Open Access Article
Open Access ArticleTracking nucleic acid nanocapsule assembly, cellular uptake and disassembly using a novel fluorescently labeled surfactant†
Md Arifuzzaman,
Alyssa K. Hartmann and
Jessica L. Rouge *
*
Department of Chemistry, University of Connecticut, 55 North Eagleville Road, Storrs, CT 06269, USA. E-mail: jessica.rouge@uconn.edu
First published on 20th November 2020
Abstract
Intracellular trafficking and delivery of nucleic acids is an area of growing interest, particularly as it relates to therapeutic applications. Spectroscopic methods have been used to observe and quantitatively measure the delivery of oligonucleotides both in vitro and in vivo. Herein we demonstrate the use of a new fluorophore labeled surfactant presenting a solvatochromatic chromophore for tracking the assembly and degradation of a hybrid biomaterial we refer to as a nucleic acid nanocapsule (NAN). We show that the surfactant enables critical micelle concentration determination, monitoring of NAN disassembly in vitro, and the ability to track the cellular movement and activity of surfactant–oligonucleotide conjugates in cells when coupled with quantitative PCR analysis.
The field of therapeutic nucleic acids (TNAs) is rapidly growing, marked by the FDA's recent approval of antisense oligonucleotides (ASOs) and short interfering RNA (siRNA) as gene regulation treatments in the last 5 years.1–3 Despite the promise TNAs hold for treating a variety of diseases, their full potential is currently hampered by challenges associated with their delivery to specific cell types and subcellular locations. The delivery hurdle for TNAs is mainly due to challenges associated with their transport through biological membranes. This is particularly true for TNAs delivered using synthetic nanocarriers. To successfully knockdown a gene, these exogenous oligonucleotides must cross the cells outer membrane, endosomal barriers, and in some cases the nuclear membrane. Therefore it is important to be able to track and ultimately determine the mechanism by which a TNA is delivered into a cell so that if specific steps to the delivery methodology are hindered by a particular aspect of the delivery pathway, improvements can be made.
Nanostructures have proven particularly useful in the delivery of oligonucleotides, improving their delivery properties4,5 and enabling enhanced stabilization of biologics.6,7 The delivery capabilities of nanoscale carriers requires that they successfully enter a cell and enable oligonucleotides to engage specific locations and targets. Much like the TNA itself, to be successful the nanostructure needs to reach the cytosol or facilitate the TNAs ability to reach the cytosol, so that it can interact with its target, often mRNA. The efficiency by which this can be accomplished determines the efficacy of a given nanostructure in aiding gene regulation applications.
Fluorescently labeled compounds are one of the most direct visual methods for determining intracellular localization of molecules in cells. However, achieving efficient intracellular trafficking of exogenous biomolecules such as DNA and RNA presents particular synthetic and stability-oriented challenges. In light of this, our group has developed a unique delivery platform that is <100 nm, which can deliver both oligonucleotides for gene regulation,8,9 and release small molecule drugs to augment intracellular therapeutic responses.10
We refer to our oligonucleotide delivery vehicle as a nucleic acid nanocapsule (NAN), as it is a micellular structure capable of encapsulating an internal cargo and is decorated with nucleic acid ligands. It is assembled from a tripropargylammonium headgroup that enables facile crosslinking and decoration of the micellar surface by both alkyne–azide and thiolyne click reactions.8 When the particle degrades it releases alkylated oligonucleotides, which we have shown may assist in endosomal escape.9 Herein we have synthesized a novel fluorescent dansyl-modified surfactant for tracking the fate of the NAN, designed and synthesized in a way that would enable us to monitor the assembled state of its individual components (Scheme 1). The use of a dansyl moiety as a fluorescent probe in proteins and other macromolecules is well known. Such compounds show characteristic changes in the location of their fluorescence maxima and fluorescence intensity, which have permitted their use to determine polar and nonpolar sites in macromolecules.11,12 The aggregation of the surfactant as it forms a micelle buries the dansyl in the interior and shields it from water. As the aggregates dissociate, the environment around the dansyl becomes more polar, resulting in a diminished fluorescent signal.
