Open Access Article
Open Access ArticleSynthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp2-iminosugar fragment†
Iris A.
Bermejo
a,
Claudio D.
Navo
 ab,
Jorge
Castro-López
c,
Ana
Guerreiro
d,
Ester
Jiménez-Moreno
a,
Elena M.
Sánchez Fernández
e,
Fayna
García-Martín
ab,
Jorge
Castro-López
c,
Ana
Guerreiro
d,
Ester
Jiménez-Moreno
a,
Elena M.
Sánchez Fernández
e,
Fayna
García-Martín
 f,
Hiroshi
Hinou
f,
Shin-Ichiro
Nishimura
f,
Hiroshi
Hinou
f,
Shin-Ichiro
Nishimura
 f,
José M.
García Fernández
f,
José M.
García Fernández
 g,
Carmen Ortiz
Mellet
g,
Carmen Ortiz
Mellet
 e,
Alberto
Avenoza
a,
Jesús H.
Busto
a,
Gonçalo J. L.
Bernardes
e,
Alberto
Avenoza
a,
Jesús H.
Busto
a,
Gonçalo J. L.
Bernardes
 dh,
Ramón
Hurtado-Guerrero
cij,
Jesús M.
Peregrina
dh,
Ramón
Hurtado-Guerrero
cij,
Jesús M.
Peregrina
 *a and
Francisco
Corzana
*a and
Francisco
Corzana
 *a
*a
aDepartamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química, E-26006 Logroño, Spain. E-mail: francisco.corzana@unirioja.es; jesusmanuel.peregrina@unirioja.es
bCIC BioGUNE, Bizkaia Technology, Park Building 800, 48170 Derio, Spain
cInstitute of Biocomputation and Physics of Complex Systems (BIFI), University of Zaragoza, Zaragoza, Spain
dInstituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal
eDepartamento de Química Orgánica, Facultad de Química, Universidad de Sevilla, E-41012 Sevilla, Spain
fGraduate School and Faculty of Advanced Life Science, Laboratory of Advanced Chemical Biology, Hokkaido University, N21 W11, Sapporo 001-0021, Japan
gInstituto de Investigaciones Químicas (IIQ), CSIC–Universidad de Sevilla, E-41092 Sevilla, Spain
hDepartment of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK
iCopenhagen Center for Glycomics, Department of Cellular and Molecular Medicine, School of Dentistry, University of Copenhagen, Copenhagen, Denmark
jFundación ARAID, Zaragoza, Spain
First published on 27th March 2020
Abstract
The Tn antigen (GalNAc-α-1-O-Thr/Ser) is a well-known tumor-associated carbohydrate determinant. The use of glycopeptides that incorporate this structure has become a significant and promising niche of research owing to their potential use as anticancer vaccines. Herein, the conformational preferences of a glycopeptide with an unnatural Tn antigen, characterized by a threonine decorated with an sp2-iminosugar-type α-GalNAc mimic, have been studied both in solution, by combining NMR spectroscopy and molecular dynamics simulations, and in the solid state bound to an anti-mucin-1 (MUC1) antibody, by X-ray crystallography. The Tn surrogate can mimic the main conformer sampled by the natural antigen in solution and exhibits high affinity towards anti-MUC1 antibodies. Encouraged by these data, a cancer vaccine candidate based on this unnatural glycopeptide and conjugated to the carrier protein Keyhole Limpet Hemocyanin (KLH) has been prepared and tested in mice. Significantly, the experiments in vivo have proved that this vaccine elicits higher levels of specific anti-MUC1 IgG antibodies than the analog that bears the natural Tn antigen and that the elicited antibodies recognize human breast cancer cells with high selectivity. Altogether, we compile evidence to confirm that the presentation of the antigen, both in solution and in the bound state, plays a critical role in the efficacy of the designed cancer vaccines. Moreover, the outcomes derived from this vaccine prove that there is room for exploring further adjustments at the carbohydrate level that could contribute to designing more efficient cancer vaccines.
Introduction
The Tn antigen is a specific human tumor-associated carbohydrate antigen (TACA) formed by N-acetylgalactosamine (GalNAc) α-O-linked to either serine (Ser) or threonine (Thr) residues.1 The overexpression of this simple structure correlates well with several types of tumors and it has been associated with metastatic behavior and tumor expansion. In general, the Tn antigen appears on the surface of epithelial cells as part of tumor-associated mucins. Mucins are heavily glycosylated proteins that play a key role in several biological processes,2 such as tissue inflammation, immune response or intercellular recognition. One of the most relevant mucins is MUC1,3,4 which is composed of 20–125 tandem-type repeats of a core sequence of 20 amino acids “HGVTSAPDTRPAPGSTAPPA”. This sequence presents five potential O-glycosylation sites made up of two Ser and three Thr residues (shown in bold letters in the tandem repeat sequence). In general, although this glycoprotein is decorated with complex O-glycans in healthy cells, in tumor tissues it usually presents simple carbohydrates, due mainly to the malfunction or relocation of GalNAc-transferases or mutations in COSMC, which is a chaperone for the follow-up glycosyltransferase C1GalT.1,3,4 Consequently, different antigens that are masked in healthy cells, such as the Tn antigen, are exposed in malignancies and can trigger an immune response. Several studies have demonstrated the occurrence of anti-MUC1 antibodies in cancer patients that can limit tumor outgrowth and dissemination.5–7 Moreover, the sequence PDTRP of MUC1 is the most immunogenic epitope recognized by anti-MUC1 antibodies,8–10 such as the SM3 antibody, which has great potential to be used in early diagnosis and treatment of breast cancer.11 For these reasons, MUC1 is considered a promising target for the development of cancer vaccines.12–14 To date, most of these vaccines have failed in clinical trials.3 An important drawback of these potential vaccines is their tendency to suffer from immunotolerance due to the presence of TACAs at low concentrations in healthy cells.15 Additionally, the short biological half-life that some of these O-glycopeptides present is related to enzymatic degradation.15,16 Thus, the use of unnatural TACA analogs that are more resistant to degradation and able to provide stronger and longer lasting immunogenicity represents an attractive and required niche of research in the development of anti-cancer vaccines.17 Given that the O-glycosidic bond is particularly sensitive to enzymatic degradation in biological media, the incorporation of GalNAc moieties chemically modified in the anomeric region into carbohydrate-based cancer vaccines has attracted much attention in recent years.18–22 These modifications included the use of C- and S-glycosides23–25 or deoxyfluoroglycosides,26–31 among others. We have recently contributed to this field by reporting the first examples of Tn antigens that feature a serine or a threonine glycosylated with a bicyclic sp2-iminosugar moiety with a substitution and configurational pattern of structural complementarity to α-GalNAc (α-sp2GalNAc),32 namely compound 1a and the corresponding mucin-related glycopeptide 2a (Fig. 1). The presence of the sp2-hybridized pseudoamide nitrogen at the position of the endocyclic oxygen in monosaccharides drastically enhances the anomeric effect in sp2-iminosugar glycomimetics, which enables access to chemically and enzymatically stable α-linked conjugates with total stereocontrol.33–35 We have also proved that the antibody scFv-SM3 exhibits a 2-fold higher affinity towards unnatural glycopeptide 2a (KD = 1.60 ± 0.16 μM) relative to the natural variant 2b (KD = 3.30 ± 0.84 μM).32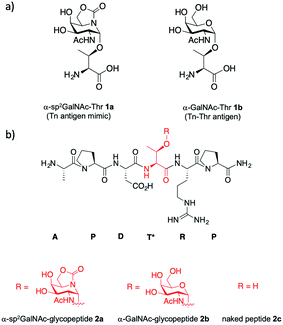 | ||
| Fig. 1 (a) Structures of α-sp2GalNAc-Thr 1a and Tn antigen with threonine 1b. (b) Structures of unnatural α-sp2GalNAc-glycopeptide 2a and the natural variants 2b and 2c. | ||
Here, we go one step further and report the synthesis and evaluation in vivo of a two-component anti-cancer vaccine (derivative 3a), composed of Tn mimic α-sp2 GalNAc-Thr (1a) in the most immunogenic domain of a MUC1 fragment and immunogenic carrier protein Keyhole Limpet Hemocyanin36 (KLH, Fig. 2). This protein was selected because it is one of the most popular protein carriers for carbohydrate-based cancer vaccines. Conjugates of this protein with several TACAs, such as Tn, GM2, GD2, Globo-H, and MUC1, have been used in clinical trials.37–42 Moreover, it has been observed that KLH-MUC1 conjugates can elicit anti-MUC1 antibodies in human patients.37,43–45 Conjugate 3b, which displays natural Tn-Thr antigen GalNAc-α-1-O-Thr in its structure, was also synthesized and tested for comparison. In addition, and with the aim of providing a rational explanation for the higher binding affinity of the unnatural derivative, glycopeptide 2a was synthesized and its structure was studied both in solution, through a combination of NMR spectroscopic experiments and molecular dynamics (MD) calculations, and in the solid state, by X-ray crystallography.
