Open Access Article
Open Access ArticleAn improved 4′-aminomethyltrioxsalen-based nucleic acid crosslinker for biotinylation of double-stranded DNA or RNA†
Kevin Wielenberga,
Miao Wanga,
Min Yanga,
Abdullah Ozerb,
John T. Lisb and
Hening Lin *ac
*ac
aDepartment of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA. E-mail: hl379@cornell.edu
bDepartment of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA
cHoward Hughes Medical Institute, Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA
First published on 2nd November 2020
Abstract
Nucleic acid crosslinkers that covalently join complementary strands of DNA/RNA have applications in both pharmaceuticals and as biochemical probes. Psoralen is a popular crosslinking moiety that reacts with double stranded DNA and RNA upon exposure to longwave UV light. The commercially available compound EZ-link psoralen-PEG3-biotin has been used in numerous studies to crosslink DNA and double-stranded RNA for genome-wide investigations. Here we present a new probe, AP3B, which uses the psoralen derivative, 4′-aminomethyltrioxsalen, to crosslink and biotinylate nucleic acids. We show that AP3B is 4 to 5 times more effective at labeling DNA in cells and produces a comparable number of crosslinks with over 100 times less compound and less exposure to UV light in vitro than EZ-link psoralen-PEG3-biotin.
Introduction
Nucleic acid crosslinkers are important in nucleic acid research and come in many varieties.1,2 These range from simple molecules such as formaldehyde to inorganic complexes3 and have been combined with moieties that can preferentially bind to a target DNA sequence.4 Crosslinkers can covalently bond to nucleotides by reacting with nucleophilic regions of the bases or the double bonds of the bases. In the latter case, activation of the compound by exposure to light is frequently required to induce pericyclic reactions.5,6Photoactivable DNA and RNA crosslinkers are particularly useful for genomic studies as they allow for spatial and temporal control of the crosslinking reactions with light.7 Psoralen is one such photoactivatable moiety and has natural affinity for DNA due to the compound's planar multicyclic structure which allows it to intercalate nucleotide base pairs. Upon exposure to UV light, psoralen undergoes 2 + 2 cycloaddition to the 5,6 double bond of pyrimidines; however, it reacts best with thymine bases of complementary 5′-TA-3′ dinucleotide stretches.8 This forms mono and bifunctional adducts,9 with bifunctional adducts making interstrand crosslinks with the two complementary strands (Fig. 1).
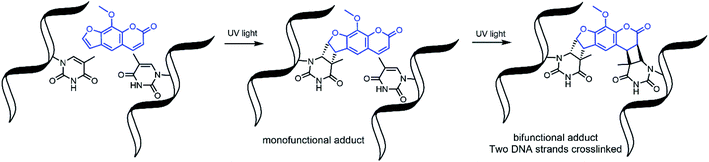 | ||
| Fig. 1 Psoralen undergoes 2 + 2 cycloaddition reactions to form mono and bifunctional adducts with DNA. | ||
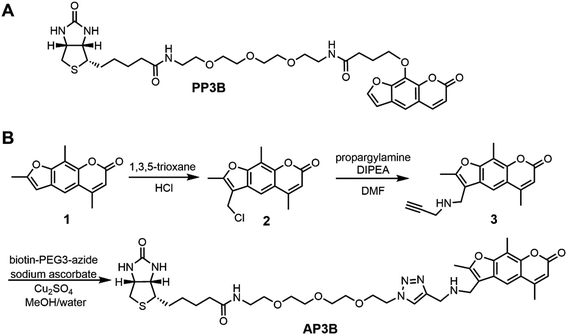
EZ-Link Psoralen-PEG3-Biotin (PP3B, Fig. 2A) is a commercially available probe that utilizes psoralen to biotinylate nucleic acids. Such crosslinkers enable biotinylation of double-stranded DNA (dsDNA) and dsRNA for experiments including sequencing of psoralen crosslinked, ligated and selected hybrids (SPLASH), studying RNA interactions, and Chem-seq to study the accessibility of DNA.10–13 Despite the prevalent use of PP3B for these applications, we noticed that other psoralen derivatives such 4′-aminomethyltrioxsalen (AMT) are more effective nucleic acid crosslinkers than the derivative used in PP3B.14 In this paper, we present the synthesis of a new psoralen derivative-biotin compound, AMT-PEG3-biotin (AP3B, Fig. 2B), and show that it is more efficient at both labeling and crosslinking DNA than the commercially available PP3B.
