Open Access Article
Open Access ArticleCarotane sesquiterpenes from Ferula vesceritensis: in silico analysis as SARS-CoV-2 binding inhibitors†
Tarik A. Mohameda,
Abdelsamed I. Elshamyb,
Mahmoud A. A. Ibrahim c,
Ammar Zellaguid,
Mahmoud F. Moustafaef,
Alaa H. M. Abdelrahmanc,
Shinji Ohtag,
Paul W. Pare
c,
Ammar Zellaguid,
Mahmoud F. Moustafaef,
Alaa H. M. Abdelrahmanc,
Shinji Ohtag,
Paul W. Pare *h and
Mohamed-Elamir F. Hegazy*a
*h and
Mohamed-Elamir F. Hegazy*a
aChemistry of Medicinal Plants Department, National Research Centre, 33 El-Bohouth St., Dokki, Giza, 12622, Egypt. E-mail: me.fathy@nrc.sci.eg; Fax: +20-23337093
bChemistry of Natural Compounds Department, National Research Centre, 33 El Bohouth St., Dokki, Giza, 12622, Egypt
cComputational Chemistry Laboratory, Chemistry Department, Faculty of Science, Minia University, Minia 61519, Egypt
dLaboratory of Biomolecule and Plant Breeding, Life Science and Nature Department, Faculty of Exact Science and Life Science and Nature, University of Larbi Ben Mhidi, 4000 Oum El Bouaghi, Algeria
eDepartment of Biology, College of Science, King Khalid University, 9004, Abha, Kingdom of Saudi Arabia
fDepartment of Botany & Microbiology, Faculty of Science, South Valley University, Qena, Egypt
gGraduate School of Integrated Sciences for Life, Hiroshima University, 1-7-1 Kagamiyama, Higashi-Hiroshima 739-8521, Japan
hDepartment of Chemistry and Biochemistry, Texas Tech University, Lubbock, TX 79409, USA. E-mail: paul.pare@ttu.edu; Fax: +1 806 742 1289
First published on 18th September 2020
Abstract
Two sesquiterpenes, 8α-anisate-dauc-4-ene-3,9-dione (webiol anisate) (1) and 10α-acetoxy-6α-benzoate-jaeschkeanadiol (2) as well as, ten known analogues (3–10), and two sesquiterpene coumarins (11–12) were isolated from an organic root extract of Ferula vesceritensis (Fam. Apiaceae). Chemical structures were elucidated based on IR, 1D- and 2D-NMR and HRMS, spectroscopic analyses. With molecular overlap observed between two protease inhibitors that are being examined as anti-COVID-19 drugs, and sesquiterpenes isolated here, metabolite molecular docking calculations were made using the main protease (Mpro), which is required for viral multiplication as well as RNA-dependent RNA polymerase (RdRp). In silico binding-inhibition analysis predicted that select F. vesceritensis sesquiterpenes can bind to these enzymes required for viral replication. Structures of the isolated constituents were also consistent with the chemo-systematic grouping of F. vesceritensis secondary metabolites with other Ferula species.
Introduction
The genus Ferula, composed of approximately 180 species, is the third-largest genus in the Apiaceae family. Over 130 species are distributed throughout the Mediterranean and Central Asia region.1,2 Many Ferula species are used as traditional medicines for treatments such as impotency and frigidity, skin infections, dysentery, neurological disorders (tranquillizer, anti-hysteric), rheumatism, headache, digestive disorders, dizziness and arthritis.3–5 Resins of Ferula are used as a febrifuge as well as a carminative agent for stomach disorders.1 Some Ferula species exhibit anticancer,6,7 anthelmintic,7 antimicrobial,6,8 antifungal,7 anticonvulsant,9 antioxidant,6,10 antiproliferative,11,12 anti-hypertensive,13 hepatoprotective7 and antibiotic-odulation6 agents. Previous phytochemical studies of Ferula species revealed that the main constituents are sesquiterpenes.14Ferula vesceritensis (Batt.), also known as F. tingitana L. var, is endemic to Algeria and Libya, where it is used as traditional medicine for the treatment of inflammatory, cancer, fever, headaches and throat infections; livestock are observed to avoid grazing on the foliage.15 Previous F. vesceritensis phytochemical studies report the accumulation of sesquiterpenes and sesquiterpene coumarins.15,16 With the recent COVID-19 pandemic, the question arises as to whether secondary metabolites from F. vesceritensis can serve as inhibitors of enzymes that participate in viral replication.
From the outset, COVID-19 was identified as a new beta coronavirus, initially referred to as SARS-CoV-2 and later named coronavirus disease-2019 (COVID-19) by the World Health Organization.17 Infections are rapidly spread by respiratory droplets with a high mortality rate in select countries; the pandemic is a significant challenge to public health. With currently no specific antiviral drugs or vaccines developed to treat COVID-19, the virus is more deadly than the SARS, H1N1, MERS, and Ebola epidemics combined with more than 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lives lost to the disease in the first half of 2020. Very recently, the U.S. Food and Drug Administration (FDA) has issued an emergency use authorization of remdesivir for treatment of suspected or laboratory-confirmed COVID-19 cases.
000 lives lost to the disease in the first half of 2020. Very recently, the U.S. Food and Drug Administration (FDA) has issued an emergency use authorization of remdesivir for treatment of suspected or laboratory-confirmed COVID-19 cases.
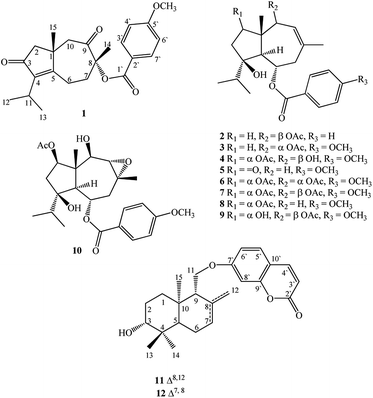
In seeking chemical inhibitors to block COVID-19 replication, the molecular docking technique was utilized to predict binding affinities for compounds isolated from F. vesceritensis against SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp), two essential enzymatic components required for viral replication. Compounds included two new sesquiterpenes (1,2) in addition to ten previously reported compounds (3–12) (Fig. 1). Chemo-systematic significance of metabolites from F. vesceritensis was also compared with other members of the Ferula genus.