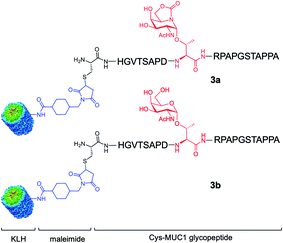 | ||
| Fig. 2 Two-component cancer vaccine candidates KLH-3a (upper panel) and KLH-3b (lower panel) synthesized in this work from glycopeptides 3a and 3b, respectively. | ||
Results and discussion
Unnatural glycopeptide 2a presents a relatively high affinity towards the SM3 antibody. To verify this as a general trend, we analysed the binding of this derivative, as well as compounds 2b and 2c, with commercially available monoclonal antibody VU-11E22,9,10 by using a microarray assay (Fig. S1 and S2†). The results derived from these experiments indicated that the antibody VU-11E2 has a preference for the glycosylated derivatives 2a and 2b relative to the naked peptide 2c, which is in good agreement with previous studies.9 However, in contrast to the SM3 antibody,32 a slightly higher affinity of the VU-11E2 antibody was observed for natural glycopeptide 2b. Therefore, the binding studies accomplished with SM3 and VU-11E2 antibodies showed that they recognize the unnatural glycopeptide 2a and the natural variant 2b with a comparable affinity, exhibiting a worse binding to the naked derivative 2c.To provide a rational explanation of the relatively high affinity of 2a towards these anti-MUC1 antibodies, we performed the conformational analysis of this glycopeptide in solution and in the SM3-bound state.
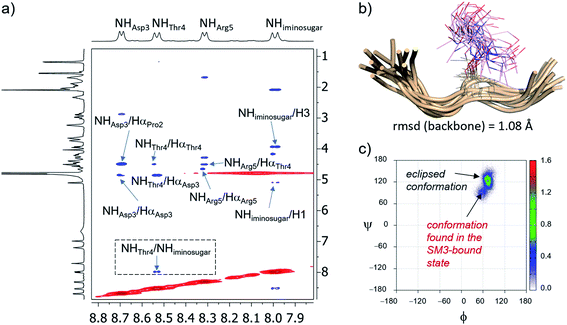
In solution, 2D NOESY experiments showed key sequentially strong NH/Hα (i, i + 1) connectivities, such as NHAsp/HαPro, NHArg/HαThr or NHThr/HαAsp, along with weak or absent NH–NH (i, i + 1) NOE interactions, which suggests a preference for extended conformations of the peptide in solution46 (Fig. 3a and Table S1†). These proton–proton distances were then used as restraints in experiment-guided MD simulations (MD simulations with time-averaged restraints; MD-tar),47 following our well-established protocol.48 These calculations were performed with AMBER 18 software,49 implemented with ff14SB,50 GLYCAM 06 (ref. 51) and general Amber force field (GAFF)52 forcefields, and provided a distribution of low-energy conformers able to reproduce NOESY data. The good agreement between the experimental and calculated distances validates the theoretical study (Table S1†). Fig. 3b shows the structural ensemble derived from 20 ns experiment-guided MD simulations. According to these calculations, and in agreement with the NOE contacts, the peptide backbone of unnatural glycopeptide 2a adopts an extended conformation in water (see also Fig. S3†). Moreover, the side-chain of the threonine derivative is quite rigid (χ1 close to 60°), in agreement with previous studies.53,54 Therefore, the conformational space sampled by the peptide fragment of compound 2a is similar to that reported for the natural variant 2b.55
Concerning the glycosidic linkage, the torsional angle ψ takes values of around 120°, which is characteristic of an eclipsed conformation (Fig. 3c).53,54 This geometry is supported by a medium-size NOE cross-peak between the NH group of the threonine (NHThr4) residue and the NH of the sugar (NHiminosugar, Fig. 3a). Of note, a low-populated conformer of the glycosidic linkage (population < 10%), characterized by values of ψ in the range of 60–90°, is also observed in solution. Markedly, this geometry of the glycosidic linkage, which is not populated by the natural variant 2b,55 is recognized by the SM3 antibody (see below).
Next, to evaluate compound 2a in the bound state, we prepared high-quality crystals of the antibody scFv-SM3 in complex with this antigen and performed the subsequent crystallographic analysis. The structure, obtained at a resolution of 2.11 Å (Table S2†), reveals that the surface groove of the antibody nicely fits all the peptide residues (Fig. 4a), as previously observed for the natural glycopeptide 2b and its naked variant (compound 2c in Fig. 1).55 Moreover, the conformation of the peptide backbone is almost identical to that displayed by 2b, indicating that the presence of the unnatural sp2-iminosugar within the GalNAc moiety does not significantly modify the overall conformation of the glycopeptide in the bound state (Fig. 4c).
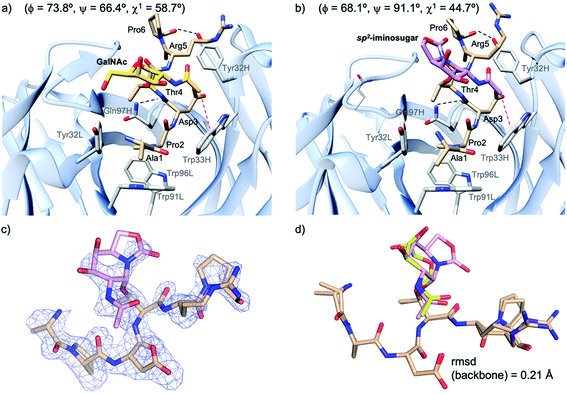 | ||
| Fig. 4 Key binding interactions of glycopeptide 2b (a) and 2a (b) with antibody scFv-SM3, as observed in the X-ray crystal structure (pdb id: 5A2K and 6TGG, respectively). Pink dashed lines indicate the hydrophobic interaction between the sugar and the scFv-SM3 surface and black dashed lines indicate hydrogen bonds. The geometry of the glycosidic linkage is shown in parentheses. (c) Electron density maps are FO–FC syntheses (blue) contoured at 2.0σ for glycopeptide 2a. (d) Superposition of the peptide backbone of glycopeptides 2a and 2b in complex with scFv-SM3. Peptide backbone carbon atoms are shown in tan. The sugar moiety in 2a and 2b is in plum and yellow, respectively. | ||
The non-covalent interactions between the antibody and the unnatural antigen are the same as for 2b and 2c, involving several hydrogen bonds, some of them water-mediated, and stacking interactions. Thus, Pro2 stacks with Tyr32L, Trp91L and Trp96L, while Asp3 and Arg5 interact hydrophobically with Trp33H and Tyr32H, respectively. In addition, the CO group of Thr4 and Pro6 interacts through a hydrogen bond with Gln97H and Tyr32H, respectively.