Results and discussion
Design and synthesis of AP3B
AMT was used as the reactive group in our probe as it has several advantages over other psoralen derivatives. Firstly, AMT has increased affinity for DNA due in part to an amino group which becomes protonated at physiological pH and interacts with the negatively charged phosphate backbone of DNA and RNA.15 The amino group also improves water solubility which we further enhanced in our new compound by including a PEG3 chain. Finally, AMT has a broader range of UV absorbance near 360 nm which allows the compound to absorb more photons near this wavelength compared to other psoralens. This results in increased probability that cycloaddition reactions will occur with AMT.16 To complete the probe, biotin is used as the affinity tag since biotin streptavidin interaction is a great affinity reagent, with 10−14 M affinity and tolerance to harsh conditions such as high temperature (up to 65 °C) and denaturing conditions (up to 0.5% SDS). The resulting compound is AMT-PEG3-Biotin (AP3B, Fig. 2B).AP3B was synthesized from commercially available starting materials (Fig. 2B). Briefly, 4′-chloromethyl-4,5′,8-trimethylpsoralen was reacted with 1,3,5-trioxane in concentrated HCl to functionalize the five-member ring with a chloromethyl group. The alkyl chloride was then substituted with propargyl amine to attach a terminal alkyne which was then reacted with biotin-PEG3-azide via click chemistry to produce AP3B.
AP3B has increased crosslinking efficiency in vitro
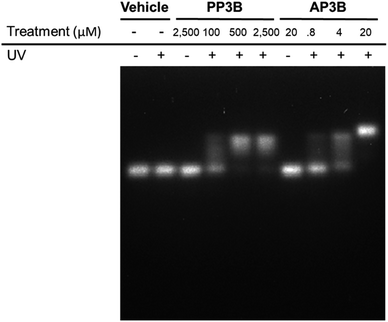
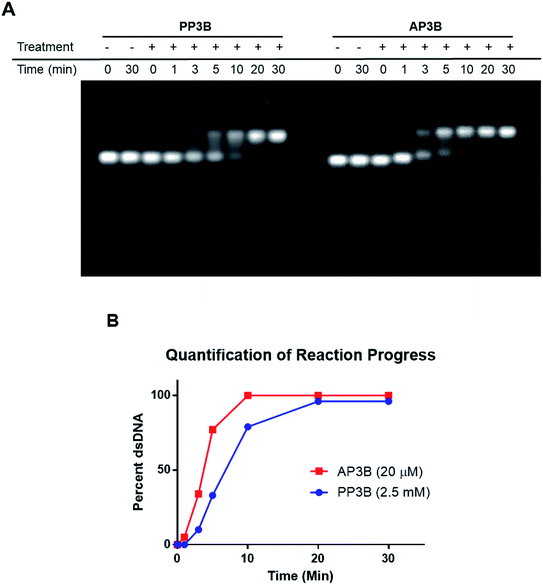
We examined and compared the efficiency of AP3B to form interstrand crosslinks in purified DNA with that of the commercially available PP3B. For this purpose, we used a denaturing agarose gel mobility shift assay. The high pH of the gel prevents base pairing by disrupting hydrogen bonding, there by producing single stranded DNA (ssDNA).17 Interstrand crosslinks formed with AP3B or PP3B prevent strands from separating, producing up shifted, higher molecular weight dsDNA bands. We compared the in vitro efficiency of AP3B and PP3B and confirmed that the reaction of either with DNA was UV light dependent as expected. The results showed that 800 nM of AP3B and 100 μM of PP3B were the two most comparable samples, producing similar amounts dsDNA (Fig. 3). Treatment with 4 μM of AP3B and 500 μM of PP3B also produced similar shifts. Thus, AP3B is about 100 times better than PP3B in vitro at producing DNA interstrand crosslinks. Additionally, PP3B did not crosslink all the available DNA even at the highest tested concentration while AP3B shifted all DNA to the double strand band at a much lower concentration (Fig. 3).The relative rates of interstrand crosslink formation for the two compounds were also compared using the mobility shift assay (Fig. 4A). In this case, the UV exposure time was varied while the concentrations of PP3B and AP3B were fixed at the highest concentration previously tested (2.5 mM and 20 μM, respectively). These concentrations produced the maximal amount of dsDNA, which served as a reference point for the end of the reaction. As expected, AP3B required less time to reach completion (5–10 min) compared to PP3B (20–30 min) (Fig. 4B).