Results and discussion
Extensive fractionation and purification of the organic extract of F. vesceritensis afforded two new sesquiterpene, 8β-anisate-dauc-4-ene-3,9-dione (webiol anisate) (1) and 10β-acetoxy-6α-benzoate-jaeschkeanadiol (2) as well as ten previously reported compounds, 10α-acetoxy-6α-anisate-jaeschkeanadiol (3),18 2α-acetoxy-10β-hydroxy-6α-anisate-jaeschkeanadiol (4),18 2-oxoferutidin (5),19 2α-acetoxy-6α-p-methoxybenzoyl-10α-acetoxy-jaeschkeanadiol (6),18 2α-acetoxy-6α-p-methoxybenzoyl-10β-acetoxy-jaeschkeanadiol (7),18 2-acetoxy-6-p-methoxybenzoyl-jaeschkeanadiol (8),20 2α-hydroxy-6αp-methoxybenzoyl-10β-acetoxy-jaeschkeanadiol (9),21 epoxyvesceritenol (10),15 coladonin (11),22 feselol (12).16,23 Structures for the known metabolites were elucidated by comparison of collected spectroscopic data (1D- and 2D-NMR as well as MS data) with literature reports.Compound 1 was obtained as a reddish amorphous powder with an optical rotation of ([α]25D +56.9) in MeOH. TOF-ESI-MS analysis showed molecular ion peak at m/z 407.1827 [M + Na ]+ (calcd for C23H28NaO5+, 407.1834) indicating a molecular formula of C23H28O5. The 13C NMR spectrum displayed 23 carbon signals that categorized to 9 quaternary carbons (comprising two keto groups at δC 205.8, 206.4), 5 methines, 4 methylenes, 5 methyls (including one methyl of methoxy). From all characterized carbons, para-anisate moiety (δC 166.3, 121.9, 132.2, 114.3 and 164.3), were clearly assigned. 1H NMR data revealed the presence of an isopropyl moiety signals at δH 1.20 (3H, d, J = 7.0 Hz), 1.22 (3H, d, J = 7.0 Hz) and 2.65 (1H, m)]. In addition to methyl groups at δH 1.04 (3H, s) and 1.69 (3H, s), an anisate moiety with characteristic aromatic protons at δH 8.24 (2H, d, J = 8.9 Hz), 7.07 (2H, d, J = 8.9 Hz) and methoxy protons at δH 3.72 (s) were observed. These structural elements suggested a carotane skeleton corresponding to a bicyclic structure with a condensed seven and five membered ring system, as previously isolated from several Ferula species.15,16,19,24,25 1H–1H COSY indicated a blocked correlation between methine proton at δH 2.65 (m) and two methyl signals at δH 1.20 (3H, d, J = 7.0 Hz), δH 1.22 (3H, d, J = 7.0 Hz), indicating that the isopropyl group was located on quaternary olefinic carbon δC 143.9. Additionally, a characteristic methyl signal at δH 1.04 (s) for H-15 showed an HMBC correlation with olefinic quaternary carbon at δC 175.2 as well as methine proton at δH 2.65 (m). Accordingly, isopropyl signals at δH 1.20d (J = 7.0 MHz), 1.22d (J = 7.0 MHz) and 2.65 m were assigned H-12, H-13, and H-11, respectively. The quaternary olefinic carbons at δC 143.9 and 175.2 were assigned to C-4 and C-5, respectively. The methine proton, H-11, showed HMBC correlations with two olefinic quaternary carbon at 143.9 (C-4), 175.2 (C-5), a keto group at δC 206.4 was assigned to C-3. H-15 showed HMBC correlations with quaternary aliphatic carbon at δC 41.1 and two aliphatic methylene carbons at δC 48.9, 43.0 assigned to C-1, C-2, and C-10 respectively that associated with the cyclopentane ring. The remaining proton signals for two methylene groups with a vicinal relationship was deduced via COSY and HSQC analyses [δH 2.74 (1H, m), 2.83 (1H, m) and δC 21.1] and [δH 1.53 (1H, m), 2.65 (1H, m) and δC 40.7] and HMBC analysis confirm localization to C-6 and C-7. Finally, the location of the anisate moiety was deduced by the correlation between the proton at δH 1.69 s (H-14) and C-7 (δC 40.7), keto group at δC 205.8 and a quaternary carbon with an oxygen function (δC 86.0) indicating that the anisate group and CH3-14 attached to a quaternary carbon with an oxygen function δC 86.0 (C-8). These correlations also confirmed the presence of the ketone at C-9. The position of the anisate at C-8 was as well deduced from the comparison of the NMR spectra with those the anisate derivative of webdiol that characterized by presence of a cycloheptane ring at C-8.15,19,25 The relative stereochemistry was deduced through coupling constants and NOESY analysis. NOESY correlations of H-15 with H-2β and H-10β, and H-10β with H-14 showed that H-15, H-14 are in a β orientation. Thus, the structure of 1 was determined as 8α-anisate-dauc-4-ene-3,9-dione.