The torsional angle ϕ of the glycosidic linkage takes a value close to 73°, in accordance with the exo-anomeric effect,56 while ψ is close to 66°. This value of ψ, which is also exhibited by glycopeptide 2a in the free state in solution, has not been detected for natural glycopeptide 2b in the SM3-bound state. This characteristic geometry of the glycosidic linkage can enhance the CH/π interaction between the methyl group of the sugar and Trp33H (Fig. 4b).
It can be concluded that glycopeptide 2a is quite rigid in solution and displays mainly a similar conformation to the natural variant 2b. In the SM3-bound state, these two glycopeptides adopt a different disposition of the sugar moiety. Of note, in derivative 2a, the N-acetyl group of the sugar is closer to a tryptophan residue, which is partly responsible for the better binding of the unnatural glycopeptide. This, together with the occurrence of this conformer in solution, may explain the better affinity reported for unnatural glycopeptide 2a relative to the natural derivative 2b.
Encouraged by the high capacity of derivative 2a to mimic the natural antigen, we carried out the synthesis of two-component cancer vaccine57KLH-3a (Fig. 2), together with the natural variant KLH-3b for comparison. Vaccine KLH-3a combines MUC1-glycopeptide 3a (Scheme 1), which features sp2-iminosugar Tn mimic 1a in the tandem repeat sequence of MUC1 and a Cys residue at its N-terminus to allow the conjugation to the immunogenic carrier protein KLH. The synthesis of 3a was accomplished by using microwave-assisted solid phase peptide synthesis (MW-SPPS). Once purified by reverse-phase (RP)-HPLC and lyophilized, compound 3a was conjugated to the KLH protein by the maleimide protocol by using succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) as a heterobifunctional linker (Scheme 1). Natural MUC1 derivative 3b and naked peptide 3c (see the ESI†), which contain antigen 1b or a threonine residue, respectively, were prepared following the same protocol as described in Scheme 1. The yield of the conjugation reactions of derivatives 3a and 3b to KLH was estimated by immunoassay experiments58 and determined by UPLC/MS (see Experimental section).
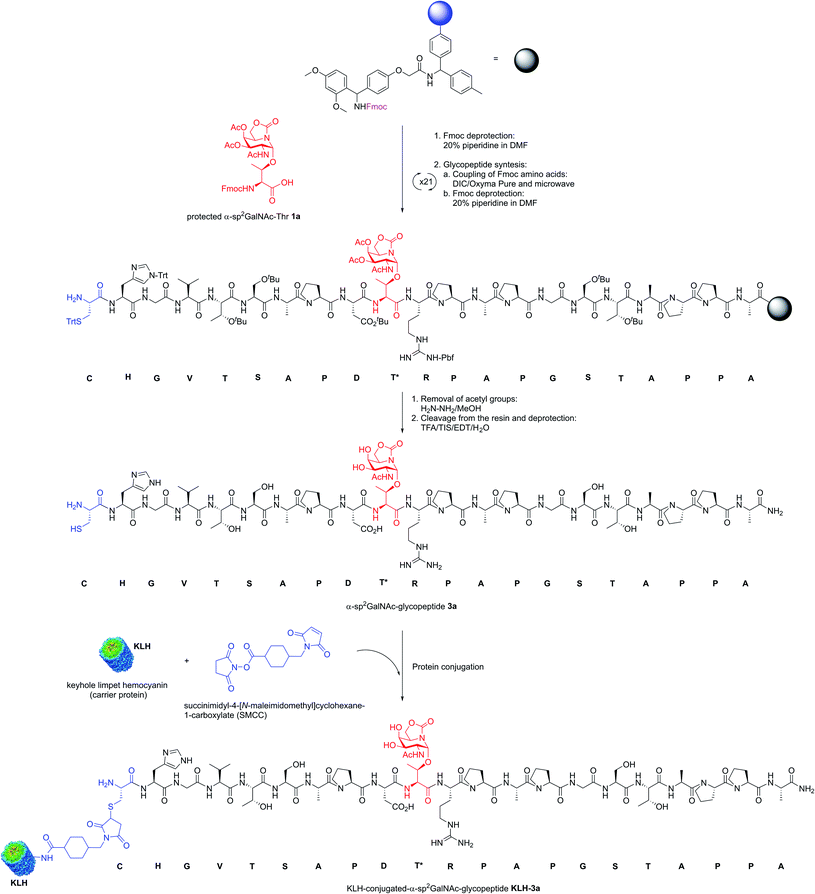 | ||
| Scheme 1 Synthesis of KLH-conjugated-α-sp2GalNAc-glycopeptide KLH-3a as a two-component cancer vaccine candidate. | ||
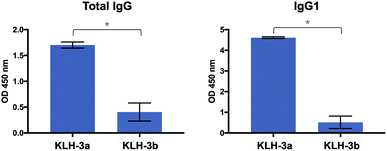
Next, Balb/c mice were immunized with KLH-3a and KLH-3b four times at biweekly intervals. The first immunizations were performed with complete Freund's adjuvant and the others were accomplished with incomplete Freund's adjuvant. One week after the last immunization, the mice were sacrificed, and serum was harvested. Murine IgG and IgM antibody responses of sera were characterized by ELISA. As can be seen in Fig. 5 and S4,† mice immunized with these vaccine candidates elicited a specific anti-MUC1 IgG antibody response and, more importantly, the unnatural vaccine KLH-3a was able to elicit higher levels of IgG antibodies in comparison to the natural variant KLH-3b. Regarding sub-typing of the IgG antibodies, both vaccines show IgG1 titers significantly higher than IgG2a (Fig. 5 and S4†), which indicates a prevalence of a Th1 response (humoral immunity).59,60 The data also reveal that a T cell-mediated class-switching takes place, because IgM antibody values were low with both vaccines (Fig. S4†).14 However, antibodies produced by both vaccines recognized likewise glycosylated and naked MUC1 epitopes (Fig. S5†). This result indicates that the elicited antibodies target mainly the peptide and not the glycan moiety.