AP3B shows higher labeling efficiency in cells than PP3B
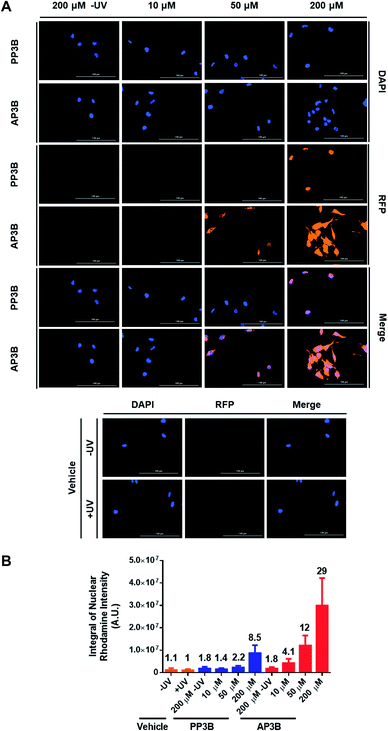
We next determined how the DNA labeling efficiency (measuring both mono-adducts and interstrand crosslinks) of AP3B compares to PP3B in cells. An immunofluorescence assay was developed to compare the two compounds. Hela cells were permeabilized with 0.1% saponin and treated with PP3B, AP3B, or DMSO and exposed to UV light. Unreacted compound was washed away, and the bound probes were recognized by a streptavidin–rhodamine conjugate which was in turn detected by florescence microscopy. Colocalization of DAPI and rhodamine signals in the cell nuclei suggested that the probes primarily labeled genomic DNA. Some cytoplasmic staining was also observed, suggesting the probes also reacted with dsRNA. Visual examination of the merged DAPI and rhodamine channels (Fig. 5A) showed increased biotinylation in the AP3B treated cells compared to the same concentration of PP3B. This observation was confirmed by quantification of the signal intensity of rhodamine in the nuclei (Fig. 5B). In all cases, probe-treated cells had higher rhodamine signal intensities than the vehicle treated cells, or non-UV exposed cells treated with the same concentration of compound. By comparing the signal intensities with the same concentrations of PP3B and AP3B or by comparing the concentrations of PP3B and AP3B that produced similar signal intensities, we concluded that the in-cell labeling efficiency of AP3B is 4 to 5 times that of PP3B.The in-cell results indicate that the labeling efficiency increase for AP3B is lower than that seen in the gel shift assay. This due at least in part to the fact that the two assays detect different adducts. While the in-cell experiment reports both mono and bifunctional adducts, the in vitro assay detects bifunctional adducts. The discrepancy in the results suggest that AP3B is moderately better than PP3B reacting with nucleic acids in general, however, when focusing on bifunctional adducts, the reaction efficiencies skew heavily in favor of AP3B. This is likely due in part to the increased reaction rate of AP3B with DNA as demonstrated in Fig. 4B. Since both ends of the crosslinking moiety must react to form interstrand crosslinks, the effect of the increased reaction rate is more obvious when only bifunctional adducts are examined. Other factors such as the accessibility of DNA to the crosslinkers in these assays may also account for some of the difference in labeling efficiency reported by the two assays. In cells, DNA is always in complex with proteins, such as histones. It is possible that the proteins associated with DNA interfere with the reaction of AP3B more than with the reaction of PP3B (AP3B has a positive charge that promotes its binding to negatively charged DNA, but in the presence of positively charged histone proteins, this feature may be significantly weakened), and thus decrease the labeling efficiency of AP3B more. With the in vitro assay however, naked DNA is used, and no proteins are present to hinder the reaction.
In summary, we have synthesized a more efficient nucleic acid crosslinking probe containing a photoactivable AMT moiety. Our experiments have shown that AP3B is more reactive with DNA than the currently available product PP3B in terms of both the amount of compound and UV exposure time required to produce similar result. Fluorescent detection of the compounds in cells shows a 5-fold increase in biotinylation for our compound compared to PP3B. The fold-increase is even higher (over 100 times) in vitro with naked DNA. These results suggest that AP3B is a highly efficient tool for dsDNA/RNA biotinylation and could potentially be used to improve the efficiency of SPLASH,10 Chem-seq,7 and other experiments.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The project described was supported by T32GM008500 from the National Institute of General Medical Sciences and the NIH common Fund 4D Nucleome Program U01HL 129958. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. This work made use of the Cornell NMR facility, which is supported, in part by, the NSF through MRI award CHE-153-1632.References
- E. Sharma, T. Sterne-Weiler, D. O'Hanlon and B. J. Blencowe, Global Mapping of Human RNA-RNA Interactions, Mol. Cell, 2016, 62, 618–626, DOI:10.1016/j.molcel.2016.04.030.
