Open Access Article
Open Access ArticlePeptidomimetics prepared by tail-to-side chain one component peptide stapling inhibit Alzheimer's amyloid-β fibrillogenesis†
Sujan
Kalita
 ,
Sourav
Kalita
,
Sourav
Kalita
 ,
Ashim
Paul
,
Ashim
Paul
 ,
Amar
Sarkar
and
Bhubaneswar
Mandal
,
Amar
Sarkar
and
Bhubaneswar
Mandal
 *
*
Laboratory of Peptide and Amyloid Research, Department of Chemistry, Indian Institute of Technology Guwahati, Assam-781039, India. E-mail: bmandal@iitg.ac.in
First published on 27th March 2020
Abstract
Alzheimer's disease (AD) is the most common form of dementia affecting the elderly population worldwide. Despite enormous efforts and considerable advancement in research, no therapeutic agents have come to light to date. However, many peptide-based and small molecule inhibitors interact efficiently with the amyloid-β (Aβ) peptide and alter its aggregation pathway. On the other hand, stapled peptides have been developed mainly to stabilize α-helix conformations and study protein–protein interactions. β-Sheet stabilization or destabilization by stapled peptides has not been explored enough. Herein, we describe the generation of a library of “tail-to-side chain” stapled peptides via lactamization and their application for the first time as modulators of Aβ1-40 self-association and fibrillogenesis. They also disrupt the preformed fibrillar aggregates into nontoxic species. Their stability in the presence of proteolytic enzymes is increased due to stapling. Therefore, the stapled peptides thus formed can be useful as potent amyloid aggregation inhibitors and pave a therapeutic pathway for combating amyloid-related diseases. Also, they may help in gaining insight into the process of aggregation.
Introduction
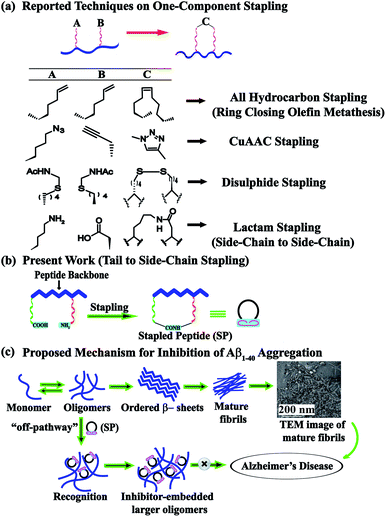
Peptide stapling is a novel strategy for constraining oligopeptides into structurally organised macrocycles by covalently linking (or stapling) the side-chains of two amino acids of a peptide.1 It dramatically improves the pharmacological properties of peptides by increasing their target binding affinity, proteolytic resistance, helicity, and cell penetration ability.2,3 Stapled peptides have received considerable attention as a potent class of stable macrocyclic α-helical peptidomimetics. The resulting constructs efficiently bind to protein targets involved in protein–protein interactions.3 A growing number of stapling techniques, using various macrocyclization chemistries, have been reported; the most widely adopted is disulfide bridge stapling.4 Early methods of peptide stapling include lactam bridge formation between lysine and aspartic/glutamic acid residues,5 ruthenium-catalyzed ring-closing metathesis using O-allyl serine or Cα-tetrasubstituted amino acids,6 and hydrocarbon stapling involving ring-closing metathesis of olefin bearing amino acids.7 Another promising candidate for peptide stapling stems from the two-component Cu(I) catalyzed azide–alkyne cycloaddition click reactions, where the resulting triazole moiety serves as the ‘staple’ in the reaction. Functionalized stapled peptides have profound applications in protein–protein interactions in human platelets.8 A stapled peptide inhibitor of HDM2/HDMX towards the activation of p53 has been reported to be under phase II clinical trials.9 Stapled peptides have also been reported as potential inhibitors of S100B(ββ), guided by the solution structure of the C-terminus of p53 complexed with S100B(ββ)10 and HIV 1 Integrase.11Inspired by such a wide range of applications, we wished to investigate the scope of the stapled peptides as modulators of amyloidosis. Among neurodegenerative disorders, Alzheimer's disease (AD) tops the list and is characterized by the deposition of amyloid-β peptide (Aβ, consisting of 40–42 residues) in interneuronal spaces.12 Aβ peptide undergoes a slow transformation from the native functional peptide to soluble Aβ oligomers and eventually to insoluble fibrillar aggregates.13 Many peptide-based molecules with the incorporation of various breaker elements have been reported to exhibit profound amyloid aggregation inhibition,14 yet there is no complete cure for the disease. Nowick et al. developed a novel class of β-sheet macrocycles, called amyloid β-sheet mimics (ABSMs) and investigated their effects on amyloid aggregation and toxicity.15 The reported macrocycles contained two strands, a heptapeptide β-strand (the upper strand) that can bind to a wide range of amyloid proteins through β-sheet interactions and a “Hao” (5-HO2CCONH-2-MeO-C6H3-CONHNH2, an unnatural amino acid composed of hydrazine, 5-amino-2-methoxybenzoic acid, and oxalic acid groups)16 molecular template flanked by two dipeptides (the lower strand) with the two strands being connected by two δ-linked ornithine residues. The “Hao” residue plays a crucial role in preventing further aggregation as it provides intramolecular hydrogen bonding with a simultaneous reduction of the exposed hydrogen bonding of the lower strand.15
Besides these, many naturally occurring products such as curcumin, resveratrol, and epigallocatechin-3-gallate (EGCG) have been reported to exhibit potential therapeutic efficacy due to their antioxidant and anti-amyloidogenic properties.17 A potent water-soluble derivative of curcumin, namely curcumin diacetate (CurDAc), not only inhibits hIAPP aggregation but also plays a significant role in lipid membrane protection, as well as its stabilization.18 Suzuki et al. reported that EGCG inhibits amyloid formation in a dose-dependent manner and diverts the hIAPP aggregation pathway towards the formation of stabilized nonfibrillar aggregates during fibrillogenesis.19
A literature survey provides several stapling techniques (Fig. 1a). However, the scope of tail-to-side-chain peptide stapling has not been explored enough. To the best of our knowledge, no such stapled peptides have been reported as inhibitors of amyloid aggregation. Herein, we report the design, synthesis, and characterization of a library of tail-to-side-chain stapled peptides (SPs) with and without anthranilic acid (Ant). Ant was used as an extra turn inducing element.20 We also report the inhibition and disruption potential of our designed stapled peptides against the aggregation of the amyloid-β peptide. Furthermore, the toxicity and in vitro proteolytic stability of the SPs are also investigated, so that they can be used for the future development of anti-amyloid therapeutics.