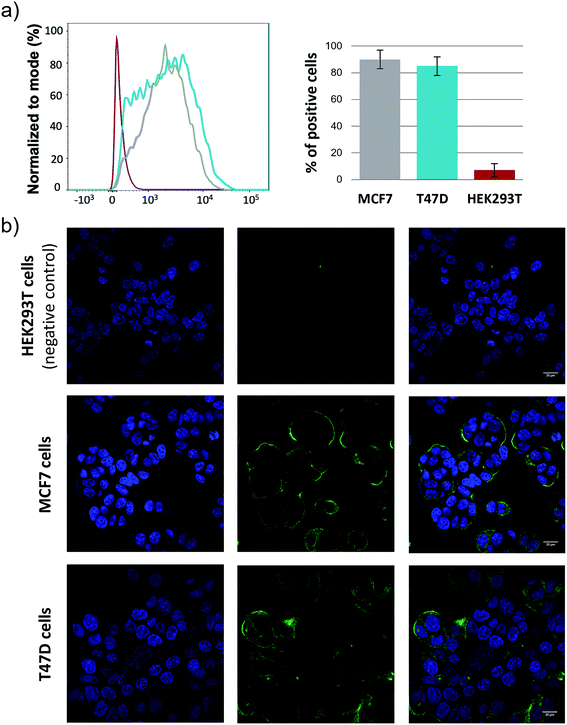
Finally, it is important to note that the elicited antibodies can selectively recognize native tumor associated MUC1 presented on the surface of human cancer cells. To demonstrate this, MCF7 and T47D cell lines, which express tumor associated MUC1 on their surface, together with a human embryonic kidney cell line (HEK293T), which was used as a negative control, were treated with antisera obtained after immunization with KLH-3a and analyzed by flow cytometry.48 The experiments showed that MCF7 and T47D strongly react with the antisera, while HEK293T cells were not targeted (Fig. 6a). These results support subsequent confocal microscopy experiments (Fig. 6b). Taken together, this result suggests that antibodies generated after the immunization of mice with unnatural vaccine KLH-3a can selectively recognize human cancer cells.
Conclusions
A MUC1-like glycopeptide with an unnatural Tn antigen that features a threonine glycosylated with an sp2-iminosugar GalNAc mimetic has been conjugated to the carrier protein KLH and tested in mice. The experiments in vivo show that this vaccine elicits specific anti-MUC1 IgG antibodies that outnumber those obtained with the natural analog. More importantly, these antibodies are able to recognize human cancer cell lines that express tumor associated MUC1 on their surface. On the one hand, the absence of the sp2-iminosugars in organisms might enhance the immunogenicity of the designed antigen. This feature, combined with the resistance of this unnatural scaffold to degradation, could contribute significantly to the effectiveness of the vaccine. On the other hand, the conformational analysis performed by NMR spectroscopy and MD simulations demonstrates that the unnatural derivative is rigid in solution and adopts a geometry similar to that found for the natural Tn-glycopeptide. According to the crystallographic analysis, the SM3-bound state reveals that the glycosidic linkage of the unnatural glycopeptide displays an orientation that facilitates the CH/π interaction between the methyl of the N-acetyl group of the sugar and a tryptophan residue, which is partly responsible for the better binding of the unnatural glycopeptide. As we have previously reported, rather flexible determinants can hinder the boosting of the immune system.61 Thus, according to the present work, when the engineered antigen is capable of copying the conformational landscape of the natural Tn antigen and exhibits a good affinity towards anti-MUC1 antibodies, this determinant is a priori a good candidate to develop effective cancer vaccines.48Therefore, the results reported in this work can contribute to exploiting the rational design of cancer vaccines.
Experimental section
Reagents and general procedures
Commercial reagents were used without further purification. Solvents were dried and redistilled prior to use in the usual way. 1H and 13C NMR spectra were measured with Bruker Avance-400 and Bruker ARX-300 spectrometers in D2O or H2O/D2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 298 K. Spectra were assigned by using COSY and HSQC. High-resolution electrospray ionization (ESI) mass spectra were recorded on a microTOF-Q-BRUKER spectrometer; accurate mass measurements were achieved by using sodium formate as an external reference.
1) at 298 K. Spectra were assigned by using COSY and HSQC. High-resolution electrospray ionization (ESI) mass spectra were recorded on a microTOF-Q-BRUKER spectrometer; accurate mass measurements were achieved by using sodium formate as an external reference.
NMR experiments
NMR experiments were performed with a 400 MHz spectrometer at 298 K. Magnitude-mode ge-2D COSY spectra were acquired with gradients by using the cosygpqf pulse program with a pulse width of 90°. Phase-sensitive ge-2D HSQC spectra were acquired by using a z-filter and selection before t1, removing the decoupling during acquisition by use of the invigpndph pulse program with CNST2 (JHC) = 145.2D NOESY experiments
NOESY experiments were performed with a Bruker Avance 400 MHz spectrometer at 298 K and pH 6.0–6.5 in H2O/D2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The experiments were conducted by using phase-sensitive ge-2D NOESY with WATERGATE for H2O/D2O (9
1). The experiments were conducted by using phase-sensitive ge-2D NOESY with WATERGATE for H2O/D2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) spectra. NOE intensities were normalized with respect to the diagonal peak at zero mixing time. Distances involving NH protons were semi-quantitatively determined by integrating the volume of the corresponding cross-peaks. The number of scans used was 16 and the mixing time was 500 ms.
1) spectra. NOE intensities were normalized with respect to the diagonal peak at zero mixing time. Distances involving NH protons were semi-quantitatively determined by integrating the volume of the corresponding cross-peaks. The number of scans used was 16 and the mixing time was 500 ms.
Microwave assisted solid-phase peptide synthesis (MW-SPPS)
This synthesis was performed automatically with Rink Amide MBHA resin (0.05 mmol) and an automated synthesizer. DIC/Oxyma Pure® was used as the coupling reagent and 20% (v/v) solution of piperidine in dimethylformamide (DMF) was used for fluorenylmethyloxycarbonyl (Fmoc) deprotection. The glycosylated amino acid building blocks were synthesized as described in the literature:32,62 the corresponding amino acid (1.5 equiv.) together with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU; 0.9 equiv.) and 0.25 mL of N,N-diisopropylethylamine [DIPEA; 2.0 M in N-methyl-2-pyrrolidone (NMP)] were dissolved in 2 mL of DMF. The reaction mixture was added to the resin and vortex mixed for 3 h, until the coupling was completed as deduced by the Kaiser test. O-Acetyl groups of the carbohydrates were deprotected with 5 mL (3 × 15 min) of a hydrazine/MeOH (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) solution. (Glyco)peptides were detached from the resin and acid-sensitive sidechain protecting groups by treatment with a solution of trifluoroacetic acid (TFA)/triisopropylsilane (TIS)/H2O/1,2-ethanedithiol (EDT; 92.5
3) solution. (Glyco)peptides were detached from the resin and acid-sensitive sidechain protecting groups by treatment with a solution of trifluoroacetic acid (TFA)/triisopropylsilane (TIS)/H2O/1,2-ethanedithiol (EDT; 92.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) for 3 h at 25 °C. (Glyco)peptides were then precipitated with cold diethyl ether and centrifuged, to afford the crude derivatives. Purification by RP-HPLC afforded the corresponding (glyco)peptides (for experimental details see the ESI†).
2.5) for 3 h at 25 °C. (Glyco)peptides were then precipitated with cold diethyl ether and centrifuged, to afford the crude derivatives. Purification by RP-HPLC afforded the corresponding (glyco)peptides (for experimental details see the ESI†).