- Z. Lu, et al., RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure, Cell, 2016, 165, 1267–1279, DOI:10.1016/j.cell.2016.04.028.
- S. Dasari and P. B. Tchounwou, Cisplatin in cancer therapy: molecular mechanisms of action, Eur. J. Pharmacol., 2014, 740, 364–378, DOI:10.1016/j.ejphar.2014.07.025.
- P. G. Baraldi, et al., Novel benzoyl nitrogen mustard derivatives of pyrazole analogues of distamycin A: synthesis and antileukemic activity, Bioorg. Med. Chem., 1999, 7, 251–262, DOI:10.1016/s0968-0896(98)00205-3.
- T. Sakamoto, Y. Tanaka and K. Fujimoto, DNA Photo-Cross-Linking Using 3-Cyanovinylcarbazole Modified Oligonucleotide with Threoninol Linker, Org. Lett., 2015, 17, 936–939, DOI:10.1021/acs.orglett.5b00035.
- C.-X. Song and C. He, Bioorthogonal labeling of 5-hydroxymethylcytosine in genomic DNA and diazirine-based DNA photo-cross-linking probes, Acc. Chem. Res., 2011, 44, 709–717, DOI:10.1021/ar2000502.
- J. Dekker, et al., The 4D nucleome project, Nature, 2017, 549, 219–226, DOI:10.1038/nature23884.
- W. P. Zhen, O. Buchardt, H. Nielsen and P. E. Nielsen, Site specificity of psoralen-DNA interstrand cross-linking determined by nuclease Bal31 digestion, Biochemistry, 1986, 25, 6598–6603, DOI:10.1021/bi00369a039.
- S. Couvé-Privat, G. Macé, F. Rosselli and M. K. Saparbaev, Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells, Nucleic Acids Res., 2007, 35, 5672–5682, DOI:10.1093/nar/gkm592.
- J. G. A. Aw, et al., In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation, Mol. Cell, 2016, 62, 603–617, DOI:10.1016/j.molcel.2016.04.028.
- B. Dadonaite, et al., The structure of the influenza A virus genome, Nat. Microbiol., 2019, 4, 1781–1789, DOI:10.1038/s41564-019-0513-7.
- X. Zhong, et al., The zinc-finger protein ZFYVE1 modulates TLR3-mediated signaling by facilitating TLR3 ligand binding, Cell. Mol. Immunol., 2020, 17, 741–752, DOI:10.1038/s41423-019-0265-6.
- L. Anders, et al., Genome-wide localization of small molecules, Nat. Biotechnol., 2014, 32, 92–96, DOI:10.1038/nbt.2776.
- S. T. Isaacs, C.-K. J. Shen, J. E. Hearst and H. Rapoport, Synthesis and characterization of new psoralen derivatives with superior photoreactivity with DNA and RNA, Biochemistry, 1977, 16, 1058–1064, DOI:10.1021/bi00625a005.
- J. E. Hearst, Psoralen Photochemistry and Nucleic Acid Structure, J. Invest. Dermatol., 1981, 77, 39–44, DOI:10.1111/1523-1747.ep12479229.
- A. Oroskar, G. Olack, M. J. Peak and F. P. Gasparro, 4′-Aminomethyl-4,5′,8-trimethylpsoralen photochemistry: the effect of concentration and UVA fluence on photoadduct formation in poly(dA-dT) and calf thymus DNA, Photochem. Photobiol., 1994, 60, 567–573, DOI:10.1111/j.1751-1097.1994.tb05149.x.
- J. Sambrook and D. W. Russell, Alkaline Agarose Gel Electrophoresis, Cold Spring Harb. Protoc., 2006, 2006, pdb.prot4027, DOI:10.1101/pdb.prot4027.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07437c |
| This journal is © The Royal Society of Chemistry 2020 |