Compound 2 was obtained as a reddish amorphous powder with an optical rotation of ([α] +20) in MeOH. TOF-ESI-MS analysis showed a molecular ion peak at m/z 400.2249 [M]+ (calcd for C24H32O5+, 400.2222) indicating a molecular formula of C24H32O5. 1H NMR data revealed the presence of an isopropyl moiety with signals at δH 0.83 (3H, d, J = 6.6 Hz), 0.96 (3H, d, J = 6.6 Hz) and 1.95 (1H, m)]. In addition to methyl groups at δH 1.18 (3H, s), 1.82 (3H, s) and 2.07 (3H, s); a benzoate moiety with characteristic aromatic protons at δH 8.00 (2H, d, J = 7.2 Hz), 7.46 (2H, t, J = 7.8 Hz) and 7.58 (1H, t, J = 7.8 Hz) was observed. The 13C NMR spectrum displayed 24 carbon signals (Table 1), which were further differentiated by DEPT to 5 methyls (1 acetate group), 3 methylenes, 10 methines (two oxygenated, 6 olefinic) and 5 quaternary carbons (1 oxygenated, 2 keto, 2 olefinic). Spectroscopic data were similar to 3 except the appearance of an additional olefinic proton at δH 7.58 (1H, t, J = 7.8) and the disappearance of methoxy protons. This methoxy substitution was confirmed by 13C-NMR analysis. 2D NMR (COSY, HMQC and HMBC) analyses (Fig. 2) and comparisons with published analogues indicated a 7/5 bicyclic cadinane-type sesquiterpene previously isolated from different Ferula species.15,16,19,24,25 The two methylene groups with a vicinal attached to the methine carbon δC 70.4 (C-6) and that the carbons signals at δC 41, 130.2 and 128.8 are assigned to C-7, C-8 and C-9, respectively. These data confirm the presence of an acetate group at C-10 attached to the methine carbon δC 70.4 (C-6) and that the carbons signals at δC 41, 130.2 and 128.8 are assigned to C-7, C-8 and C-9, respectively. These data confirm the presence of an acetate group at C-10. The two methylene groups with a vicinal relationship deduced via COSY and HSQC analysis [δH 1.68 (m), 1.24 (m) and δC 37.5] and [H-3 at δH 1.57 m, 1.93 m and δC 31.2] and a blocked correlation between methine proton at δH 1.95 (m) and two methyl signals at δH 0.83 (3H, d, J = 6.6 Hz), δH 0.96 (3H, J = 6.6 Hz) were observed. Long-range 1H–13C correlations associated with two methyl groups (δH 0.83 and 0.96) to the carbon signals at δC 37.3 and 86.5 as well as methylene protons H-2 δH 1.68 (m), 1.24 (m) and H-3 at δH 1.57 m, 1.93 m were also observed. Additionally, a characteristic methyl signal at δH 1.20 s for H-15 showed correlation with the carbon signals at δC 48.2 (C-1), 37.7 (C-2), 57.7 (C-5) and 80.7 (C-10) indicating that the isopropyl group and acetate groups were located on quaternary oxygenated carbon δC 86.5 for C-4 and oxygenated methine carbon at δC 80.7 for C-10 respectively. Finally, the location of the benzoate moiety was deduced by the correlation between the olefinic methyl at δH 1.82 s for H-14 via HMBC correlations with carbons signals at δC 130.2, 128.8 and the carbon of methylene at δC 41.2 as well as H-5 at δH 2.11 d (10.8). In addition, correlations of H-5 with carbon of oxygenated methine at δC 70.4 were observed; these correlations indicate that the benzoate group is attached to the methine carbon δC 70.4 (C-6) and that the carbons signals at δC 41, 130.2 and 128.8 are assigned to C-7, C-8 and C-9, respectively. These data confirm the presence of an acetate group at C-10.
| 1 | 2 | |||
|---|---|---|---|---|
| 1H NMR (C5D5N, J MHz) | 13C NMR (C5D5N) | 1H NMR (CDCl3, J MHz) | 13C NMR (CDCl3) | |
| 1 | — | 41.1 | — | 48.2 |
| 2 | 2.24 br d (17.4), 3.00 br d (17.4) | 48.9 | 1.68 m, 1.24 m | 37.5 |
| 3 | — | 206.4 | 1.57 m, 1.93 m | 31.2 |
| 4 | — | 143.9 | — | 86.5 |
| 5 | — | 175.2 | 2.11 d (10.8) | 57.7 |
| 6 | 2.74 m, 2.83 m | 21.1 | 5.30 td (10.8, 3.0) | 70.4 |
| 7 | 1.53 m, 2.65 m | 40.7 | 2.53 t (12.0), 2.30 dd (14.4, 3.0) | 41.2 |
| 8 | — | 86.0 | — | 130.2 |
| 9 | — | 205.8 | 5.25 br s | 128.8 |
| 10 | 2.44 d (12.4), 3.11 d (12.4) | 43.0 | 5.22 br s | 80.7 |
| 11 | 2.65 m | 25.5 | 1.95 m | 37.3 |
| 12 | 1.20 d (7.0) | 20.2 | 0.83 d (6.6) | 17.4 |
| 13 | 1.22 d (7.0) | 20.3 | 0.96 d (6.6) | 18.5 |
| 14 | 1.69 s | 21.4 | 1.82 s | 26.4 |
| 15 | 1.04 s | 29.3 | 1.18 s | 15.7 |
| 1′ | — | 166.3 | — | 166.4 |
| 2′ | — | 121.9 | — | 131.5 |
| 3′,7′ | 8.24 d (8.9) | 132.2 | 8.00 d (7.2) | 129.6 |
| 4′,6′ | 7.07 d (8.9) | 114.3 | 7.46 t (7.8) | 128.6 |
| 5′ | — | 164.3 | 7.58 t (7.8) | 133.3 |
| OCH3 | 3.72 s | 55.4 | — | — |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, OAc O, OAc |
— | — | — | 170.8 |
| CH3, OAc | — | — | 2.07 s | 21.2 |
The relative stereochemistry was deduced via a coupling constant, a td for proton at δH 5.30 (1H, J = 10.8, 3.0, H-6) is characteristic for the C-6β geminal proton of trans-fused daucane skeletons.18 A small coupling between protons at δH 5.25 (1H, s, H-9) and 5.22 (1H, s, H-10) was only possible when the proton at C-10 in α orientation; therefore acylated group at C-10 in the β orientation.18,21 The structure of 2 was therefore identified as 10β-acetoxy-6α-benzoate-jaeschkeanadiol.
Molecular docking
Utilizing AutoDock molecular docking software, binding affinities were predicted for isolated compounds 1–12 with SARS-CoV-2 Mpro and RdRp to inhibit SARS-CoV-2 replication. The predicted binding affinities are listed in Table 2 and compared to two human immunodeficiency virus (HIV) protease inhibitors that have recently been subjected to clinical investigations as promising anti-COVID-19 drugs:26 darunavir (DrugBank code: DB01264) and lopinavir (DrugBank code: DB01601). Two-dimensional representations for binding modes of 1–12 as well as darunavir and lopinavir inside the active sites of SARS-CoV-2 Mpro and RdRp are depicted in Fig. S2 and S3,† respectively. The assayed natural products exhibited intermediate binding affinities towards SARS-CoV-2 Mpro, and RdRp with docking scores ranged from −9.9 to −6.7 and from −7.6 to −6.4 kcal mol−1, respectively (Table 2). The observed high affinities are attributed to multiple hydrogen bonds, van der Waals and hydrophobic interactions between the natural products and proximal amino acids in the enzyme active site for Mpro and RdRp. Compound 1 demonstrated the highest binding affinities of −9.9 and −7.6 kcal mol−1, forming three hydrogen bonds with HIS163 (1.81 Å) and GLU166 (2.10, 2.08 Å), and six hydrogen bonds with TYR619 (1.99 Å), LYS621 (1.84, 2.05, 2.39 Å), CYS622 (1.78 Å) and LYS798 (2.21 Å) inside the active sites of Mpro and RdRp, respectively (Fig. 3 and 4). Compared to 1, the docking scores of lopinavir are similar to binding affinities of −9.8 and −7.5 kcal mol−1 towards Mpro and RdRp, respectively. In contrast, darunavir showed lower docking scores of −8.2 and −4.4 kcal mol−1 with Mpro and RdRp, respectively. Together these results provide quantitative data of the binding affinities of 1 as promising SARS-CoV-2 Mpro and RdRp inhibitor.| Compound | Main protease (Mpro) | RNA-dependent RNA polymerase (RdRp) | ||
|---|---|---|---|---|
| Docking score (kcal mol−1) | Binding features (hydrogen bond length in Å) | Docking score (kcal mol−1) | Binding features (hydrogen bond length in Å) | |
| 1 | −9.9 | HIS163 (1.81 Å), GLU166 (2.10, 2.08 Å) | −7.7 | TYR619 (1.99 Å), LYS621 (1.84, 2.05, 2.39 Å), CYS622 (1.78 Å), LYS798 (2.21 Å) |
| 2 | −8.8 | GLY143 (1.95 Å), GLU166 (2.06 Å) | −7.5 | LYS621 (1.96, 2.11 Å) |
| 3 | −6.6 | — | −6.9 | ASP618 (2.23 Å), ASP623 (2.86 Å), CYS622 (2.02 Å), TRP800 (1.96 Å) |
| 4 | −7.4 | GLU166 (2.45 Å) | −6.2 | TRP619 (2.21, 2.05 Å), CYS622 (2.10 Å) |
| 5 | −8.8 | GLY143 (2.08 Å), CYS145 (2.67 Å), GLU166 (2.27 Å) | −6.4 | ASP618 (2.19 Å), TRP800 (1.96 Å), LYS621 (2.77 Å) |
| 6 | −8.5 | HIS163 (2.12 Å), GLU166 (2.70 Å), THR190 (2.41 Å), GLN192 (1.79, 2.20 Å) | −6.4 | ASP618 (2.29 Å), LYS621 (2.09 Å), TRP800 (1.96 Å) |
| 7 | −8.0 | GLU166 (2.24 Å), GLN192 (2.00 Å) | −7.1 | ASP618 (2.26 Å), LYS621 (2.25, 2.52 Å), TRP800 (2.14 Å) |
| 8 | −9.0 | GLY143 (2.47 Å), CYS145 (1.87, 2.67 Å), THR190 (2.89 Å) | −6.4 | ASP618 (2.22 Å), LYS621 (2.20 Å), TRP800 (2.07 Å) |
| 9 | −6.4 | — | −6.5 | ASP618 (2.33 Å), ASP623 (2.95 Å), CYS622 (1.90 Å), TRP800 (2.02 Å) |
| 10 | −9.7 | CYS145 (1.86 Å), GLU166 (2.61 Å), GLN189 (2.80 Å), THR190 (2.75 Å) | −6.9 | LYS621 (1.98 Å), ASP760 (1.81 Å), TRP800 (1.81 Å) |
| 11 | −8.0 | GLU166 (2.20 Å) | −6.4 | TYR619 (2.34 Å), LYS621 (2.74 Å), GLU811 (1.88 Å) |
| 12 | −8.1 | MET49 (3.04 Å), SER144 (2.21 Å), GLN189 (2.92 Å) | −6.6 | ASP623 (2.11 Å), GLU811 (1.74 Å) |
| Darunavir | −8.2 | GLU166 (1.94, 2.88 Å), LEU167 (1.96 Å) | −4.4 | TYR619 (2.11 Å), ASP760 (1.83 Å), GLU811 (2.24, 2.26 Å) |
| Lopinavir | −9.8 | LEU141 (1.96 Å), GLY143 (2.01 Å), SER144 (3.09 Å), HIS164 (2.62 Å) | −7.5 | ARG553 (2.65 Å), LYS621 (3.03 Å), CYS622 (2.59 Å), ASP623 (2.40 Å) |
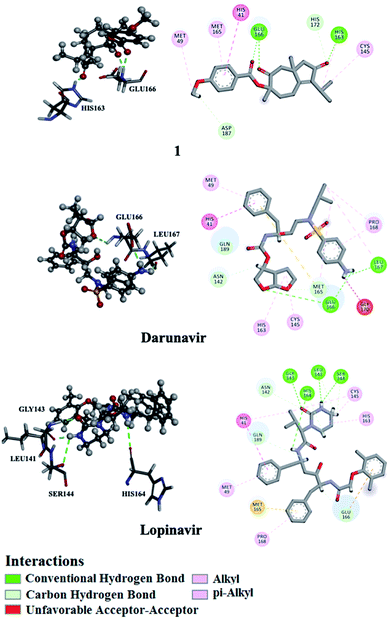 | ||
| Fig. 3 2D and 3D representations of interactions of 1, darunavir, and lopinavir with proximal amino acid residues of the SARS-CoV-2 main protease (Mpro). | ||
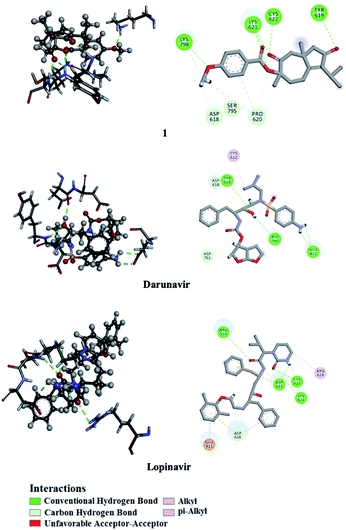 | ||
| Fig. 4 2D and 3D representations of interactions of 1, darunavir, and lopinavir with proximal amino acid residues of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). | ||
Molecular target prediction and network analysis
Using Swiss Target Prediction software, gene overlap between predicted responses activated by 1 and protein targets associated with severe acute respiratory syndrome diseases showed EGFR, MAPK14, and CTSL, as illustrated in the Venn diagram comparison (Fig. 5). Epidermal growth factor receptor (EGFR) inhibition may prevent an extreme fibrotic response to SARS-CoV and other respiratory viral infections and modulate the wound healing response to SARS-CoV.27 MAPK14 inhibition is predicted to block the ACE2 signaling pathway, and in turn, reduce cell internalization of SARS-CoV-2.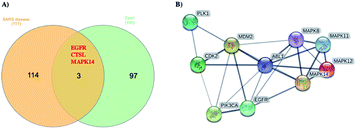 | ||
| Fig. 5 (A) Venn diagram analysis of 1 and SARS disease genes and (B) STRING PPI network for the top 10 targets identified by network analyzer for 1 as potent SARS-CoV-2 inhibitor. | ||
Angiotensin-converting enzyme 2 (ACE2) is a host protein and the receptor for SARS-CoV-2 entry.28 The cysteine protease cathepsin L (CTSL) is implicated in several types of pathology, and its inhibition plays a role in controlling inflammation, counterproductive immune responses. Besides, CTSL is an alternate molecular marker for drug design against SARS.29 The development of protease inhibitors able to inhibit CTSL, CTSB, and related proteases would be an excellent starting point for the development of broad-spectrum antiviral therapies. Targets genes activated by compound 1 were also analyzed via a STRING-PPI network and visualized by Cytoscape 3.8.0. EGFR and MAPK14 were observed among the top 10 scored genes for 1 (Table S1†).
Chemosystematic significance
From F. vesceritensis collected from the Algerian Sahara, our team and others isolated and identified several compounds mainly sesquiterpenes such as feselol, 13-hydroxyfeselol, 3-angeloxycoladonin, ferulsinaic acid, (−)-samarcandone, 1-methoxy-β-L-glucopyranoside, lapiferin, 10-hydroxylancerodiol-6-anisate, 2,10-diacetyl-8-hydroxyferutriol-6-anisate, 10-hydroxylancerodiol-6-benzoate, vesceritenone and epoxy-vesceritenol, farnesiferol A, 2-acetyl-jaechkeanadiol-6-anisate, lasidiol-10-anisate, 10-oxojaesckeanadiol-6-anisate, lapidol, coladin, coladonin, lancerodiol p-hydroxybenzoate and jaeschkeanin.15,16,30 In the present study, 12 secondary metabolites including two new carotene sesquiterpenes 1 and 2 were identified.The chemotaxonomic significance of F. vesceritensis was established depending upon the comparison of the described chemical compounds including our isolates and the isolated compounds from other Ferula ecospecies around the world. Our results as well as previous studies15,16,30 have found that isolated sesquiterpenes comprise mainly of daucane and/or carotene type sesquiterpenes in addition to some sesquiterpene coumarins. This first overview showed complete agreement between the isolates of our plant and all described metabolites from other Ferula species.14 In addition to daucane-type sesquiterpenes, sesquiterpene lactone and glycosides are commonly described sesquiterpenes from the genus such as F. hermonis,4,25,31 F. communis subsp. communis21, F. varia,32 F. Diversivittata,33 F. Sinaica,24 and F. jaeschkeana.34 A genetic correlation between F. vesceritensis and other Ferula species were deduced via an overlap of sesquiterpenes and sesquiterpenes esters and more specifically with the diversity of carotene type sesquiterpenes.
F. vesceritensis also exhibited a presence of sesquiterpene coumarins. Survey of the plants belonging to Ferula genus, sesquiterpene coumarins were found as one of the main characteristic metabolites for this genus such as F. teterrima, F. sinkiangensis,8 F. narthex,35 F. assa-foetida,36 F. tunetana,37 F. fukanensis,38 F. sinaica,23 F. assa-foetida24 and others.39 From these reports F. vesceritensis was deduced to be closely related with other Ferula species based on sesquiterpene type.
Eudesmanolide sesquiterpenes, their esters, and glycosides are present in some Ferula species. For examples, F. sinaica,24 and F. varia32,40,41 biosynthesize eudesmanolide sesquiterpenes. Also, F. ferulioides is observed to synthesize a diversity of unusual sesquiterpenes, including four resacetophenones, in addition to sesquiterpene coumarins with special skeleton alongside of the common sesquiterpenes in Ferula plants42
A final chemotaxonomical observation was that there are direct genetic relationships between the Algerian plant, F. vesceritensis, and the other Ferula ecospecies via the ability for biosynthetically building of the sesquiterpenes in general. More specific mapping of the Ferula species, F. vesceritensis was strongly correlated with all Ferula species via the ability to biosynthesize carotene and/or daucane sesquiterpenes esters and sesquiterpene coumarins. Also, F. sinaica, and F. varia were characterized by special types of sesquiterpenes, eudesmanolides while F. ferulioides was characterized with respect to phenones sesquiterpenes and sesquiterpenes coumarins with special skeletons.
Experimental
General experimental procedures
Optical rotations were recorded on a JASCO P-2300 polarimeter (Tokyo, Japan). NMR spectra were measured on a Bruker 500 NMR spectrometer (USA)-500 spectrometer (500 MHz for 1H and 125 MHz for 13C, respectively). All chemical shifts (δ) are given in ppm units with reference to TMS as an internal standard, and coupling constants (J) are reported in Hz. HRMS experiments were performed on Fourier transform ion cyclotron mass spectrometer. High-performance liquid chromatography (HPLC) was performed on an Agilent pump equipped with an Agilent-G1314 variable wavelength UV detector at 254 nm and a semi-preparative reverse-phase column (Econosphere™, RP-C18, 5 μm, 250 × 4.6 mm, Alltech, Deerfield, IL, USA). Silica gel 60 (230–400 mesh) was used for column chromatography. Pre-coated silica gel plates (Kieselgel 60 F254, 0.25 mm) were used for TLC analyses. Spots were visualized by heating after spraying with 10% H2SO4.Plant material
Roots of F. vesceritensis were collected during the flowering stage in March 2017 near Biskra, approximately 300 miles southeast of Algiers, Algeria by Prof. Dr Amar Zellagui, Department of Chemistry, Constantine University, where a voucher specimen has been deposited (AM#112).Extraction and separation
Root of F. vesceritensis (1 kg) was crushed and extracted with CH2Cl2–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at room temperature. The extract was concentrated in vacuo to obtain a residue (30 g). The residue was fractionated by silica gel CC (6 × 120 cm) eluted with n-hexane (3 L), followed by a gradient of n-hexane–CH2Cl2 up to 100% CH2Cl2 and CH2Cl2–MeOH up to 15% MeOH (2 L of each solvent mixture).
1) at room temperature. The extract was concentrated in vacuo to obtain a residue (30 g). The residue was fractionated by silica gel CC (6 × 120 cm) eluted with n-hexane (3 L), followed by a gradient of n-hexane–CH2Cl2 up to 100% CH2Cl2 and CH2Cl2–MeOH up to 15% MeOH (2 L of each solvent mixture).
The n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 (1
CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) fraction (3.5 g) was subjected to a second silica gel column (3 × 120 cm) eluted with n-hexane
1) fraction (3.5 g) was subjected to a second silica gel column (3 × 120 cm) eluted with n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 (6
CH2Cl2 (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) generating two subfractions. Subfraction 1A (0.8 g) was further purified by HPLC eluted with MeOH
1) generating two subfractions. Subfraction 1A (0.8 g) was further purified by HPLC eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (80
H2O (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20). The flow rate was set to 1.5 mL min−1 and was at 0–70 min to afford 1 (10 mg, purity >98% by HPLC), (eluent hexane/EtOAc 2
20). The flow rate was set to 1.5 mL min−1 and was at 0–70 min to afford 1 (10 mg, purity >98% by HPLC), (eluent hexane/EtOAc 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Rf = 0.45) and 2 (15 mg purity >96% by HPLC), (eluent hexane/EtOAc 2
1, Rf = 0.45) and 2 (15 mg purity >96% by HPLC), (eluent hexane/EtOAc 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Rf = 0.40). Subfraction 2A (1 g) was also purified by HPLC eluted with MeOH
1, Rf = 0.40). Subfraction 2A (1 g) was also purified by HPLC eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (75
H2O (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) The flow rate was set at 1.5 mL min−1 was at 0–60 min to afford 3 (25 mg, purity >98% by HPLC), (eluent hexane/EtOAc 1
25) The flow rate was set at 1.5 mL min−1 was at 0–60 min to afford 3 (25 mg, purity >98% by HPLC), (eluent hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.25), 4 (25 mg, purity >97% by HPLC), (eluent n-hexane/EtOAc 1
2, Rf = 0.25), 4 (25 mg, purity >97% by HPLC), (eluent n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Rf = 0.45) and 10 (35 mg, purity >98% by HPLC), (eluent hexane/EtOAc 1
1, Rf = 0.45) and 10 (35 mg, purity >98% by HPLC), (eluent hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.30). An n-hexane CH2Cl2 (1
2, Rf = 0.30). An n-hexane CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) fraction (3.3 g) was subjected to a silica gel fractionation (3 × 120 cm) eluted with n-hexane–CH2Cl2–MeOH to give two subfractions. Subfraction 1B (1.2 g) was further purified by HPLC eluted with MeOH
3) fraction (3.3 g) was subjected to a silica gel fractionation (3 × 120 cm) eluted with n-hexane–CH2Cl2–MeOH to give two subfractions. Subfraction 1B (1.2 g) was further purified by HPLC eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (70
H2O (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30). The flow rate was set at 2.0 mL min−1 was at 0–60 min to afford 5 (25 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1
30). The flow rate was set at 2.0 mL min−1 was at 0–60 min to afford 5 (25 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.25), 6 (15 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1
2, Rf = 0.25), 6 (15 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.33), 7 (20 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1
2, Rf = 0.33), 7 (20 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.30) and 8 (10 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1
2, Rf = 0.30) and 8 (10 mg, purity >98% by HPLC), eluded with n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.35); subfraction 2B (1.5 g) was also purified by HPLC eluted with MeOH
2, Rf = 0.35); subfraction 2B (1.5 g) was also purified by HPLC eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (70
H2O (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30). The flow rate was set at 1.5 mL min−1 was at 0–60 min to afford 9 (12 mg, purity >98% by HPLC), (eluent n-hexane/EtOAc 1
30). The flow rate was set at 1.5 mL min−1 was at 0–60 min to afford 9 (12 mg, purity >98% by HPLC), (eluent n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Rf = 0.40). The 100% CH2Cl2 fraction was subjected to HPLC eluted with MeOH
1, Rf = 0.40). The 100% CH2Cl2 fraction was subjected to HPLC eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (65
H2O (65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35). The flow rate was set at 2.0 mL min−1 was at 0–60 min to afford 11 (17 mg, purity >96% by HPLC), (eluent n-hexane/EtOAc 1
35). The flow rate was set at 2.0 mL min−1 was at 0–60 min to afford 11 (17 mg, purity >96% by HPLC), (eluent n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.45) and 12 (17 mg, purity >97% by HPLC), (eluent n-hexane/EtOAc 1
2, Rf = 0.45) and 12 (17 mg, purity >97% by HPLC), (eluent n-hexane/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, Rf = 0.40).
2, Rf = 0.40).
Molecular docking calculations
Autodock4.2 software was used to perform all molecular docking calculations.43 The crystal structures of SARS-CoV-2 main protease (Mpro; PDB code: 6LU7 (ref. 44)) and RNA-dependent RNA polymerase (RdRp; PDB code: 6M71 (ref. 45)) were taken as templates. Water molecules, ions, and ligand, if exist, were deleted. H++ server was chosen to study the protonation state of Mpro and RdRp, and all missing hydrogen atoms were accordingly added.46 Default docking parameters were employed, except the number of genetic algorithm (GA) run and the maximum number of energy evaluation (eval). The GA and eval values were set to 250 and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. The docking grid was set to 60 Å × 60 Å × 60 Å with a grid spacing value of 0.375 Å. The grid center was placed at the center of the active site of Mpro and RdRp. Prior to molecular docking, 3D structures of the isolated compounds were minimized using SZYBKI software with MMFF94S force field software (SZYBKI, 2016). The partial atomic charges for the compounds were calculated using Gasteiger method.47 The predicted binding poses for each compound were processed by the built-in clustering analysis (1.0 Å RMSD tolerance), and the lowest energy conformation from the largest cluster was selected as representative.
000, respectively. The docking grid was set to 60 Å × 60 Å × 60 Å with a grid spacing value of 0.375 Å. The grid center was placed at the center of the active site of Mpro and RdRp. Prior to molecular docking, 3D structures of the isolated compounds were minimized using SZYBKI software with MMFF94S force field software (SZYBKI, 2016). The partial atomic charges for the compounds were calculated using Gasteiger method.47 The predicted binding poses for each compound were processed by the built-in clustering analysis (1.0 Å RMSD tolerance), and the lowest energy conformation from the largest cluster was selected as representative.
Protein–protein interactions
The online web-based tools of Swiss Target Prediction (http://www.swisstargetprediction.ch) were applied to predict the biological targets for the most promising isolated compounds as SARS-CoV-2 inhibitors. The Dis Ge NET online database (https://www.disgenet.org) was utilized to collect the available database for SARS diseases. Venn diagram was designed using Interacti-Venn online tool.48 Protein–protein interaction (PPI) network was generated using a functional database of STRING for top predicted targets.49 Cytoscape 3.8.0 was employed to investigate target-function relation based on the network topological.50Conclusion
Ferula vesceritensis root extract afford two new sesquiterpenes, 8α-anisate-dauc-4-ene-3,9-dione (webiol anisate) (1) and 10α-acetoxy-6α-benzoate-jaeschkeanadiol (2) and ten known secondary metabolites. All compounds were in silico tested as anti-COVID-19 drugs using the main protease (Mpro) and RNA-dependent RNA polymerase (RdRp). The binding affinities indicated that 1 was a promising SARS-CoV-2 Mpro and RdRp inhibitor.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work under grant no. (R.G.P2/90/41).Notes and references
- L. Boulos, Medicinal Plants of North Africa, Reference Publications, Algonac, 1983, p. 183 Search PubMed.
- U. Yaqoob and I. A. Nawchoo, J. King Saud Univ., Sci., 2017, 29, 19–27 CrossRef.
- A. H. Al-Ja'fari, R. Vila, B. Freixa, F. Tomi, J. Casanova, J. Costa and S. Canigueral, Phytochemistry, 2011, 72, 1406–1413 CrossRef.
- A. A. Auzi, A. I. Gray, M. M. Salem, A. A. Badwan and S. D. Sarker, J. Asian Nat. Prod. Res., 2008, 10, 711–717 CrossRef.
- K. Tamemoto, Y. Takaishi, B. Chen, K. Kawazoe, H. Shibata, T. Higuti, G. Honda, M. Ito, Y. Takeda, O. K. Kodzhimatov and O. Ashurmetov, Phytochemistry, 2001, 58, 763–767 CrossRef CAS.
- M. Paydar, Y. L. Wong, B. A. Moharam, E. Movahed, W. F. Wong and C. Y. S. Looi, J. Med. Sci., 2013, 13, 236 CrossRef CAS.
- P. K. Upadhyay, S. Singh and G. Agrawal, Int. J. Green Pharm., 2017, 11(2), 240–247 Search PubMed.
- J.-R. Yang, Z. An, Z.-H. Li, S. Jing and H.-L. Qina, Chem. Pharm. Bull., 2006, 54, 1595–1598 CrossRef CAS.
- M. Sayah and A. Mandegari, Iran. Biomed. J., 2003, 7, 139–144 Search PubMed.
- H. Zhang, J. Lu, L. Zhou, L. Jiang and M. Zhou, Int. J. Clin. Exp. Med., 2015, 8, 20845–20852 CAS.
- M. Moradzadeh, H. R. Sadeghnia, S. H. Mousavi, M. Mahmoodi and A. Hosseini, Cell. Mol. Biol., 2017, 63, 17–22 CrossRef.
- F. Poli, G. Appendino, G. Sacchetti, M. Ballero, N. Maggiano and F. O. Ranelletti, Phytother. Res., 2005, 19, 152–157 CrossRef CAS.
- M. Ghanbari, M. Zahedi Khorasani and A. Vakili, Journal of Medicinal Plants, 2012, 3, 62–68 Search PubMed.
- M. Mohammadhosseini, A. Venditti, S. D. Sarker, L. Nahar and A. Akbarzadeh, Ind. Crops Prod., 2019, 129, 350–394 CrossRef CAS.
- K. Oughlissi-Dehak, P. Lawton, S. Michalet, C. Bayet, N. Darbour, M. Hadj-Mahammed, Y. A. Badjah-Hadj-Ahmed, M. G. Dijoux-Franca and D. Guilet, Phytochemistry, 2008, 69, 1933–1938 CrossRef CAS.
- A. A. Ahmed, M. E. Hegazy, A. Zellagui, S. Rhouati, T. A. Mohamed, A. A. Sayed, M. A. Abdella, S. Ohta and T. Hirata, Phytochemistry, 2007, 68, 680–686 CrossRef CAS.
- WHO, J World Health Organization, Geneva, available via https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020, accessed, 2020, 10.
- M. Miski and T. J. Mabry, Phytochemistry, 1985, 24, 1735–1741 CrossRef CAS.
- G. Appendino, J. Jakupovic, S. Alloatti and M. Ballero, Phytochemistry, 1997, 45, 1639–1643 CrossRef CAS.
- D. Lamnaouer, M.-T. Martin, D. Molho and B. Bodo, Phytochemistry, 1989, 28, 2711–2716 CrossRef CAS.
- S. Dall'Acqua, M. A. Linardi, F. Maggi, M. Nicoletti, V. Petitto, G. Innocenti, G. Basso and G. Viola, Bioorg. Med. Chem., 2011, 19, 5876–5885 CrossRef.
- M. Pinar and B. Rodríguez, Phytochemistry, 1977, 16, 1987–1989 CrossRef CAS.
- A. A. Ahmed, Phytochemistry, 1999, 50, 109–112 CrossRef CAS.
- A. A. Ahmed, M. H. Abdel-Razek, M. I. Nassar, S. Izumi, S. Ohta and T. Hirata, Phytochemistry, 2001, 57, 513–515 CrossRef CAS.
- A. Lhuillier, N. Fabre, E. Cheble, F. Oueida, S. Maurel, A. Valentin, I. Fouraste and C. Moulis, J. Nat. Prod., 2005, 68, 468–471 CrossRef CAS.
- C. Harrison, Nat. Biotechnol., 2020, 38, 379–381 CrossRef.
- T. Venkataraman and M. B. Frieman, Antiviral Res., 2017, 143, 142–150 CrossRef CAS.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280 CrossRef CAS.
- Y. Zhou, P. Vedantham, K. Lu, J. Agudelo, R. Carrion Jr, J. W. Nunneley, D. Barnard, S. Pöhlmann, J. H. McKerrow and A. R. Renslo, Antiviral Res., 2015, 116, 76–84 CrossRef CAS.
- A. M. Gamal-Eldeen and M. E. F. Hegazy, Nat. Prod. Res., 2010, 24, 246–257 CrossRef CAS.
- Z. Z. Ibraheim, W. M. Abdel-Mageed, H. Dai, H. Guo, L. Zhang and M. Jaspars, Phytother. Res., 2012, 26, 579–586 CrossRef CAS.
- S.-i. Kurimoto, K. Suzuki, M. Okasaka, Y. Kashiwada, O. K. Kodzhimatov and Y. Takaishi, Phytochem. Lett., 2012, 5, 729–733 CrossRef CAS.
- M. Iranshahi, A. Sahebkar, S. Hosseini, M. Takasaki, T. Konoshima and H. Tokuda, Phytomedicine, 2010, 17, 269–273 CrossRef CAS.
- M. Singh, A. Agnihotri, S. Garg, S. Agarwal, D. Gupta, G. Keshri and V. Kamboj, Planta Med., 1988, 54, 492–494 CrossRef CAS.
- A. Amin, E. Tuenter, P. Cos, L. Maes, V. Exarchou, S. Apers and L. Pieters, Molecules, 2016, 21, 1287 CrossRef.
- M. H. Abd El-Razek, S. Ohta, A. A. Ahmed and T. Hirata, Phytochemistry, 2001, 58, 1289–1295 CrossRef CAS.
- A. Jabrane, H. B. Jannet, Z. Mighri, J. F. Mirjolet, O. Duchamp, F. Harzallah-Skhiri and M. A. Lacaille-Dubois, Chem. Biodivers., 2010, 7, 392–399 CrossRef CAS.
- T. Motai and S. Kitanaka, Chem. Pharm. Bull., 2004, 52, 1215–1218 CrossRef CAS.
- Z. E. Nazari and M. Iranshahi, Phytother. Res., 2011, 25, 315–323 CrossRef CAS.
- S. Kurimoto, K. Suzuki, M. Okasaka, Y. Kashiwada, O. K. Kodzhimatov and Y. Takaishi, Chem. Pharm. Bull., 2012, 60, 913–919 CrossRef CAS.
- K. Suzuki, M. Okasaka, Y. Kashiwada, Y. Takaishi, G. Honda, M. Ito, Y. Takeda, O. K. Kodzhimatov, O. Ashurmetov and M. Sekiya, J. Nat. Prod., 2007, 70, 1915–1918 CrossRef CAS.
- T. Liu, S. Wang, L. Xu, W. Fu, S. Gibbons and Q. Mu, Chem. Biodivers., 2015, 12, 599–614 CrossRef CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao and H. Yang, bioRxiv, 2020 DOI:10.1101/2020.02.26.964882.
- Y. Gao, L. Yan, Y. Huang, F. Liu, Y. Zhao, L. Cao, T. Wang, Q. Sun, Z. Ming, L. Zhang, J. Ge, L. Zheng, Y. Zhang, H. Wang, Y. Zhu, C. Zhu, T. Hu, T. Hua, B. Zhang, X. Yang, J. Li, H. Yang, Z. Liu, W. Xu, L. W. Guddat, Q. Wang, Z. Lou and Z. Rao, Science, 2020, 368, 779–782 CrossRef CAS.
- J. C. Gordon, J. B. Myers, T. Folta, V. Shoja, L. S. Heath and A. Onufriev, Nucleic Acids Res., 2005, 33, W368–W371 CrossRef CAS.
- J. Gasteiger and M. Marsili, Tetrahedron, 1980, 36, 3219–3228 CrossRef CAS.
- H. Heberle, G. V. Meirelles, F. R. da Silva, G. P. Telles and R. Minghim, BMC Bioinf., 2015, 16, 169 CrossRef.
- R. Li, X. Ma, Y. Song, Y. Zhang, W. Xiong, L. Li and L. Zhou, J. Cell. Biochem., 2019, 120, 11265–11273 CrossRef CAS.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06901a |
| This journal is © The Royal Society of Chemistry 2020 |