Microarrays
Anti-MUC1 mouse monoclonal antibody VU-11E2 (0.86 mg mL−1) and FluoroLinkTM CyTM3-labeled goat anti-mouse IgG were used in these experiments. The microarray printing and the microarray monoclonal antibody binding assay were performed following the same protocols previously reported by us63 (for experimental details see the ESI and Fig. S1 and S2†).Molecular dynamics simulations with time averaged restraints (MD-tar)
The simulations were carried out with the AMBER 18 package,49 and implemented with ff14SB (ref. 50) and GAFF.52 Parameters for the unnatural sugar mimic were generated with the antechamber module of AMBER 18, with partial charges set to fit the electrostatic potential generated with HF/6-31G(d) by RESP.64 The charges were calculated according to the Merz–Singh–Kollman scheme with Gaussian 16.65 The system was neutralized by adding explicit counter ions (Cl−). Prior to MD-tar47 productive simulation on glycopeptide 2a, we performed an equilibration protocol consisting of an initial minimization of the water box of 5000 steps, followed by a 2500-step minimization of the whole system. Then, the TIP3P66 water box was heated at constant volume until 298 K by using a time constant for a heat bath coupling of 1 ps. The equilibration finished with 200 ps of MD simulation without restraints, at a constant pressure of 1 bar and temperature of 298 K, turning on the Langevin temperature scaling with a collision frequency of 1 ps. An 8 Å cutoff was applied to Lennard-Jones interactions. Periodic boundary conditions and the particle mesh Ewald method67 were turned on in every step of the equilibration protocol to evaluate the long-range electrostatic forces, using a grid spacing of 1 Å. 20 ns MD-tar47 simulations were run with the same settings used in the last step of the equilibration protocol. A time step of 1 fs was used. The NOE-derived distances shown in Table S1† were imposed as time-averaged restraints, applying an r−6 averaging. The equilibrium distance range was set to rexp − 0.2 Å ≤ rexp ≤ rexp + 0.2 Å. The force constants rk2 and rk3 used in each case were 10 kcal mol−1·Å−2. A decay constant of 2000 ps was used throughout the MD simulations. The coordinates were saved each 1 ps, thus obtaining MD trajectories of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames. The analysis of the MD-tar trajectories has been carried out with the cpptraj module of AMBER 18.
000 frames. The analysis of the MD-tar trajectories has been carried out with the cpptraj module of AMBER 18.
Crystallization
Expression and purification of scFv-SM3 has been described previously by us.55 Crystals were grown by sitting drop diffusion at 18 °C. The drops were prepared by mixing 0.5 μL of protein solution, which contained 15 mg mL−1 scFv-SM3 and 10 mM glycopeptide 2a with 0.5 μL of the mother liquor. Crystals of scFv-SM3 with the peptide above were grown in 20% PEG 3350 in 0.2 M disodium hydrogen phosphate. The crystals were cryoprotected in mother liquor containing 15% ethylene glycol and frozen in a nitrogen gas stream cooled to 100 K.Structure determination and refinement
Diffraction data were collected on synchrotron beamline I24 of the Diamond Light Source (Harwell Science and Innovation Campus, Oxfordshire, UK) at a wavelength of 0.97 Å and a temperature of 100 K. Data were processed and scaled with the XDS package68 and CCP4 software.69,70 Relevant statistics are given in Table S2.† The crystal structures were solved by molecular replacement with Phaser69,70 and by using the PDB entry 1SM3 as the template. Initial phases were further improved by cycles of manual model building in Coot71 and refinement with REFMAC5.72 The final models were validated with PROCHECK.73 Coordinates and structure factors have been deposited in the Worldwide Protein Data Bank (wwPDB; PDB id 6TGG).Conjugation
Conjugation of MUC1 derivatives was conducted by INYCOM Biotech (Zaragoza, Spain) following a standard procedure as follows. First, SMCC (2 mg) was dissolved in 10 μL of DMF and then added to a KLH solution (50 μL, 20 mg mL−1 in phosphate-buffered saline (PBS), pH 7.2). The mixture was diluted by adding 25 μL of PBS. After 1 h at room temperature, the mixture was diluted with 175 μL of PBS and purified by using dialysis (14 kDa MWCO) against PBS for 24 h. Glycopeptides (2 mg dissolved in 500 μL of PBS) were treated with KLH-SMCC (2 mg in 200 μL of PBS). After 2 h at room temperature, the mixture was purified by using dialysis (14 kDa MWCO) against PBS for 24 h. The total protein concentration (absorbance at 280 nm) was 1.1 and 1.0 mg mL−1 for KLH-3a and KLH-3b, respectively. The glycopeptide/KLH ratio was determined by UPLC/MS,74 using glycopeptides 3a and 3b as standards (Fig. S6†). The experimentally calculated values were 3a/KLH = 3305 and 3b/KLH = 3420.Immunizations
Immunizations were carried out by INYCOM Biotech (Zaragoza, Spain). Three female MUC1.Tg mice (Balb/c) that express human MUC1 at the physiological level (14-week-old, average weight = 24 g) were immunized four times at biweekly intervals at the base of the tail intradermally with vaccine KLH-3a or KLH-3b (25 μg), together with complete or incomplete Freund's adjuvant. The endpoint was one week after the 4th immunization.Serologic assays
Anti-MUC1 IgG, IgG1, IgG2a, IgG2b, IgG3 and IgM antibody titers were determined by enzyme-linked immunosorbent assay, which was conducted by INYCOM Biotech (Zaragoza, Spain) as follows. ELISA plates (JetBioFil, China) were coated with 75 μL of a 5 μg mL−1 solution of either 3b or 3c in PBS and incubated for 1 h at 37 °C. Unreacted sites were blocked with 225 μL of BSA 1% in washing buffer (0.05% Tween-20 in PBS 1×) for 1 h at 37 °C. Afterwards, wells were washed twice with washing buffer. 75 μL of serum samples (blood taken from each mouse's tail diluted in 800 μL of BSA 1% solution) were allowed to bind to immobilized antigens for 30 min at 37 °C. Wells were then washed twice with washing buffer, prior to incubation with 75 μL of biotinylated anti-mouse antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution in conjugated solvent) for 30 min at 37 °C. After washing three times with washing buffer, incubation with 75 μL streptavidin-HRP (Sigma), 1
1000 dilution in conjugated solvent) for 30 min at 37 °C. After washing three times with washing buffer, incubation with 75 μL streptavidin-HRP (Sigma), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution in conjugation solvent, was carried out for 30 min at 37 °C. Wells were washed five times with washing buffer. 75 μL of TMB were then incubated for 5 min at room temperature. For the final color-developing step, 75 μL of 0.2 M HCl were added to stop the reaction. Absorbance at 450 nm was read, using an iMark™ BIO RAD Microplate Reader. Comparisons were performed by using an unpaired t test with equal SD. Differences were considered significant when P < 0.05.
500 dilution in conjugation solvent, was carried out for 30 min at 37 °C. Wells were washed five times with washing buffer. 75 μL of TMB were then incubated for 5 min at room temperature. For the final color-developing step, 75 μL of 0.2 M HCl were added to stop the reaction. Absorbance at 450 nm was read, using an iMark™ BIO RAD Microplate Reader. Comparisons were performed by using an unpaired t test with equal SD. Differences were considered significant when P < 0.05.