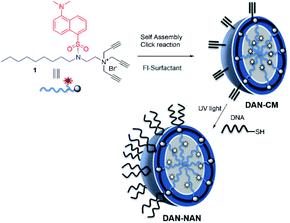 | ||
| Scheme 1 Stepwise assembly of dansyl modified nucleic acid nanocapsules (DAN-NANs). The tri-alkyl modified dansyl-surfactant 1 shown at top left is placed in water and quickly self assembles into a micelle structure presenting alkynes at its surface. An esterified diazido crosslinker 2 is used to stabilize the structure using copper I catalyzed click chemistry. Remaining alkynes are used as a point of attachment for a thiolated DNA molecules using a photodriven crosslinking step.8 | ||
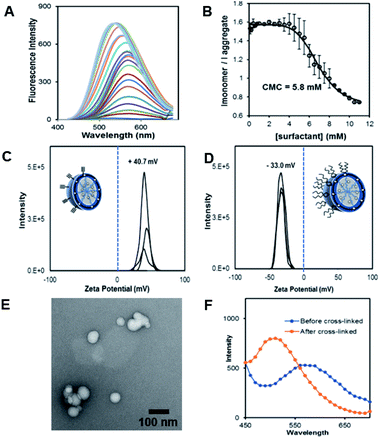
The fluorescent dansyl surfactant unit was synthesized from commercially available ethylenediamine and 1-bromononane (Scheme S1†). In addition to an easier synthesis from less expensive starting materials, the new surfactant has a fluorophore on the hydrophobic backbone. An important property of a surfactant is its critical micelle concentration (CMC), the concentration above which micelles begin to form. To determine the CMC, we used fluorescence spectroscopy methods.13 In a typical assay, we prepared aqueous solutions of dansyl surfactant at concentrations ranging from 0.5–10 mM in water. The formation of the micelle from the dansyl surfactant causes a blue shift in the emission maximum of the dansyl moiety from its initial fluorescence maxima ∼560 nm, while greatly enhancing the emission intensity (Fig. 1A). Dansyl is an environmentally sensitive fluorescent probe which fluoresces weakly in water but strongly in nonpolar environments; its emission wavelength typically increases with increasing environmental polarity.14–16 The blue shift and stronger emission indicates that the dansyl is in a nonpolar microenvironment and thus that micelles have likely formed. Using the dansyl surfactant the CMC was determined to be 5.8 mM (Fig. 1B).
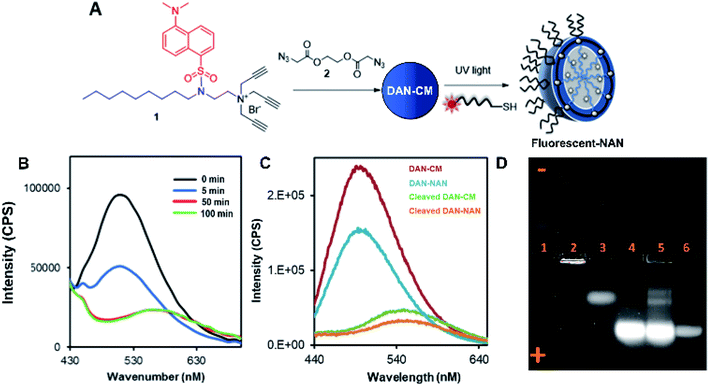
Scheme 1 shows the preparation of our NAN. The synthesis of the crosslinked micelle (CM) from the dansyl surfactant prior to DNA conjugation exhibited a uniform size distribution (20 ± 3 nm) and positive charge (+41 ± 4 mV). After DNA attachment, the particles exhibited a shift in surface charge to −33 mV (Fig. 1C and D). The particle size also increased from 20 nm to 40 nm as would be expected for the attachment of a 22 mer polyT20 DNA strand (Table S1†). Nanoscale characterization of the particles post DNA functionalization by DLS exhibited size values similar to those observed for traditional NANs,14 with a net negative charge of −41 mV to −33 mV for ester and PEG cross-linked Fl-NANs, respectively (Table S1†). The NANs were also analyzed by TEM indicating 40 nm sized particles (Fig. 1E). The formation of the crosslinked micelles was also apparent from the large blue shift in the emission maximum of dansyl from 560 nm to 510 nm as well as the enhanced emission intensity (Fig. 1F). In our previous studies, a dye was encapsulated within the micelle to track its cleavage and disassembly by enzymes.8 Here we show that an external dye is not necessary, rather, the resulting CMs built from DAN–surfactants can be subjected to treatment with base (NaOH), catalyzing the hydrolysis of the ester linkages within the nanocapsule and is directly monitored by fluorescence spectroscopy. As seen in Fig. 2, the CM containing the ester linkage (2) was successfully cleaved by base as indicated by a red shift in the emission and a decrease in fluorescence over time (Fig. 2B). As a control, CMs crosslinked with a non-hydrolyzable PEG crosslinker (3) were prepared and treated with base (NaOH) with little to no cleavage observed (Fig. S10†). The affect of various pH conditions was also investigated to determine whether the micelles are stable at pH conditions relevant to cellular uptake/endocytosis (pH 7) and endosomal maturation (pH 5). Fluorescence studies indicated the dansyl CMs are stable over a range of biologically relevant pHs (Fig. S11†).
After incorporation of DNA ligands (polyT20), the complete fluorescent nucleic acid nanocapsule (DAN-NAN) is formed. Previous syntheses of NANs exclusively utilized UV driven thiol-yne click chemistry to attach the DNA, however for these studies azide-modified polyT20 DNA was also explored as an alternative to thiol-yne chemistry due to concerns over UV damage to the dansyl signal that may result in a weaker probe signal. Post NAN synthesis it was found that the charge on NANs synthesized via a copper catalyzed azide–alkyne cycloaddition (CuAAC) reaction had a lower net negative charge than that of NANs synthesized using thiolated DNA (Table S2†). This is thought to be due to the fact that the thiolated DNA can create 2 new sulfur carbon bonds per surface alkyne available whereas the CuAAC click reaction results in the attachment of a single DNA ligand via the formation of a triazole ring. In light of these results, the fluorescence cleavage study of NANs was carried out using azide-modified DNA, whereas for all other studies, including cell studies, the thiol-yne reaction was used to maintain DNA density. Using NaOH we were able to monitor the disassembly of ester crosslinked NANs containing the dansyl surfactant as seen by the red shift in the emission and a decrease in the sample's overall fluorescence (Fig. 2C).
Next a 1% agarose gel was run to analyze the various assembled and disassembled NAN components. As seen in Fig. 2D, the free dansyl surfactant signal is weakly observed in lane 1 under the UV excitation with emission filter between 500–550 nm. However, upon assembly into a crosslinked micelle CM (lane 2), a stronger signal is observed, attributed to the dansyl dye being buried within the hydrophobic micelle. In lane 3, the degradation of the CM by NaOH results in the release of a dansyl surfactant that travels towards the positive electrode. This shift in mobility is likely due to the flanking carboxylic groups on the surfactant – a product of the ester crosslinkers degradation. Lane 4 contains the dansyl modified NANs where we see the CM band move further towards the positive electrode due to the presence of DNA at the surface which is more negatively charged. After treatment of the NANs in lane 5 with esterase, we see the reappearance of smaller molecular weight conjugates consistent with the release of fluorescent oligonucleotide surfactant conjugates. Lastly lane 6 indicates where fluorescein labeled DNA travels to, at an equal concentration to the DNA on the NAN. The stronger intensity of the bands in lanes 4 and 5 may be due to either the dansyl signal from the NANs core or the fluorescent DNA at the surface of the NAN. The appearance of the bands in lane 5 post esterase treatment that were not present in lane 4 indicate the release of the danysl modified surfactant and its conjugates. Lane 6 indicates that NANs and free DNA run to roughly the same location in a 1% gel.
We next introduced dansyl labeled NANs into cells via endocytosis where esterases are present. First, HeLa cells were treated with free 1 μM dansyl surfactant as a control and monitored for evidence of uptake by confocal microscopy. Within the first hour, the dansyl surfactant can be seen in late endosomes as indicated by its colocalization with lysotracker red (Fig. 3a). The cells were excited at a wavelength of 405 nm, close to the absorbance maxima of the free dansyl surfactant and its emission monitored over wavelengths in which we would expect to see the dansyl dye if not aggregated (560–600 nm). After 6 h, it is evident that some of the signal from the surfactant moved into the cytosol as seen by the diffuse nature of the fluorescent signal throughout the cell (Fig. 3B), thus indicating its ability to permeate the endosome. This mobility is attributed to the surfactants cationic and lipophilic character. Although 1 μM is far below the CMC of the surfactant, the punctated signal in the green channel seen in Fig. 3A suggests some degree of aggregation, likely due to the high ionic strength of the culturing media and the cytoplasm. The strong signal at 560 nm suggests however that there is dynamic interaction of the surfactant with its aqueous environment, enough so that they are still able to fluoresce.
Next, we set out to test the ability of the fluorescently tagged surfactant to aid in tracking the movement of oligonucleotide–surfactant conjugates. In order to do this, a DNA labeled with a TYE 665 red dye was functionalized on a CM constructed from the dansyl surfactant and ester crosslinker, generating a fluorescent NAN (FL-NAN) (Fig. 2A). TYE 665 was chosen as its wavelength is far enough away from dansyl's emission such that there should not be any spectral overlap. HeLa cells were incubated for 24 hours with 1 μM FL-NANs. The cells were dually monitored at the wavelengths needed for visualizing both the surfactant's dansyl signal (Exc. 405 nm, Em. 540–600 nm, green channel) and the TYE 665 dye on the oligonucleotide (Exc. 633 nm, Em. 650–700 nm, red channel).
The results show that when the FL-NANs are initially taken up into cells via endocytosis, the dansyl signal and TYE 665 dye signal colocalize, indicating that the oligonucleotide and surfactant have traveled to the same area. This is to be expected at early time points if the FL-NAN is still intact (Fig. 4C and D). After 6 h of incubation it is apparent that the oligonucleotide signal and surfactant remain partially colocalized (Fig. 4) white boxes – see ESI† for full analysis (Fig. S12 & S13†). After 24 h, it is observed that the signal from the dansyl surfactant (green channel) is localized to specific spots within the cell that are distinctly separate from areas that have the TYE 665 labeled oligonucleotide signal. We interpreted this to be due to either the DNA surfactant conjugate had escaped the endosome and is able to access the cytosol, or that the DNA degraded, releasing the TYE 665 dye label allowing it to diffuse out of the endosome. As a control for the microscopy analysis, DAN–surfactants were crosslinked with a nondegradable PEG crosslinker to investigate if changes could be observed in the localization of the surfactant relative to the oligonucleotide based on the biodegradability of the NAN. The results in Fig. S14† show similar images to the ester crosslinked NANs, indicating that either the resolution of the experiment was not sufficient to see differences in colocalization between the PEG and ester crosslinked NANs or that both the ester and PEG crosslinked NANs succumb to the same fate.
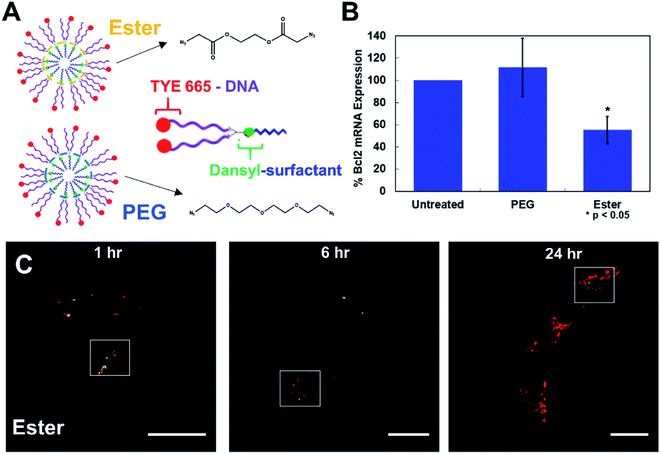 | ||
| Fig. 4 Cells treated with 1 μM fluorescent-NANs constructed from DAN–surfactant and TYE 665 dye labeled DNA. (A) Ester and PEG crosslinked NAN designs and schematic of an individual surfactant–DNA conjugate with the location of each dye indicated. (B) Gene silencing efficacy of a PEG vs. ester crosslinked NAN. NANs were functionalized with an anti-Bcl2 DNAzyme and incubated with HeLa cells; results shown as percent expression of Bcl2 mRNA by qPCR quantification. Error bars represent n = 3 trials with standard deviation. (C) Confocal microscopy time course of HeLa cells treated with 1 μM ester NANs. Boxes indicate representative areas analyzed for colocalization using ImageJ software (Fig. S12 & S13†). Here it can be seen that there are areas of colocalization of the TYE 665 dye and the dansyl dye at early time points, however by later time points there is a clear separation of the surfactant label from the oligonucleotide label. | ||
To further investigate this question, we conducted a gene knockdown experiment involving an anti-Bcl2 (ref. 17) DNAzyme ligand (Table S3†) on either a PEG or an ester crosslinked NAN. DNAzymes need to directly engage their mRNA targets in the cytosol to catalyze their degradation and achieve gene silencing. As the DNAzyme ligand is covalently attached to the surfactants that make up the NAN, and the DNAzyme needs to escape the endosome and enter the cytosol to engage its mRNA target, DNAzyme-mediated mRNA knockdown can serve as an excellent readout of endosomal escape. We hypothesized that the enzymatic hydrolysis of the ester crosslinker would enable better knockdown efficiency than the PEG crosslinked NAN as the DNAzyme–surfactant conjugates of the ester NAN could be released by esterases in the endosome. To investigate this, we conducted RT–qPCR quantification of mRNA expression for a target gene, Bcl2, in HeLa cells. As seen in Fig. 4B, the ester crosslinked NANs performed more effectively than the PEG crosslinked NANs, resulting in a roughly 40% decrease in Bcl2 expression compared to the limited change seen with the PEG crosslinker NAN. This indicates the importance of a biodegradable crosslinker for the DNAzyme NANs activity as well as the need for the surfactant tail on the DNAzyme. Using this information, we could better interpret the resulting microscopy data. As the DNAzyme is not chemically modified for enhanced stability in our experiments, degradation is to be expected over time. In general it is likely that in both cases there is degradation of the native DNA ligand of the DNA–surfactant conjugate, resulting in TYE 665 dye escaping the endosome and entering the cytosol as the DNAzyme is not chemically modified for enhanced stability in our experiments. However, when coupled with the highly sensitive detection method of PCR – we were able to detect that ester crosslinked NANs have a population of DNA–surfactant conjugates that are entering the cytosol and effectively achieving gene knockdown.
The results presented here show that the use of a dansyl probe on the surfactant, the building block of the NAN, enables a new way to track its assembly and stability of its inner crosslinked micelle. When coupled with PCR for monitoring gene regulation, the release of individual DNA–surfactant conjugates from the NAN can be detected efficiently in cells. Using this new surfactant, a facile approach to determining the critical micelle concentration is shown, introducing a versatile strategy that can aid our understanding of DNA micelle dynamics and nucleic acid delivery.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support by the University of Connecticut PATH award and NSF grant 1847869 is gratefully acknowledged.Notes and references
- E. E. Neil and E. K. Bisaccia, J. Pediatr. Pharmacol. Ther., 2019, 24, 194–203 Search PubMed.
- H. Ledford, Nature, 2018, 560, 291–292 CrossRef CAS PubMed.
- J. Ruger, S. Ioannou, D. Castanotto and C. A. Stein, Trends Pharmacol. Sci., 2019, 41, 27–41 CrossRef PubMed.
- J. Hrkach and R. Langer, Drug Delivery Transl. Res., 2020, 10, 567–570 CrossRef PubMed.
- S. Gudipati, K. Zhang and J. L. Rouge, Trends Biotechnol., 2019, 37, 983–994 CrossRef CAS PubMed.
- Y. Mei, R. Wang, W. Jiang, Y. Bo, T. Zhang, J. Yu, M. Cheng, Y. Wu, J. Cheng and W. Ma, Biomater. Sci., 2019, 7, 2640–2651 RSC.
- Y. Weng, Q. Huang, C. Li, Y. Yang, X. Wang, J. Yu, Y. Huang and X.-J. Liang, Mol. Ther.--Nucleic Acids, 2019, 19, 581–601 CrossRef PubMed.
- J. K. Awino, S. Gudipati, A. K. Hartmann, J. J. Santiana, D. F. Cairns-Gibson, N. Gomez and J. L. Rouge, J. Am. Chem. Soc., 2017, 139, 6278–6281 CrossRef CAS PubMed.
- A. K. Hartmann, D. F. Cairns-Gibson, J. J. Santiana, M. Q. Tolentino, H. M. Barber and J. L. Rouge, ChemBioChem, 2018, 19, 1734–1739 CrossRef CAS PubMed.
- A. K. Hartmann, S. Gudipati, A. Pettenuzzo, L. Ronconi and J. L. Rouge, Bioconjugate Chem., 2020, 31, 1063–1069 CrossRef CAS PubMed.
- M. Lundberg and M. Johansson, Biochem. Biophys. Res. Commun., 2002, 291, 367–371 CrossRef CAS PubMed.
- L. D. Hughes, R. J. Rawle and S. G. Boxer, PLoS One, 2014, 9, e87649 CrossRef PubMed.
- Y. Zhao, J. Org. Chem., 2009, 74, 834–843 CrossRef CAS PubMed.
- A. K. Tripathi, M. Mohapatra and A. K. Mishra, Phys. Chem. Chem. Phys., 2015, 17, 29985–29994 RSC.
- Y. H. Li, L. M. Chan, L. Tyer, R. T. Moody, C. M. Himel and D. M. Hercules, J. Am. Chem. Soc., 1975, 97, 3118–3126 CrossRef CAS.
- M. D. Arifuzzaman and Y. Zhao, J. Org. Chem., 2016, 81, 7518–7526 CrossRef CAS PubMed.
- X. Yang, Z. Li, J. He and L.-Q. Sun, Biochem. Biophys. Res. Commun., 2016, 479, 544–550 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09472b |
| This journal is © The Royal Society of Chemistry 2020 |