Results and discussion
Peptide design
For the tail-to-side-chain peptide stapling, we have attached dicarboxylic acids, COOH–(CH2)n–COOH, (n = 2, 3 and 4) as linkers at the N-terminus and Fmoc-Lys(Boc)-OH at the C-terminus, keeping LVFF in the middle of the peptide sequence, for proper recognition and binding with Aβ1-40 peptide (Table 1). The guanidine headgroup of arginine has been proposed to form putative bidentate hydrogen-bonding interactions with the phosphates present in lipid bilayers.21 Therefore, Fmoc-Arg-(Pbf)-OH was inserted at the C-terminus for enhanced membrane interaction. An analog of SPs devoid of Arg was insoluble in phosphate buffer solution (PBS, pH 7.4). Arg improves the solubility of SPs as well. Before attaching the dicarboxylic acids in the peptide sequence, N-methylated glycine was inserted to avoid the side reaction that would otherwise have occurred between the lone pair of –NH– of glycine and the free –COOH group of the attached dicarboxylic acids. We have used commercially available Aβ1-40 as the aggregating peptide. We synthesized three SPs without Ant, the turn inducing element (SP1, SP2, and SP3), and three SPs with Ant (SP4, SP5, and SP6) otherwise differing only in the length of the attached dicarboxylic acids. A linear control peptide, LP1, was also prepared (Table 1). The sequence of LP1 has previously been reported and it is known to be a better aggregation inhibitor than its analog which is devoid of Ant.20 The peptides were designed maintaining sequence homology with the crucial aggregating part of Aβ1-40 peptide. We investigated the effect of the chain length of the dicarboxylic acids, if any, on the process of inhibition and disruption. Before the stapling step, the Boc group in the side chain of the Lys residue was deprotected using five eq. of ZnBr2 in DCM,22 followed by coupling with BOP and DIPEA. The amide bond formation between the carboxylic acid group of the N-terminal dicarboxylic acid and the side chain of Lys resulted in the formation of tail-to-side-chain stapled peptides (SPs, Fig. 1b).| Peptide code | Sequence | Functions |
|---|---|---|
| a Standard amino acids are represented by their one-letter code, Ant = anthranilic acid, N-Me = N-methylation, Ac = acetyl group. Dicarboxylic acids, COOH–(CH2)n–COOH, used are adipic acid (n = 4), glutaric acid (n = 3) and succinic acid (n = 2). | ||
| SP1 |
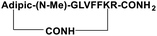
|
Inhibitor |
| SP2 |
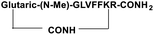
|
Inhibitor |
| SP3 |
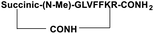
|
Inhibitor |
| SP4 |
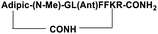
|
Inhibitor |
| SP5 |
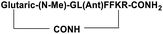
|
Inhibitor |
| SP6 |
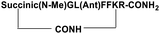
|
Inhibitor |
| LP1 | Ac-L(Ant)FFK-CONH2 | Control |
Non-amyloidogenicity of the synthesized SPs
We first examined the amyloidogenic nature of the synthesized SPs using various biophysical methods. The fibrillar morphology detected by Transmission Electron Microscopy (TEM) is a characteristic feature of an amyloidogenic peptide.23 However, none of the SPs were found to exhibit such fibrillar structures when examined by TEM. The appearance of green-gold birefringence under cross-polarized light upon staining with Congo-red dye is another characteristic feature of amyloid fibrils.24 However, no such green-gold birefringence was observed for the Congo-red stained SPs when examined under cross-polarized light, indicating the non-amyloidogenicity of the SPs. Further, the conformational changes of the SPs were monitored by Circular Dichroism (CD) and Fourier Transform Infrared Spectroscopy (FT-IR) and it was observed that none of the SPs exhibited a β-sheet abundant conformation.Therefore, it was concluded that our synthesized SPs do not form β-sheets and are non-amyloidogenic in nature.
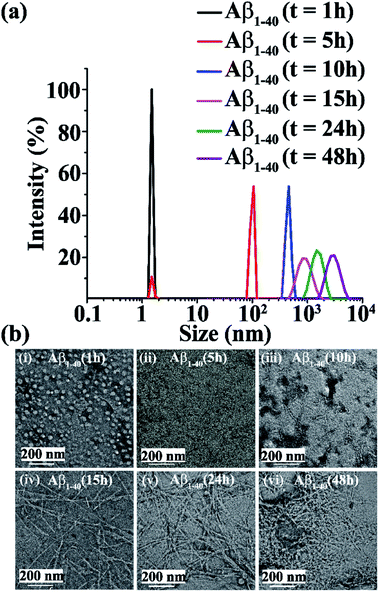
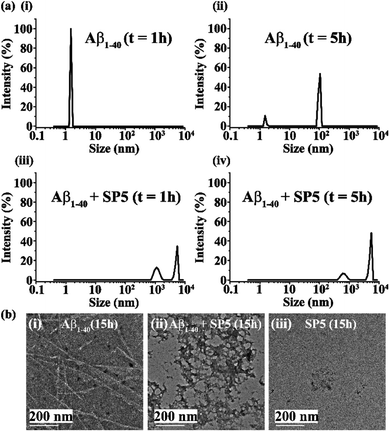
Early events of Aβ1-40 aggregation monitored by DLS and TEM
Upon incubation of Aβ1-40 in PBS at pH 7.4 and 37 °C, the kinetics of aggregation was monitored by Dynamic Light Scattering (DLS) and Transmission Electron Microscopy (TEM) and the morphology of the different species formed with time was examined. The size distribution of the various species formed was studied by DLS and the corresponding images were obtained by TEM in parallel. Aβ1-40 samples exhibited a size distribution in the 1–10 nm region after 1 h of incubation, which is consistent with the monomeric species (black, Fig. 2a and b(i)). After 5 h, the size of the particles increased to yield a size distribution centered at 100 nm with the simultaneous reduction of the species in the region of 1–10 nm, indicating the formation of oligomeric intermediate species when observed by TEM25,26 (red, Fig. 2a and b(ii)). Extending the incubation time up to 10 h further increased the sizes of the particles with an increased diameter of 100–1000 nm, which displayed protofibrillar amyloid intermediates when examined by TEM25,26 (blue, Fig. 2a and b(iii)). After 15 h of incubation, the size distribution of the particles was centered at 1000 nm, with the simultaneous growth of long and thin fibrils when observed by TEM (magenta, Fig. 2a and b(iv)). As amyloid aggregation progressed, the size distribution exceeded 1000 nm after 24 h of incubation of Aβ1-40, which exhibited thick and matured fibrils observed by TEM (olive, Fig. 2a and b(v)). With incubation time, t > 24 h, Aβ1-40 displayed a dense fibrous network of aggregates having a diameter range from 1000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm (purple, Fig. 2a and b(vi)).
000 nm (purple, Fig. 2a and b(vi)).
Inhibition of amyloid aggregation by the SPs
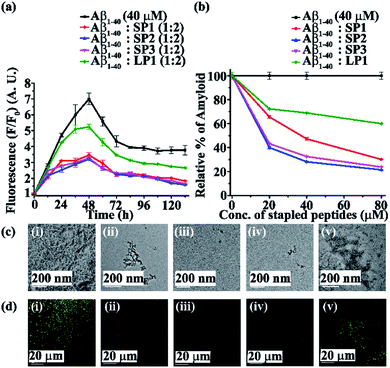
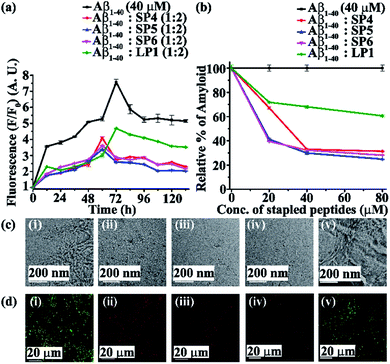
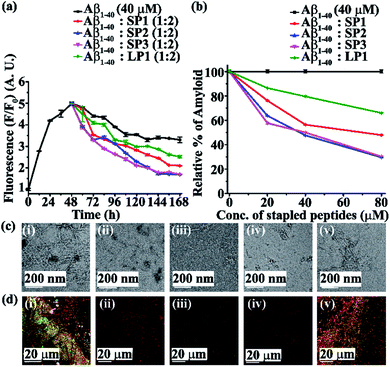
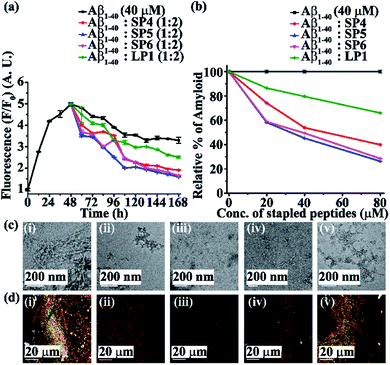
Next, we investigated the inhibitory potential of the SPs in inhibiting amyloid fibrils. The aggregating peptide, Aβ1-40, was co-incubated in the absence and presence of a 0.5-, 1- and 2- fold molar excess of the SPs and LP1 for six days under physiological conditions in vitro (PBS of pH 7.4 at 37 °C). The kinetics of amyloid aggregation of Aβ1-40 was monitored by time-dependent Thioflavin T (ThT) fluorescence assay. For this, two different sets of fluorescence experiments were performed, one with SPs without Ant, i.e., SP1, SP2 and SP3 (Fig. S15, ESI†), and one with SPs with Ant, i.e., SP4, SP5 and SP6 (Fig. S16, ESI†), in the mentioned doses. The fluorescence intensity decreased with the increase in doses of the SPs. In both the sets, the fluorescence intensity of Aβ1-40 was found to increase with time when incubated alone at pH 7.4 and 37 °C (black, Fig. 3a and 4a). But, in the presence of a 2-fold molar excess of SP1 (red, Fig. 3a), SP2 (blue, Fig. 3a), SP3 (magenta, Fig. 3a), SP4 (red, Fig. 4a), SP5 (blue, Fig. 4a), and SP6 (magenta, Fig. 4a), it was suppressed significantly, indicating the inhibitory potential of the SPs.The 2-fold molar excess of the control peptide LP1 also reduced the fluorescence intensity of Aβ1-40 (olive, Fig. 3a and 4a), but to a lesser extent as compared to the SPs. Hence, SPs are found to be better inhibitors of amyloid aggregation than the linear control peptide LP1. From the dose-dependent study, the 2-fold molar excess of SP1, SP2, SP3, SP4, SP5, SP6 and LP1 resulted in 70% (red, Fig. 3b), 78.5% (blue, Fig. 3b), 76.1% (magenta, Fig. 3b), 69% (red, Fig. 4b), 75.1% (blue, Fig. 4b), 72% (magenta, Fig. 4b) and 40% (olive, Fig. 3b and 4b) inhibition, respectively. The above results suggest that the inhibition potential of all the SPs is similar and independent of the chain length. Further, when observed by TEM, Aβ1-40 alone exhibited fibril rich morphology, indicating amyloid formation (Fig. 3c(i) and 4c(i)). However, when a 2-fold molar excess of SP1 (Fig. 3c(ii)), SP2 (Fig. 3c(iii)), SP3 (Fig. 3c(iv)), SP4 (Fig. 4c(ii)), SP5 (Fig. 4c(iii)) and SP6 (Fig. 4c(iv)) was co-incubated separately with Aβ1-40, no such fibrillar assembly was observed, indicating amyloid inhibition by the SPs. Again, upon staining with Congo red, Aβ1-40 alone exhibited green-gold birefringence when observed under cross-polarized light (Fig. 3d(i) and 4d(i)). However, when a 2-fold molar excess of SP1, SP2, SP3 (Fig. 3d(ii), (iii), and (iv), respectively), SP4, SP5, and SP6 (Fig. 4d(ii), (iii), and (iv), respectively) were co-incubated separately with Aβ1-40, no such green-gold birefringence was observed, indicating significant inhibition of amyloid formation. However, when 2 equivalents of LP1 were co-incubated with Aβ1-40, some fibrillar structures were observed by TEM (Fig. 3c(v) and 4c(v)) and green-gold birefringence was observed under cross-polarized light (Fig. 3d(v) and 4d(v)), indicating the inefficiency of 2 equivalents of LP1 to inhibit Aβ1-40 fibrillization.
Further, after six days of incubation, Aβ1-40 alone exhibited β-sheet rich conformation in both CD and FT-IR analyses; however, in the presence of a 2-fold molar excess of SP1, SP2, SP3 (Fig. S17, ESI†), SP4, SP5, and SP6 (Fig. S18, ESI†), β-sheet rich conformations were found to decrease, indicating inhibition of amyloid aggregation. All these results collectively indicate that the SPs are efficient inhibitors of amyloid aggregation of Aβ1-40, exhibiting significant inhibition even in a 2-fold molar ratio.
In both sets of experiments, SP2 and SP5 have shown better efficacy out of all the six SPs. Hence, out of the three dicarboxylic acids used at the N-terminus for stapling, SPs with glutaric acid have exhibited efficient inhibition of aggregation. Moreover, the SPs without Ant exhibited similar inhibitory potential as compared to those with Ant, indicating the absence of any synergistic effect.
Furthermore, although the SPs inhibited Aβ aggregation efficiently, two equivalents of them were not enough for complete inhibition of Aβ aggregation. Hence, to achieve better inhibition, we increased the dose of the SPs up to a 5-fold molar excess against Aβ1-40, and the effect of the increased dose on aggregation was monitored by ThT fluorescence assay. Only SP2 and SP5 were tested, as they performed relatively better than the other SPs in the previous experiments. It was observed that 5-fold molar excesses of SP2 and SP5 performed better (Fig. S19(a), ESI†) than the 2-fold molar excesses, inhibiting aggregation approximately up to ∼95% (Fig. S19(b), ESI†). Thus, a 5-fold molar excess of the SPs with respect to Aβ1-40 was required for almost complete inhibition of aggregation.
Disruption of preformed amyloid fibrillar aggregates by the SPs
Next, we investigated the ability of the SPs to disrupt the preformed amyloid fibrils in vitro. The fibrillization process by Aβ1-40 reaches its maximum at around 48–72 h. Therefore, as before, two experiments were carried out separately, where the SPs were added to the preformed fibrillar assembly at 48 h of incubation. Aβ1-40 was incubated alone in PBS at pH 7.4 and 37 °C for two days and after this, various molar excesses (0.5-, 1- and 2-fold) of the SPs (SP1, SP2, and SP3, Fig. S20, ESI†, and SP4, SP5, and SP6, Fig. S21, ESI†) were added to it. The ability of the SPs to disrupt the fibrillar assembly was monitored using various biophysical tools. Here again, the fluorescence intensity was found to reduce with the increase in doses of the SPs. From the time-dependent ThT fluorescence assay, the fluorescence intensity of Aβ1-40 was found to increase with time when it was alone in the solution (black, Fig. 5a and 6a), but in the presence of a 2-fold molar excess of SP1, SP2, SP3 (red, blue, and magenta, respectively, Fig. 5a), SP4, SP5, and SP6 (red, blue, and magenta, respectively, Fig. 6a), it was suppressed, indicating disruption of preformed fibres. A 2-fold excess of the control peptide LP1 also suppressed the Aβ1-40 fluorescence intensity (olive, Fig. 5a and 6a) but to a lesser extent than the SPs. From the dose-dependent study, the 2-fold molar excess of SP1, SP2, and SP3 exhibited 52.1%, 70.3%, and 70% (red, blue and magenta, respectively, Fig. 5b), whereas, that of SP4, SP5, and SP6 exhibited 60%, 74% and 71.5% (red, blue and magenta, respectively Fig. 6b), and that of LP1 exhibited 34% (olive, Fig. 5b and 6b) disruption.Moreover, Aβ1-40 appeared fibrillar (Fig. 5c(i) and 6c(i)) when observed by TEM. However, when Aβ1-40 was incubated in the presence of a 2-fold molar excess of SP1, SP2, and SP3 (Fig. 5c(ii), (iii), and (iv), respectively) and SP4, SP5, and SP6 (Fig. 6c(ii), (iii), and (iv), respectively), no such fibre was observed, indicating significant disruption of preformed-amyloid by the SPs. Further, the results from the Congo-red stained birefringence studies indicated that Aβ1-40 alone exhibited green-gold colour when examined under cross-polarized light (Fig. 5d(i) and 6d(i)), but for Aβ1-40 incubated in the presence of a 2-fold molar excess of SP1, SP2, SP3 (Fig. 5d(ii), (iii), and (iv), respectively), SP4, SP5, and SP6 (Fig. 6d(ii), (iii), and (iv), respectively), no such green-gold birefringence was observed which indicated disruption of amyloid fibrils by the SPs. However, when two equivalents of LP1 were co-incubated with Aβ1-40, some fibrillar structures were observed by TEM (Fig. 5c(v) and 6c(v)) and green-gold birefringence was observed under cross-polarized light (Fig. 5d(v) and 6d(v)), indicating less efficiency of 2 equivalents of LP1 to disrupt preformed Aβ1-40 fibrils.
After six days of incubation, Aβ1-40 alone exhibited a β-sheet abundant conformation in both CD and FT-IR analyses, but the presence of a 2-fold molar excess of each of the SPs (Fig. S22 and S23, ESI†) decreased the β-sheet conformation, indicating their amyloid disruption capability. All the above results indicated that the SPs disrupted preformed amyloid fibrils in a dose-dependent manner, with a 2-fold molar excess of them showing the most efficient disruption ability.
Here, again, the SPs without Ant exhibited almost similar disruption abilities as compared to those with Ant, which indicated that peptide stapling plays a vital role in the disruption of the preformed fibrils too. Out of all the six SPs, those with glutaric acid being used as linkers at the N-terminus (i.e. SP2 and SP5) exhibited the most efficient disruption abilities.
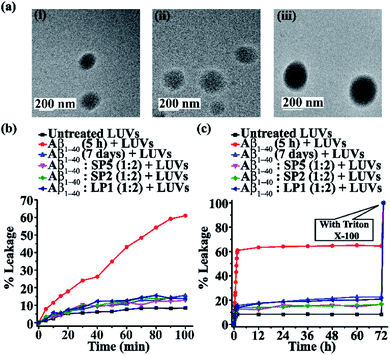
Vesicle leakage assay27
The aggregation pathway of amyloidogenic peptides not only involves the amyloid fibre state, but also sometimes the ‘on-pathway’ oligomeric intermediate state that is converted into amyloid fibres or sometimes the ‘off-pathway’ species that are generated independently of fibre formation.28 Aβ1-40 soluble oligomers have been reported to be more toxic than the matured amyloid fibrils as they cause cell damage by pore formation on the cell membranes.29 These pores, in turn, may associate with ion channels and disturb the cellular homeostasis. The factors responsible for membrane fragmentation are not necessarily related to amyloidogenesis or, in other words, membrane disruption can occur irrespective of amyloid fibrillogenesis.28To check whether our designed SPs disrupt amyloid fibrils to generate non-toxic species, we performed a dye leakage assay. For this purpose, we prepared large unilamellar vesicles (Text on page S15, ESI LUVs, Fig. 7a(i)–(iii))30 which entrap the carboxyfluorescein dye. To perform the vesicle leakage study, we prepared six sets of different samples, including untreated LUVs (without any peptide) as the control. The peptides (SP2, SP5, and LP1) were added to the Aβ1-40 solution after 48 h of incubation, and then all the solutions were kept incubated for seven days. All the peptide solutions were added separately to the prepared LUVs, where 25 μL of the dye-loaded LUVs and 50 μL of the peptide solutions were taken and diluted up to 1000 μL to obtain a final concentration of 50 μM for the lipid solution and 2.5 μM for the Aβ1-40 solution, with the peptide and the lipid maintaining a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and leakage studies were performed. The samples prepared for the leakage study are named as follows.
20, and leakage studies were performed. The samples prepared for the leakage study are named as follows.
Sample 1- untreated LUVs
Sample 2- Aβ1-40 (incubated for 5 h) + LUVs
Sample 3- Aβ1-40 (incubated for 7 days) + LUVs
Sample 4- Aβ1-40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SP5 (1
SP5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) + LUVs
2) + LUVs
Sample 5- Aβ1-40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SP2 (1
SP2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) + LUVs
2) + LUVs
Sample 6- Aβ1-40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LP1 (1
LP1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) + LUVs
2) + LUVs
To obtain complete dye leakage (100% dye release) from the vesicles, 10 μL of Triton X-100 was added at the end of the experiment, and the total fluorescence was measured. The percentage of dye leakage was calculated as follows31
It was observed from the dye leakage experiment that for sample 2 (i.e. Aβ1-40 incubated for 5 h), there was a rapid increase of dye leakage of LUVs from 0 to 100 min, but such an increase was not observed after 12 h, while the natural leakage of LUVs (untreated LUVs) was saturated after 60 min. The % of dye leakage is shown in Fig. 7b and c, with the untreated LUVs; sample 1 is shown in black, sample 2 in red, sample 3 in blue, sample 4 in magenta, sample 5 in olive and sample 6 is shown in dark blue. Therefore, from the results of the dye leakage experiment, it was evident that the oligomers formed from Aβ1-40 after 5 h of incubation were more toxic than the matured fibrils formed from Aβ1-40 after seven days of incubation, as is evident from the % of dye leakage of the former. Since the oligomers are relatively more toxic, they cause pore formation on the artificial cells or LUVs, thereby releasing the carboxyfluorescein dye from the LUVs, causing the enhancement of fluorescence intensity.
In vitro cellular membrane disruption by Aβ1-40 is initiated by a two-step mechanism. Firstly, prior to fibril formation, soluble oligomers are formed that bind to the membranes to form small ion-selective channel-like pores. The pores formed by Aβ1-40 are unstable and merge into larger aggregates, thereby converting into fibres during the membrane disruption process and eventually detaching from the membrane. Secondly, Aβ1-40 fibril growth causes nonselective physical membrane disruption through a detergent-like process.28,32
On the other hand, fibrils disrupted by the designed SPs (SP2 and SP5) and the control peptide (LP1) did not form pores on the LUVs significantly, as the increase of their fluorescence intensity (sample 4, magenta, sample 5, olive & sample 6, dark blue, Fig. 7b and c) was almost similar to that of the untreated LUVs (sample 1, black, Fig. 7b and c). Thus, the SPs disrupt the preformed amyloid fibrils, but no evidence of the presence of toxic pore-forming species was detected even in 7 day old peptide solutions (Fig. 7b and c). Hence, this result indicates that SPs disrupt the pre-formed amyloid but do not produce toxic soluble oligomers.
Preliminary investigation of the mechanism of inhibition of aggregation by SPs
To gain insight into the mechanism of the inhibitory action of the SPs on Aβ1-40 aggregation, we studied the changes in the size distribution of Aβ1-40 alone as well as in the presence of 2 equivalents of SPs with time by using dynamic light scattering. Since the inhibition efficiencies of all the six SPs were nearly the same, here we considered only one SP (SP5) for the DLS study. Aβ1-40 (40 μM) and the SP (80 μM) were incubated at pH 7.4 and 37 °C. At time t = 1 h, Aβ1-40 species exhibited size distribution in the range of 1–10 nm (Fig. 8a(i)). Again, 5 h of incubation of Aβ1-40 in the absence of the SP produced oligomeric intermediate species with size distribution centered at 100 nm with the simultaneous reduction of the species in the region 1–10 nm (Fig. 8a(ii)). In contrast, when Aβ1-40 was incubated for 1 h in the presence of the SP in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, two types of species, one exhibiting size distribution centered at ∼1000 nm and the other in the range of 1000–10
2, two types of species, one exhibiting size distribution centered at ∼1000 nm and the other in the range of 1000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm appeared instantly. This implies that Aβ1-40 after mixing with the SP was assembled immediately due to which it exhibited an increased size distribution (Fig. 8a(iii)). After 5 h of incubation of Aβ1-40 in the presence of the SP, it was observed that the former species diminished, and the latter increased simultaneously (Fig. 8a(iv)). The drastic change in the size distribution of Aβ1-40 suggests that in the presence of the SP, Aβ1-40 aggregation proceeded without the formation of toxic oligomeric species that were produced during the native aggregation pathway. In other words, the presence of the SP alters the aggregation pathway of Aβ1-40 which proceeds towards an off-pathway aggregation pathway.
000 nm appeared instantly. This implies that Aβ1-40 after mixing with the SP was assembled immediately due to which it exhibited an increased size distribution (Fig. 8a(iii)). After 5 h of incubation of Aβ1-40 in the presence of the SP, it was observed that the former species diminished, and the latter increased simultaneously (Fig. 8a(iv)). The drastic change in the size distribution of Aβ1-40 suggests that in the presence of the SP, Aβ1-40 aggregation proceeded without the formation of toxic oligomeric species that were produced during the native aggregation pathway. In other words, the presence of the SP alters the aggregation pathway of Aβ1-40 which proceeds towards an off-pathway aggregation pathway.
Further, to gain morphological insight into the off-pathway oligomers, TEM was performed after 15 h of incubation of the peptide samples. When incubated alone at pH 7.4 and 37 °C, Aβ1-40 exhibited long and thin fibrils (Fig. 8b(i)), but in the presence of 2 equivalents of the SP, such fibrils were not observed, and instead some off-pathway aggregated species were predominantly formed (Fig. 8b(ii)). Furthermore, when the SP (80 μM) alone was incubated at pH 7.4 and 37 °C, no such aggregated species were observed by TEM (Fig. 8b(iii)). These results indicate the aggregation of Aβ1-40 in the presence of the SP might have proceeded through an “off-pathway” aggregation mechanism (Fig. 1c). When Aβ1-40 was mixed with the SP, inhibitor-embedded off-pathway oligomeric species were generated predominantly, which further associated with larger species to form larger particles (Fig. 8a(iii) and (iv)). The off-pathway oligomers correspond to non-toxic species that cannot progress to toxic amyloid fibrils, thereby preventing the native aggregation pathway.33 However, further investigations are required to understand the mechanism better.
Proteolytic stability study
One of the most striking features of stapled peptides is their ability to withstand a proteolytic environment.34,35 To check the stability of our synthesized stapled peptides, we performed an in vitro stability study in the presence of RPMI (Roswell Park Memorial Institute) 1640 media supplemented with 10% FBS serum (v/v) and 1% penicillin/streptomycin antibiotic, obtained from GIBCO. From the results of all the performed biophysical studies, it was evident that two SPs, SP2 and SP5, gave the best results among all the six SPs. Hence, we selected both SP2 and SP5 for the stability study and compared their stability with that of the linear peptide LP1. For performing the stability study, 1 mL of the RPMI medium was taken in three different Eppendorf tubes and incubated for 15 min at 37 °C. After this, 10 μL of the peptide solutions (SP2, SP5, and LP1) in DMSO (5 mg mL−1) were added separately to the RPMI medium with suitable sonication and vortexing and incubated at 37 °C. Thereafter, at different time intervals (0 h, 1 h, 5 h, 10 h, 15 h and 25 h), 100 μL of each aliquot was taken from the three Eppendorf vials and added to 200 μL of ethanol solution (96% aqueous ethanol) to precipitate the FBS serum. The precipitate was allowed to cool at 4 °C for 15 min and then centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The supernatant was collected and analyzed by RP-HPLC and MALDI-TOF mass analyses. A linear gradient of 5–100% CH3CN for 18 min, followed by 100% CH3CN until 20 min was used at a flow rate of 5 mL min−1 using a semi-preparative C18 μ-Bondapak column. A binary solvent system comprising solvent A (0.1% TFA in H2O) and solvent B (0.1% TFA in CH3CN) was used.
000 rpm for 5 min. The supernatant was collected and analyzed by RP-HPLC and MALDI-TOF mass analyses. A linear gradient of 5–100% CH3CN for 18 min, followed by 100% CH3CN until 20 min was used at a flow rate of 5 mL min−1 using a semi-preparative C18 μ-Bondapak column. A binary solvent system comprising solvent A (0.1% TFA in H2O) and solvent B (0.1% TFA in CH3CN) was used.
From time-dependent RP-HPLC, for the stapled peptides, SP2 (Fig. 9a) and SP5 (Fig. 9b), we did not observe any trace of degradation in the presence of proteolytic enzymes, instead complete retention of the peaks of the pure compounds was observed up to 25 hours. On the other hand, the linear peptide LP1 showed some degradation after 1 hour, though it was not completely degraded (Fig. 9c). We have previously reported that the presence of one breaker element (Ant) in the peptide sequence confers some stability, but after some time, in the presence of proteolytic enzymes, some degradation was observed.14
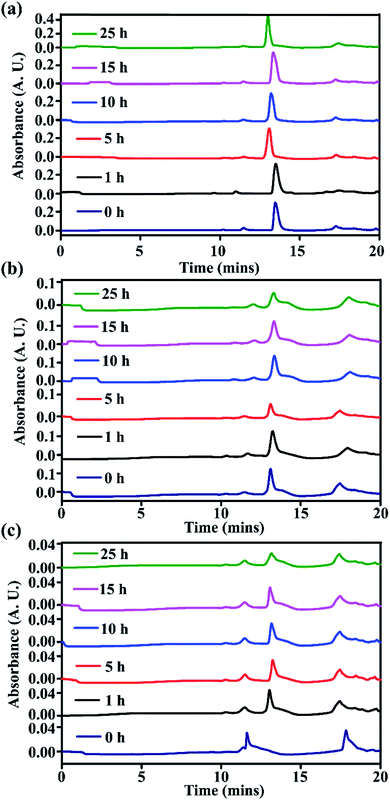 | ||
| Fig. 9 Kinetics of stability study of stapled peptides (a) SP2, (b) SP5, and (c) LP1 in the presence of proteolytic enzymes (FBS serum) monitored by RP-HPLC. | ||
Further, from MALDI-TOF analyses, for the stapled peptides, complete retention was observed from 0 h to 25 h (for SP2, Fig. S24 and S25, ESI† and SP5, Fig. S26 and S27, ESI†). On the other hand, for the linear peptide, LP1, retention was observed at 0 h (Fig. S28, ESI†), but after 1 h, some trace of degradation were observed which remained constant up to 25 h. When the 10 h and 25 h fragments were analysed (Fig. S29 and S30, ESI†), some other mass fragments were also observed. Hence, from these results, it can be inferred that compared to the linear peptide, stapled peptides are quite resistant to degradation in the presence of proteolytic enzymes, which indicates that stapling enhances the proteolytic stability of peptides.
Conclusions
We have carried out tail-to-side chain peptide stapling using different dicarboxylic acids at the N-terminus. This stapling resulted in the formation of macrocycles both in the absence and in the presence of a turn inducing element (Ant). The results indicate that the tail to side-chain stapling does not require any turn inducing element in the peptide sequence to form the macrocycle. It was observed that the dicarboxylic acids, COOH–(CH2)n–COOH for n = 0 and 1, did not form any macrocycles. We did not extend the value of ‘n’ beyond four, as increasing the length of the chain would provide more flexibility to the macrocycle. We have demonstrated the potential of the SPs in inhibiting Aβ1-40 aggregation under physiological conditions and disrupting the preformed fibrillar aggregates into non-toxic species, as confirmed from the dye leakage assay. The efficacy towards inhibition and disruption was found to be independent of the chain length of the diacid used for the ease of stapling. Herein, we have shown two comparative analyses of efficacy of interfering fibrillogenesis: the first one is the stapled versus linear peptide, and the second one is that of the stapled peptide without any turn inducer versus that with a turn inducer. From the first comparison, we observed that the stapled peptides are far better inhibitors and disruptors of amyloid aggregation compared to the linear counterparts. On the other hand, the stapled peptides are remarkably efficient towards both inhibition and disruption, even in the absence of any turn inducer. Their potential was almost comparable to that of the stapled peptides with a turn inducer. In other words, the factor of ‘peptide stapling’ solely exhibits a similar effect over the turn-inducer factor combined with the stapling. Macrocycles resulting from peptide stapling are structurally rigid and conformationally restricted, because of which probably the lower equivalents of the SPs (two-fold molar ratios) were active, compared to the linear peptide in both inhibition of aggregation as well as disruption of the preformed fibrillar assembly. However, a five-fold molar excess of SPs was required for almost complete inhibition of aggregation; a more thoughtful design may improve the efficacy. Such macrocycles may emerge as a potential class of therapeutic tools for fighting against amyloidogenesis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are thankful to the DBT twinning program (NER-BPMC BT/PR16164/NER/95/88/2015) for the financial support, IIT Guwahati for the necessary facilities, and the CIF, IIT Guwahati, for TEM analysis.Notes and references
- Y. H. Lau, P. de. Andrade, Y. Wu and D. R. Spring, Chem. Soc. Rev., 2015, 44, 91–102 RSC.
- G. L. Verdine and G. J. Hilinski, Methods Enzymol., 2012, 503, 3–33 CAS.
- Y. S. Tan, D. P. Lane and C. S. Verma, Drug Discovery Today, 2016, 21, 1642–1653 CrossRef CAS PubMed.
- P. Barthe, S. Rochette, C. Vita and C. Roumestand, Protein Sci., 2000, 9(5), 942–955 CrossRef CAS PubMed.
- S. R. Tala, S. M. Schnell and C. H. Luevano, Bioorg. Med. Chem. Lett., 2015, 25(24), 5708–5711 CrossRef CAS PubMed.
- S. L. Mangold, D. J. O'Leary and R. H. Grubbs, J. Am. Chem. Soc., 2014, 136, 12469–12478 CrossRef CAS PubMed.
- C. E. Schafmeister, J. Po and G. L. Verdine, J. Am. Chem. Soc., 2000, 122, 5891–5892 CrossRef CAS.
- J. Legre, N. S. Ahmed, J. S. Gaynord, Y. Wu, K. M. Herlihy, Y. S. Tan, M. E. L. Pires, R. Jha, Y. H. Lau, H. F. Sore, C. Verma, D. H. O'Donovan, N. Pugh and D. R. Spring, Chem. Sci., 2018, 9, 4638–4643 RSC.
- F. M. M Bernstam, et al. , J. Clin. Oncol., 2017, 35, 2505 Search PubMed.
- S. Kannan, P. G. A. Aronica, Y. S. Tan and C. S. Verma, ACS Omega, 2019, 4, 5335–5344 CrossRef CAS.
- Y. Q. Long, S. X. Huang, Z. Zawahir, Z. L. Xu, H. Li, T. W. Sanchez, Y. Zhi, S. D. Houwer, F. Christ, Z. Debyser and N. Neamati, J. Med. Chem., 2013, 56, 5601–5612 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- E. Gazit, Angew. Chem., 2002, 114, 267–269 CrossRef.
- A. Paul, S. Kalita, S. Kalita, P. Sukumar and B. Mandal, Sci. Rep., 2017, 7, 40095, DOI:10.1038/srep40095.
- P. N. Cheng, C. Liu, M. Zhao, D. Eisenberg and J. S. Nowick, Nat. Chem., 2012, 4, 927–933 CrossRef CAS PubMed.
- J. S. Nowick, D. M. Chung, K. Maitra, S. Maitra, K. D. Stigers and Y. Sun, J. Am. Chem. Soc., 2000, 122, 7654–7661 CrossRef CAS.
- H. F. Ji and H. Y. Zhang, Acta Pharmacol. Sin., 2008, 29, 143–151 CrossRef CAS PubMed.
- A. S. Pithadia, A. Bhunia, R. Sribalan, V. Padmini, C. A. Fierke and A. Ramamoorthy, Chem. Commun., 2016, 52, 942–945 RSC.
- Y. Suzuki, J. R. Brender, K. Hartman, A. Ramamoorthy and E. N. G. Marsh, Biochemistry, 2012, 51, 8154–8162 CrossRef CAS PubMed.
- A. Paul, K. C. Nadimpally, T. Mondal, K. Thalluri and B. Mandal, Chem. Commun., 2015, 51, 2245–2248 RSC.
- J. B. Rothbard, E. Kreider, C. L. VanDeusen, L. Wright, B. L. Wylie and P. A. Wender, J. Med. Chem., 2002, 45, 3612–3618 CrossRef CAS PubMed.
- R. Kaul, Y. Brouillette, Z. Sajjadi, K. A. Hansford and W. D. Lubell, J. Org. Chem., 2004, 69, 6131–6133 CrossRef CAS PubMed.
- (a) M. R. Nilsson, Methods, 2004, 34, 151–160 CrossRef CAS PubMed; (b) P. Walsh, J. Yau, K. Simonetti and S. Sharpe, Biochemistry, 2009, 48, 5779–5781 CrossRef CAS PubMed.
- C. Wu, J. Scott and J. E. Shea, Biophys. J., 2012, 103, 550–557 CrossRef CAS PubMed.
- K. G. Malmos, L. M. B. Mejia, B. Weber, J. Buchner, M. R. Alvarado, H. Naiki and D. Otzen, Amyloid, 2017, 24, 1–16 CrossRef PubMed.
- S. A. Hudson, H. Ecroyd, T. W. Kee and J. A. Carver, FEBS J., 2009, 276, 5960–5972 CrossRef CAS PubMed.
- T. Sheynis, A. Friediger, W. F. Xue, A. L. Hellewell, K. W. Tipping, E. W. Hewitt, S. E. Radford and R. Jeilinek, Biophys. J., 2013, 105, 745–755 CrossRef CAS PubMed.
- J. R. Brender, E. L. Lee, M. A. Cavitt, A. Gafni, D. G. Steel and A. Ramamoorthy, J. Am. Chem. Soc., 2008, 130, 6424–6429 CrossRef CAS PubMed.
- P. Cao, et al. , Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 19279–19284 CrossRef CAS PubMed.
- (a) T. L. Williams, I. J. Day and L. C. Serpell, Langmuir, 2010, 26, 17260–17268 CrossRef CAS PubMed; (b) M. Traikia, et al. , Eur. Biophys. J., 2000, 29, 184–195 CrossRef CAS PubMed.
- J. McLaurin and A. Chakrabartty, J. Biol. Chem., 1996, 271, 26482–26489 CrossRef CAS PubMed.
- M. F. M. Sciacca, S. A. Kotler, J. R. Brender, J. Chen, D. K. Lee and A. Ramamoorthy, Biophys. J., 2012, 103, 702–710 CrossRef CAS PubMed.
- T. Arai, D. Sasaki, T. Araya, T. Sato, Y. Sohma and M. Kanai, ChemBioChem, 2014, 15, 2577–2583 CrossRef CAS PubMed.
- L. D. Walensky and G. H. Bird, J. Med. Chem., 2014, 57, 6275–6288 CrossRef CAS PubMed.
- H. Jenssen and S. I. Aspmo, Methods Mol. Biol., 2008, 494, 177–186 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization data of the stapled peptides, experimental methods and complementary data. See DOI: 10.1039/c9sc06076f |
| This journal is © The Royal Society of Chemistry 2020 |