Culture of cell lines
Three different cell lines were used for the in vitro studies, namely HEK293T (a human embryonic kidney cell line), MCF7 (a human breast adenocarcinoma cell line) and T47D cells (a human breast ductal carcinoma cell line). The cells were maintained in a humidified incubator at 37 °C under 5% CO2 and grown by using 1× DMEM (Dulbecco's modified Eagle's medium) with sodium pyruvate and without L-glutamine (Invitrogen, Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Life Technologies), 1× MEM NEAA (Gibco, Life Technologies), 1× GlutaMAX (Gibco, Life Technologies), 200 units per mL penicillin and 200 μg mL−1 streptomycin (Gibco, Life Technologies) and 10 mM HEPES (Gibco, Life Technologies). The medium of the T47D cells was supplemented with 10 μg mL−1 insulin (Gibco, Life Technologies).Reactivity of antibodies towards human cancer cell lines determined by flow cytometry analysis
The reactivity of the antibodies elicited by KLH-3a towards breast cancer cell lines was determined by staining the cells with the antisera followed by flow cytometry analysis. For this purpose, MCF7 and T47D cells (with high expression of TA-MUC1) and HEK293T cells (with no expression of TA-MUC1) were fixed with an ice-cold solution of 4% paraformaldehyde in PBS (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per FACS tube) for 10 min. After fixation, the cells were subjected to a permeabilization step with 0.1% Triton-X100 in PBS for 15 min, followed by a blocking step with 10% FBS in PBS for 30 min. The cells were then incubated with 50 μL of 1
000 cells per FACS tube) for 10 min. After fixation, the cells were subjected to a permeabilization step with 0.1% Triton-X100 in PBS for 15 min, followed by a blocking step with 10% FBS in PBS for 30 min. The cells were then incubated with 50 μL of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution of mice sera. After 1 h of incubation, cells were incubated with 50 μL per·well of goat anti-mouse polyclonal IgG H&L Alexa Fluor 488 (1
50 dilution of mice sera. After 1 h of incubation, cells were incubated with 50 μL per·well of goat anti-mouse polyclonal IgG H&L Alexa Fluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibody from Abcam, for an additional 1 h. All the incubation periods were completed at rt and were followed by washing steps with PBS (with 4 min centrifugation at 4000 rpm to remove the supernatant). Acquisition was done with a BD LSR Fortessa setup with a 488 nm laser and a 530/30 nm band-pass filter (combination used for Alexa488 detection). Data analysis was done with FlowJo (version 6.3.4, FlowJo) software.
2000) secondary antibody from Abcam, for an additional 1 h. All the incubation periods were completed at rt and were followed by washing steps with PBS (with 4 min centrifugation at 4000 rpm to remove the supernatant). Acquisition was done with a BD LSR Fortessa setup with a 488 nm laser and a 530/30 nm band-pass filter (combination used for Alexa488 detection). Data analysis was done with FlowJo (version 6.3.4, FlowJo) software.
Reactivity of antibodies towards human breast cancer cell lines analyzed by confocal microscopy
Antiserum staining of cell lines MCF7, T47D and HEK293T (negative control) was investigated by confocal microscopy. To this end, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded in 8-well μ-Slide Ibidi Plates and grown for 24 h. The cells were then incubated with CellMask Deep Red membrane dye (1
000 cells per well were seeded in 8-well μ-Slide Ibidi Plates and grown for 24 h. The cells were then incubated with CellMask Deep Red membrane dye (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) for 10 min at 37 °C, followed by a fixation step with 4% paraformaldehyde in PBS for 10 min at 37 °C. The cells were then washed with PBS and permeabilized with 1% Triton X-100 in PBS for 15 min at rt. The cells were then incubated with the mice sera (1
1000) for 10 min at 37 °C, followed by a fixation step with 4% paraformaldehyde in PBS for 10 min at 37 °C. The cells were then washed with PBS and permeabilized with 1% Triton X-100 in PBS for 15 min at rt. The cells were then incubated with the mice sera (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution) at 4 °C overnight. The cells were washed with PBS and incubated with 200 μL per well of goat anti-mouse polyclonal IgG H&L Alexa Fluor 488 (1
100 dilution) at 4 °C overnight. The cells were washed with PBS and incubated with 200 μL per well of goat anti-mouse polyclonal IgG H&L Alexa Fluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibody from Abcam, for 2 h at rt. Finally, the cells were washed and incubated with Hoechst (1 μg mL−1) for 10 min at rt to stain the nuclei and analyzed with a Zeiss LSM 710 confocal laser point-scanning microscope with a 40× oil objective and numerical aperture = 1.3. The secondary antibody was visualized with an argon laser source (488 nm, emission 500–550 nm), while CellMask Deep Red stained membranes were visualized upon excitation with a DPSS 561-10 laser (561 nm, emission 570–640 nm), and nuclei were visualized with a diode 405-30 laser (450 nm, emission 420–470 nm).
2000) secondary antibody from Abcam, for 2 h at rt. Finally, the cells were washed and incubated with Hoechst (1 μg mL−1) for 10 min at rt to stain the nuclei and analyzed with a Zeiss LSM 710 confocal laser point-scanning microscope with a 40× oil objective and numerical aperture = 1.3. The secondary antibody was visualized with an argon laser source (488 nm, emission 500–550 nm), while CellMask Deep Red stained membranes were visualized upon excitation with a DPSS 561-10 laser (561 nm, emission 570–640 nm), and nuclei were visualized with a diode 405-30 laser (450 nm, emission 420–470 nm).
Author contributions
I. A. B., C. D. N. and A. A. synthesized, purified and characterized the glycopeptides. J. H. B. and J. M. P. and F. C. performed and analyzed the NOESY experiments. J. C.-L. and R. H.-G. purified antibody scFv-SM3, crystallized the complex and refined the crystal structure. A. G. and G. J. L. B. carried out the flow cytometry and the confocal microscopy experiments. E. M. S. F, J. M. G. F. and C. O. M. performed the synthesis, purification and characterization of the unnatural building bock with the sp2-iminosugar to use in solid-phase peptide synthesis. F. G.-M., H. H. and S.-I. N. carried out the microarray assays. I. A. B. and E. J.-M. determined the glycopeptide/KLH ratio by using UPLC/MS. F. C. performed the conformational analysis of the antigen in solution. F. C. and J. M. P. wrote the article with the other authors' contributions. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Ministerio de Ciencia, Innovación y Universidades and the Agencia Estatal de Investigación (projects RTI2018-099592-B-C21 and RTI2018-097609-B-C21), the Ministerio de Economía y Competitividad (SAF2016-76083-R), European Regional Development Funds (FEDER-UE), the Royal Society (URF\R\180019 to G. J. L. B.) and FCT Portugal (PhD studentship, SFRH/BD/115932/2016 to A. G. and FCT Investigator IF/00624/2015 to G. J. L. B.). M. C. O further thanks CITIUS for technical support. I. A. B. thanks the Asociación Española Contra el Cancer en La Rioja for a grant. E. J.-M. thanks Universidad de La Rioja for a postdoctoral fellowship. R. H.-G. acknowledges Diamond Light Source (Oxford, UK) synchrotron beamline I04 (experiment numbers mx10121-19). He also acknowledges ARAID, MEC (CTQ2013-44367-C2-2-P and BFU2016-75633-P) and Gobierno de Aragón (E34_R17 and LMP58_18) with FEDER (2014–2020) funds for ‘Building Europe from Aragón’ for financial support. The research leading to these results has also received funding from FP7 442 (2007–2013) under BioStruct-X (grant agreement no. 283570 and BIOSTRUCTX_5186). F. G.-M. of Hokkaido University acknowledges grant from the Naito Foundation. INYCOM Biotech (Zaragoza, Spain) has the required approval statements to perform studies with animals. The authors thank Dr Vikki Cantrill for her help with the editing of this manuscript.References
- T. Ju, V. I. Otto and R. D. Cummings, Angew. Chem., Int. Ed., 2011, 50, 1770–1791 CrossRef CAS PubMed.
- D. W. Kufe, Nat. Rev. Cancer, 2009, 9, 874–885 CrossRef CAS PubMed.
- J. Taylor-Papadimitriou, J. M. Burchell, R. Graham and R. Beatson, Biochem. Soc. Trans., 2018, 46, 659–668 CrossRef CAS PubMed.
- V. Apostolopoulos, L. Stojanovska and S. E. Gargosky, Cell. Mol. Life Sci., 2015, 72, 4475–4500 CrossRef CAS PubMed.
- O. Blixt, D. Bueti, B. Burford, D. Allen, S. Julien, M. Hollingsworth, A. Gammerman, I. Fentiman, J. Taylor-Papadimitriou and J. M. Burchell, Breast Cancer Res., 2011, 13, R25 CrossRef CAS PubMed.
- H. Chen, S. Werner, S. Tao, I. Zörnig and H. Brenner, Cancer Lett., 2014, 346, 178–187 CrossRef CAS PubMed.
- Z. M. Tang, Z. G. Ling, C. M. Wang, Y. Bin Wu and J. L. Kong, PLoS One, 2017, 12, e0182117 CrossRef PubMed.
- H. Takeuchi, K. Kato, K. Denda-Nagai, F. G. Hanisch, H. Clausen and T. Irimura, J. Immunol. Methods, 2002, 270, 199–209 CrossRef CAS PubMed.
- U. Karsten, N. Serttas, H. Paulsen, A. Danielczyk and S. Goletz, Glycobiology, 2004, 14, 681–692 CrossRef CAS PubMed.
- Y. Yoshimura, K. Denda-Nagai, Y. Takahashi, I. Nagashima, H. Shimizu, T. Kishimoto, M. Noji, S. Shichino, Y. Chiba and T. Irimura, Sci. Rep., 2019, 9, 16641 CrossRef PubMed.
- P. Dokurno, P. A. Bates, H. A. Band, L. M. D. Stewart, J. M. Lally, J. M. Burchell, J. Taylor-Papadimitriou, D. Snary, M. J. E. Sternberg and P. S. Freemont, J. Mol. Biol., 1998, 284, 713–728 CrossRef CAS PubMed.
- N. Gaidzik, U. Westerlind and H. Kunz, Chem. Soc. Rev., 2013, 42, 4421–4442 RSC.
- M. A. Wolfert and G.-J. Boons, Nat. Chem. Biol., 2013, 9, 776 CrossRef CAS PubMed.
- R. M. Wilson and S. J. Danishefsky, J. Am. Chem. Soc., 2013, 135, 14462–14472 CrossRef CAS PubMed.
- G. L. Beatty and W. L. Gladney, Clin. Cancer Res., 2015, 21, 687–692 CrossRef CAS PubMed.
- C. Nativi, F. Papi and S. Roelens, Chem. Commun., 2019, 55, 7729–7736 RSC.
- N. Martínez-Sáez, J. M. Peregrina and F. Corzana, Chem. Soc. Rev., 2017, 46, 7154–7175 RSC.
- Z. Guo and Q. Wang, Curr. Opin. Chem. Biol., 2009, 13, 608–617 CrossRef CAS PubMed.
- Q. Wang, S. A. Ekanayaka, J. Wu, J. Zhang and Z. Guo, Bioconjugate Chem., 2008, 19, 2060–2067 CrossRef CAS PubMed.
- Q. Wang and Z. Guo, ACS Med. Chem. Lett., 2011, 2, 373–378 CrossRef CAS PubMed.
- C.-C. Liu and X.-S. Ye, Glycoconjugate J., 2012, 29, 259–271 CrossRef CAS PubMed.
- B. Richichi, B. Thomas, M. Fiore, R. Bosco, H. Qureshi, C. Nativi, O. Renaudet and L. BenMohamed, Angew. Chem., Int. Ed., 2014, 53, 11917–11920 CrossRef CAS PubMed.
- B. Kuberan, S. A. Sikkander, H. Tomiyama and R. J. Linhardt, Angew. Chem., Int. Ed., 2003, 42, 2073–2075 CrossRef CAS PubMed.
- L. Awad, R. Madani, A. Gillig, M. Kolympadi, M. Philgren, A. Muhs, C. Gérard and P. Vogel, Chem.–Eur. J., 2012, 18, 8578–8582 CrossRef CAS PubMed.
- D. R. Bundle, J. R. Rich, S. Jacques, H. N. Yu, M. Nitz and C.-C. Ling, Angew. Chem., Int. Ed., 2005, 44, 7725–7729 CrossRef CAS PubMed.
- C. Mersch, S. Wagner and A. Hoffmann-Röder, Synlett, 2009, 2167–2171 CAS.
- M. Johannes, M. Reindl, B. Gerlitzki, E. Schmitt and A. Hoffmann-Röder, Beilstein J. Org. Chem., 2015, 11, 155–161 CrossRef PubMed.
- A. Hoffmann-Röder, A. Kaiser, S. Wagner, N. Gaidzik, D. Kowalczyk, U. Westerlind, B. Gerlitzki, E. Schmitt and H. Kunz, Angew. Chem., Int. Ed., 2010, 49, 8498–8503 CrossRef PubMed.
- F. Yang, X.-J. Zheng, C.-X. Huo, Y. Wang, Y. Zhang and X.-S. Ye, ACS Chem. Biol., 2011, 6, 252–259 CrossRef CAS PubMed.
- A. Hoffmann-Röder and M. Johannes, Chem. Commun., 2011, 47, 9903–9905 RSC.
- T. Oberbillig, C. Mersch, S. Wagner and A. Hoffmann-Röder, Chem. Commun., 2012, 48, 1487–1489 RSC.
- E. M. Sánchez Fernández, C. D. Navo, N. Martínez-Sáez, R. Gonçalves-Pereira, V. J. Somovilla, A. Avenoza, J. H. Busto, G. J. L. Bernardes, G. Jiménez-Osés, F. Corzana, J. M. García Fernández, C. Ortiz Mellet and J. M. Peregrina, Org. Lett., 2016, 18, 3890–3893 CrossRef PubMed.
- E. M. Sánchez-Fernández, R. Rísquez-Cuadro, M. Chasseraud, A. Ahidouch, C. Ortiz Mellet, H. Ouadid-Ahidouch and J. M. García Fernández, Chem. Commun., 2010, 46, 5328–5330 RSC.
- E. M. Sánchez-Fernández, R. Rísquez-Cuadro, C. Ortiz Mellet, J. M. García Fernández, P. M. Nieto and J. Angulo, Chem.–Eur. J., 2012, 18, 8527–8539 CrossRef PubMed.
- R. Rísquez-Cuadro, R. Matsumoto, F. Ortega-Caballero, E. Nanba, K. Higaki, J. M. García Fernández and C. Ortiz Mellet, J. Med. Chem., 2019, 62, 5832–5843 CrossRef PubMed.
- J. R. Harris and J. Markl, Micron, 1999, 30, 597–623 CrossRef CAS PubMed.
- S. Adluri, T. Gilewski, S. Zhang, V. Ramnath, G. Ragupathi and P. Livingston, Br. J. Cancer, 1999, 79, 1806–1812 CrossRef CAS PubMed.
- A. M. M. Eggermont, S. Suciu, P. Rutkowski, J. Marsden, M. Santinami, P. Corrie, S. Aamdal, P. A. Ascierto, P. M. Patel, W. H. Kruit, L. Bastholt, L. Borgognoni, M. G. Bernengo, N. Davidson, L. Polders, M. Praet and A. Spatz, J. Clin. Oncol., 2013, 31, 3831–3837 CrossRef CAS PubMed.
- D. Miles, H. Roché, M. Martin, T. J. Perren, D. A. Cameron, J. Glaspy, D. Dodwell, J. Parker, J. Mayordomo, A. Tres, J. L. Murray and N. K. Ibrahim, Oncologist, 2011, 16, 1092–1100 CrossRef CAS PubMed.
- J. Zhu, Q. Wan, D. Lee, G. Yang, M. K. Spassova, O. Ouerfelli, G. Ragupathi, P. Damani, P. O. Livingston and S. J. Danishefsky, J. Am. Chem. Soc., 2009, 131, 9298–9303 CrossRef CAS PubMed.
- P. J. Sabbatini, V. Kudryashov, G. Ragupathi, S. J. Danishefsky, P. O. Livingston, W. Bornmann, M. Spassova, A. Zatorski, D. Spriggs, C. Aghajanian, S. Soignet, M. Peyton, C. O'Flaherty, J. Curtin and K. O. Lloyd, Int. J. Cancer, 2000, 87, 79–85 CrossRef CAS PubMed.
- S. J. Danishefsky and J. R. Allen, Angew. Chem., Int. Ed., 2000, 39, 836–863 CrossRef CAS PubMed.
- T. Gilewski, S. Adluri, G. Ragupathi, S. Zhang, T.-J. Yao, K. Panageas, M. Moynahan, A. Houghton, L. Norton and P. O. Livingston, Clin. Cancer Res., 2000, 6, 1693–1701 CAS.
- P. J. Sabbatini, G. Ragupathi, C. Hood, C. A. Aghajanian, M. Juretzka, A. Iasonos, M. L. Hensley, M. K. Spassova, O. Ouerfelli, D. R. Spriggs, W. P. Tew, J. Konner, H. Clausen, N. Abu Rustum, S. J. Dansihefsky and P. O. Livingston, Clin. Cancer Res., 2007, 13, 4170–4177 CrossRef CAS PubMed.
- C. Musselli, P. O. Livingston and G. Ragupathi, J. Cancer Res. Clin. Oncol., 2001, 127, R20–R26 CrossRef CAS PubMed.
- H. J. Dyson and P. E. Wright, Curr. Opin. Struct. Biol., 1993, 3, 60–65 CrossRef CAS.
- D. A. Pearlman, J. Biomol. NMR, 1994, 4, 1–16 CAS.
- I. Compañón, A. Guerreiro, V. Mangini, J. Castro-López, M. Escudero-Casao, A. Avenoza, J. H. Busto, S. Castillón, J. Jiménez-Barbero, J. L. Asensio, G. Jiménez-Osés, O. Boutureira, J. M. Peregrina, R. Hurtado-Guerrero, R. Fiammengo, G. J. L. Bernardes and F. Corzana, J. Am. Chem. Soc., 2019, 141, 4063–4072 CrossRef PubMed.
- D. A. Case, S. R. Brozell, D. S. Cerutti, T. E. Cheatham III, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, D. Ghoreishi, H. Gohlke, A. W. Goetz, D. Greene, R. Harris, N. Homeyer, S. Izadi, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D. J. Mermelstein, K. M. Merz, Y. Miao, G. Monard, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, J. Smith, J. Swails, R. C. Walker, J. Wang, H. Wei, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, AMBER 2018, University of California, San Francisco, 2018 Search PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- K. N. Kirschner, A. B. Yongye, S. M. Tschampel, J. González-Outeiriño, C. R. Daniels, B. L. Foley and R. J. Woods, J. Comput. Chem., 2008, 29, 622–655 CrossRef CAS PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- F. Corzana, J. H. Busto, G. Jiménez-Osés, M. García de Luis, J. L. Asensio, J. Jiménez-Barbero, J. M. Peregrina and A. Avenoza, J. Am. Chem. Soc., 2007, 129, 9458–9467 CrossRef CAS PubMed.
- I. A. Bermejo, I. Usabiaga, I. Compañón, J. Castro-López, A. Insausti, J. A. Fernández, A. Avenoza, J. H. Busto, J. Jiménez-Barbero, J. L. Asensio, J. M. Peregrina, G. Jiménez-Osés, R. Hurtado-Guerrero, E. J. Cocinero and F. Corzana, J. Am. Chem. Soc., 2018, 140, 9952–9960 CrossRef CAS PubMed.
- N. Martínez-Sáez, J. Castro-López, J. Valero-González, D. Madariaga, I. Compañón, V. J. Somovilla, M. Salvadó, J. L. Asensio, J. Jiménez-Barbero, A. Avenoza, J. H. Busto, G. J. L. Bernardes, J. M. Peregrina, R. Hurtado-Guerrero and F. Corzana, Angew. Chem., Int. Ed., 2015, 54, 9830–9834 CrossRef PubMed.
- A. García-Herrero, E. Montero, J. L. Muñoz, J. F. Espinosa, A. Vián, J. L. García, J. L. Asensio, F. J. Cañada and J. Jiménez-Barbero, J. Am. Chem. Soc., 2002, 124, 4804–4810 CrossRef PubMed.
- L. Cipolla, F. Peri and C. Airoldi, Adv. Anticancer Agents Med. Chem., 2008, 8, 92–121 CrossRef CAS PubMed.
- A. Kaiser, N. Gaidzik, U. Westerlind, D. Kowalczyk, A. Hobel, E. Schmitt and H. Kunz, Angew. Chem., Int. Ed., 2009, 48, 7551–7555 CrossRef CAS PubMed.
- V. Lakshminarayanan, P. Thompson, M. A. Wolfert, T. Buskas, J. M. Bradley, L. B. Pathangey, C. S. Madsen, P. A. Cohen, S. J. Gendler and G.-J. Boons, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 261–266 CrossRef CAS PubMed.
- F. Nimmerjahn and J. V Ravetch, Science, 2005, 310, 1510–1512 CrossRef CAS PubMed.
- N. Martínez-Sáez, N. T. Supekar, M. A. Wolfert, I. A. Bermejo, R. Hurtado-Guerrero, J. L. Asensio, J. Jiménez-Barbero, J. H. Busto, A. Avenoza, G.-J. Boons, J. M. Peregrina and F. Corzana, Chem. Sci., 2016, 7, 2294–2301 RSC.
- C. Plattner, M. Höfener and N. Sewald, Org. Lett., 2011, 13, 545–547 CrossRef CAS PubMed.
- V. J. Somovilla, I. A. Bermejo, I. S. Albuquerque, N. Martínez-Sáez, J. Castro-López, F. García-Martín, I. Compañón, H. Hinou, S.-I. Nishimura, J. Jiménez-Barbero, J. L. Asensio, A. Avenoza, J. H. Busto, R. Hurtado-Guerrero, J. M. Peregrina, G. J. L. Bernardes and F. Corzana, J. Am. Chem. Soc., 2017, 139, 18255–18261 CrossRef CAS PubMed.
- C. I. Bayly, P. Cieplak, W. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- N. 4 Collaborative Computational Project, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1994, 50, 760–763 CrossRef PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- G. N. Murshudov, P. Skubák, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
- R. A. Laskowski, M. W. MacArthur, D. S. Moss and J. M. Thornton, J. Appl. Crystallogr., 1993, 26, 283–291 CrossRef CAS.
- J. K. Gathuru, F. Koide, G. Ragupathi, J. L. Adams, R. T. Kerns, T. P. Coleman and P. O. Livingston, Vaccine, 2005, 23, 4727–4733 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc06334j |
| This journal is © The Royal Society of Chemistry 2020 |