- L. M. Stevers, P. J. de Vink, C. Ottmann, J. Huskens and L. Brunsveld, J. Am. Chem. Soc., 2018, 140, 14498–14510 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, supporting figures and supporting discussion. See DOI: 10.1039/d0sc00074d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |