Open Access Article
Open Access ArticleSynthesis, biological evaluation, and in silico studies of novel chalcone- and pyrazoline-based 1,3,5-triazines as potential anticancer agents†
Leydi M.
Moreno
a,
Jairo
Quiroga
ab,
Rodrigo
Abonia
ab,
Antonino
Lauria
c,
Annamaria
Martorana
c,
Henry
Insuasty
 d and
Braulio
Insuasty
d and
Braulio
Insuasty
 *ab
*ab
aHeterocyclic Compounds Research Group, Department of Chemistry, Universidad del Valle, A.A. 25360 Cali, Colombia. E-mail: braulio.insuasty@correounivalle.edu.co
bCenter for Bioinformatics and Photonics-CIBioFI, A.A. 25360 Cali, Colombia
cDipartimento di Scienze e Tecnologie Biologiche Chimiche e Farmaceutiche “STEBICEF”, Università di Palermo, Viale delle Scienze Ed. 17, I-90128 Palermo, Italy
dHeterocyclic Compounds Research Group, Department of Chemistry, Universidad de Nariño, A.A. 1175 Pasto, Colombia
First published on 15th September 2020
Abstract
A novel series of triazin-chalcones (7,8)a–g and triazin-N-(3,5-dichlorophenyl)pyrazolines (9,10)a–g were synthesized and evaluated for their anticancer activity against nine different cancer strains. Triazine ketones 5 and 6 were synthesized from the cyanuric chloride 1 by using stepwise nucleophilic substitution of the chlorine atom. These ketones were subsequently subjected to a Claisen–Schmidt condensation reaction with aromatic aldehydes affording chalcones (7,8)a–g. Then, N-(3,5-dichlorophenyl)pyrazolines (9,10)a–g were obtained by cyclocondensation reactions of the respective chalcones (7,8)a–g with 3,5-dichlorophenylhydrazine. Among all the evaluated compounds, chalcones 7d,g and 8g exhibited more potent in vitro anticancer activity, with outstanding GI50 values ranging from 0.422 to 14.9 μM and LC50 values ranging from 5.08 μM to >100 μM. In silico studies, for both ligand- and structure-based, were executed to explore the inhibitory nature of chalcones and triazine derivatives. The results suggested that the evaluated compounds could act as modulators of the human thymidylate synthase enzyme.
1. Introduction
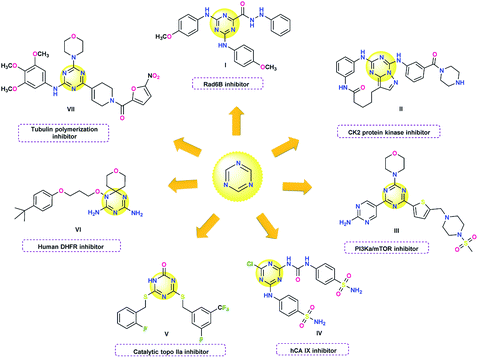
Heterocycles constitute a structural scaffold in a wide range of drugs and have occupied a prominent place in medicinal chemistry due to their ability to imitate and interact with biological molecules thus exhibiting remarkable pharmacological properties.1–4 The utility of heterocyclic rings, such as triazines and pyrazolines in different areas of medicine, particularly as potential anticancer drugs continue to be investigated.5–8The 1,3,5-triazine is a versatile ring that can act as a scaffold to carry three functionalized branches at 2,4 and 6-positions and this property allows to easily modulate the physicochemical and biological properties of these derivatives.5 The anticancer activity mechanisms of these systems can be associated with different targets. Thus, 1,3,5-triazine-2-carbohydrazides exhibited Rad6B inhibitory activity,9 while macrocyclic pyrazolo[1,5-a] [1,3,5]triazines showed potent inhibition of CK2 protein kinase.10 Other mechanisms of action are related to the inhibition of phosphatidylinositol 3-kinase α/mammalian target of rapamycin (PI3Kα/mTOR),11 carbonic anhydrase (CA),12–14 human topoisomerase IIα,15 dihydrofolate reductase (hDHFR)16 and tubulin polymerization17 (Fig. 1).
Similarly, pyrazoline rings are quite promising fragments due their anticancer properties. Pyrazoline hybrids with heterocyclic rings such as imidazopyridine,8 thiazole,18 triazine,19,20 4-thiazolidinone-indole21 and dihydroquinolone22 have been reported as potential anticancer agents. Pyrazoline rings can be easily obtained by a cyclocondensation reaction of chalcones with hydrazine derivatives.23 Chalcones have also shown marked biological activity as anticancer agents,24–26 that allows comparative studies of antiproliferative activity to be carried out between the precursor chalcones and the pyrazolinic derivatives and at the same time expand the mosaic of compounds evaluated.
The enzyme thymidylate synthase (TS) is an E2F1-regulated enzyme that is essential for DNA synthesis and repair. TS and mRNA levels are elevated in many human cancers, and high TS levels have been correlated with poor prognosis in patients with colorectal, breast, cervical, bladder, kidney, and non-small cell lung cancers.27 Inhibition of TS causing cells incapable of undergoing accurate DNA replication, ultimately leading to cell death.28,29 Owing to their important role in cell, TS represent a natural target for anticancer therapies.
In a preliminary work we reported the synthesis and anticancer activity of chalcone- and pyrazoline-based 1,3,5-triazines.20 Several of these tested molecules showed low toxicity and outstanding antiproliferative activity against a wide range of cancer cell lines with GI50 values in the range of 0.569–16.6 μM. A raw SAR analysis showed that N-(3,5-dichlorophenyl)pyrazolines and chalcones were the most active molecules. According to the previously described, we focused on the promising trisubstituted-1,3,5-triazinic systems and decided to synthesize and evaluate a new serie of N-(3,5-dichlorophenyl)pyrazoline and chalcone based 1,3,5-triazines as anticancer agents. Additionally, in silico studies were done to help understand the possible mode of action of the new active compounds.
2. Results and discussion
2.1. Chemistry
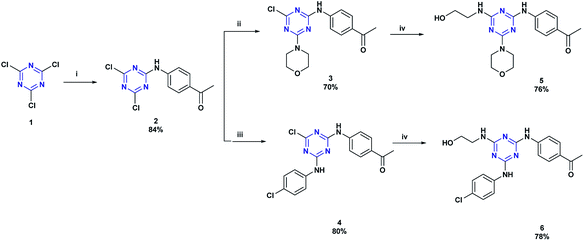
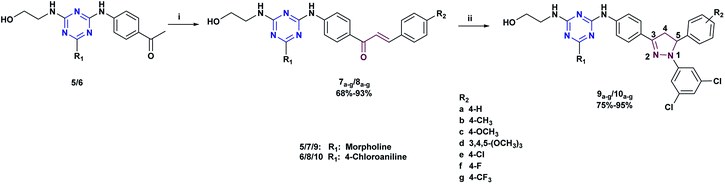
The target compound 9a–g and 10a–g were obtained by a multi-stage synthesis as described in Schemes 1 and 2. Initially, 2,4,6-trichloro-1,3,5-triazine 1 was submitted to consecutive nucleophilic substitution reactions: first with 4-aminoacetophenone, then with morpholine or 4-chloroaniline and finally with ethanolamine, under the conditions described in Scheme 1, to afford the respective 2,4,6-trisubstituted-1,3,5-triazines 5 and 6 in excellent yields. These precursors were characterized by FTIR, 1H-NMR, 13C-NMR, and mass spectrometry (see Experimental section).(E)-1-(4-((4-((2-hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-(substituted)chalcones 7a–g and (E)-1-(4-((4-((4-chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(substituted)chalcones 8a–g were obtained from triazine-ketones 5 and 6, respectively, by Claisen–Schmidt condensation reaction with substituted benzaldehydes in the presence of 20% KOH solution in EtOH (Scheme 2). The structures of chalcones 7a–g and 8a–g were confirmed by spectroscopic techniques (FTIR, 1H-NMR, 13C-NMR and mass spectrometry). These compounds showed wide FTIR absorption bands in the range of 3277–3315 cm−1 assigned to O–H groups. The IR spectra also showed absorption bands at 1641–1651, 1560–1587 and 1506–1558 cm−1 assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C functionalities, respectively. In the 1H-NMR spectrum of chalcone 7e, for example, the signal of the vinylic proton Hβ of the α,β-unsaturated system appears as a doublet at 7.68 ppm with a coupling constant of 3J = 15.6 Hz, which agrees with a E-configuration. The signal of other vinyl proton Hα appears overlapped with signals of aromatic protons at 7.89–7.99 ppm. In the 13C-NMR spectrum the signals of α and β carbon atoms were observed at 118.4 and 141.4 ppm, respectively. The mass spectrum shows molecular ion peak at m/z 480 and with a isotopic profile of 12
C functionalities, respectively. In the 1H-NMR spectrum of chalcone 7e, for example, the signal of the vinylic proton Hβ of the α,β-unsaturated system appears as a doublet at 7.68 ppm with a coupling constant of 3J = 15.6 Hz, which agrees with a E-configuration. The signal of other vinyl proton Hα appears overlapped with signals of aromatic protons at 7.89–7.99 ppm. In the 13C-NMR spectrum the signals of α and β carbon atoms were observed at 118.4 and 141.4 ppm, respectively. The mass spectrum shows molecular ion peak at m/z 480 and with a isotopic profile of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ([M]+
4 ([M]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+), characteristic of a compound bearing one chlorine atom in its structure.
[M + 2]+), characteristic of a compound bearing one chlorine atom in its structure.
Reaction of the synthesized chalcones 7a–g and 8a–g with 3,5-dichlorophenylhydrazine hydrochloride in ethanol under reflux afforded N-(3,5-dichlorophenyl)pyrazolines 9a–g and 10a–g (Scheme 2), respectively, in racemic mixtures. Taking as example compound 9e, in the 1H-NMR spectrum, the formation of the pyrazoline ring is confirmed by the appearance of two double doublets at 3.15 ppm (with 2JAM = 17.7 Hz and 3JAX = 4.4 Hz), and at 3.92 ppm (with 2JAM = 17.7 Hz, and 3JMX = 11.7 Hz) corresponding to protons on the diastereotopic center C-4, while the H-5 proton is observed as a double doublet at 5.63 ppm (with 3JMX = 11.7 Hz and 3JAX = 4.4 Hz), confirming the existence of an AMX coupling system in the pyrazoline ring. In the 13C-NMR, the absence of the carbonyl carbon signal and the appearance of signals at 43.3 and 61.6 ppm corresponding to C-4 and C-5, respectively, also confirmed the formation of pyrazolic ring. In general, mass spectra of N-(3,5-dichlorophenyl)pyrazolines 9a–g and 10a–g show well-defined molecular ion peaks.
It must be highlighted that the synthetic pathway used to obtain the triazine derivatives was efficient, the formation of by-products was not observed, and high yields were achieved. The purity was verified by thin-layer chromatography (TLC), mass spectrometry and proton NMR spectroscopy.
2.2. Anticancer activity evaluation
The in vitro anticancer activities of all the synthesized triazine derivatives 5, 6, (7–10)a–g were evaluated by the National Cancer Institute (NCI). Initially, compounds were subjected to preliminary screening at one-dose (10 μM) against 60 human cancer cell lines consisting of nine tumor panels, which include leukemia, lung, colon, melanoma, renal, prostate, CNS, ovarian, and breast cancer cell lines and the results are reported as NCI mean graphs. According to the data analysis of the one-dose mean graphs low GP (growth percent) values indicates better growth inhibition percentages (%GI) (i.e. %GI = 100 − GP). The negative GP values correspond to lethal activity, hence, more negative values represent higher activity of the assayed compound. Additionally, low mean values represent better activity and is used as a criteria for further studies. Indeed, mean values ≤ 50 indicate that the compound is active. According to the higher (%GI and lethality) and lower mean values for one-dose criteria, among the evaluated componds, chalcones 7d, 7g and 8g were actives and displayed the better %GI, lethality and mean values than the remaining assayed compounds, Table 1.| Compound | Mean growth | Most sensitive cell line | Growth inhibition percent (%GI)a/lethalityb |
|---|---|---|---|
| a %GI (growth inhibition percentage) = 100 − GP (growth percentage). b Negative values mean lethality of the respective cancer cell line. c Compounds with the most relevant inhibitory activity against all cancer cell lines in terms of their mean values. | |||
| 5 | 97.18 | SNB-75 (CNS cancer) | 33.36 |
| 6 | 89.48 | SR (leukemia) | 35.27 |
| 7a | 81.57 | SR (leukemia) | 87.24 |
| 7b | 95.32 | UACC-62 (melanoma) | 27.15 |
| 7c | 87.49 | MCF7 (breast cancer) | 70.68 |
| 7d | 17.60 | UACC-62 (melanoma) | −31.70b,c |
| 7e | 77.12 | MCF7 (breast cancer) | 90.27 |
| 7f | 79.95 | MCF7 (breast cancer) | 88.77 |
| 7g | 34.86 | U251 (CNS cancer) | −51.34b,c |
| 8a | 71.26 | HCT-15 (colon cancer) | 91.45 |
| 8b | 77.30 | MCF7 (breast cancer) | 78.50 |
| 8c | 68.77 | RPMI-8226 (leukemia) | 91.94 |
| 8d | 69.89 | MOLT-4 (leukemia) | 84.74 |
| 8e | 58.28 | HCT-116 (colon cancer) | −78.88a |
| 8f | 67.99 | RPMI-8226 (leukemia) | 98.22 |
| 8g | 42.92 | HCT-116 (colon cancer) | −85.59b,c |
| 9a | 82.34 | UACC-62 (melanoma) | 45.95 |
| 9b | 93.57 | MCF7 (breast cancer) | 33.41 |
| 9c | 87.26 | MCF7 (breast cancer) | 45.02 |
| 9d | 50.41 | RXF 393 (renal cancer) | 94.92 |
| 9e | 84.61 | MCF7 (breast cancer) | 43.41 |
| 9f | 74.76 | 786-0 (renal cancer) | 67.01 |
| 9g | 72.39 | RXF 393 (renal cancer) | 80.30 |
| 10a | 95.13 | HS 578T (breast cancer) | 29.35 |
| 10b | 96.74 | HS 578T (breast cancer) | 31.73 |
| 10c | 90.95 | UO-31 (renal cancer) | 35.19 |
| 10d | 74.43 | MOLT-4 (leukemia) | 65.16 |
| 10e | 95.42 | HS 578T (breast cancer) | 35.59 |
| 10f | 94.58 | HS 578T (breast cancer) | 38.64 |
| 10g | 99.72 | HS 578T (breast cancer) | 26.04 |
Based on the mean growth data reported in Table 1 and Fig. S1,† it can be observed that:
• The chalcones with substituent R1 = 4-chloroaniline 8a–g showed better activity than the respective pyrazolines 10a–g. This pattern of behavior was not observed in all cases when the substituent was R1 = morpholine.
• Pyrazolines with substituent R1 = morpholine showed better activity than those with substituent R1 = 4-chloroaniline.
• The presence of the 3,4,5-(OCH3)3 (R2 = d) and CF3 (R2 = g) groups favor the anticancer properties in both chalcones and pyrazolines, with the exception of chalcone 8d and pyrazoline 10g.
• The chalcone 7d with substituent R1 = morpholine and R2 = 3,4,5-(OCH3)3 was the leading structure among the series of synthesized compounds, with the best anticancer property.
Due to compounds 7d,g and 8g exhibited the broadest spectrum and the highest inhibitory activity among the 30 evaluated compounds against all nine panels of human cancer cell lines at one-dose assay, they were subjected to evaluation at five concentrations of dilution (i.e. 100, 10, 1.0, 0.1 and 0.01 μM), in order to determine their antiproliferative activity (GI50 and LC50) (Table 2). Chalcone 7d showed GI50 values in the range of 0.422–3.05 μM and LC50 values of 6.03 to >100 μM, being the SR cell line (leukemia, GI50 = 0.422 μM and LC50 > 100 μM) the most sensitive strain. Compound 7g exhibited GI50 values in the range of 1.25–8.66 μM and LC50 values of 5.08 to >100 μM, being the MCF7 cell line (breast cancer, GI50 = 1.25 μM) the most sensitive strain, while compound 8g showed GI50 values in the range of 1.48–14.9 μM and LC50 values of 5.41 to >100 μM, being specially effective against the HCT-116 cell line (colon cancer) with GI50 = 1.48 μM. The best cytotoxicity value was shown by compound 7g against UO-31 (renal cancer, LC50 = 5.08 μM). Should be noted that compounds 7d,g and 8g showed better antiproliferative activity than 5-fluorouracil (5-FU) (standard drug) in several cancer cell lines (Table 2, bold entries).
| Panel name | Cell name | Compounds | 5-FU NS 18893d | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 7d | 7g | 8g | |||||||
| GI50a | LC50b | GI50 | LC50 | GI50 | LC50 | GI50 | LC50 | ||
| a GI50 was the drug concentration resulting in a 50% reduction in the net protein increase (as measured by SRB staining) in control cells during the drug incubation, determined at five concentration levels (100, 10, 1.0, 0.1, and 0.01 μM). Italics entries are the most relevant GI50 values of each compound and bold entries are GI50 values (of our compounds) lower than GI50 values of 5-FU. b LC50 is a parameter of cytotoxicity that reflects the molar concentration needed to kill 50% of the cells. c Data obtained from NCI's in vitro disease-oriented human cancer cell lines screen in μM. d The values of activity against human cancer cell lines displayed by 5-FU correspond to that reported by. Please visit: https://dtp.cancer.gov/dtpstandard/cancerscreeningdata/index.jsp. | |||||||||
| Leukemia | CCRF-CEM | 0.935 | >100 | 2.41 | >100 | 2.75 | >100 | 10.00 | >100 |
| HL-60(TB) | 1.79 | >100 | 2.92 | >100 | 12.2 | >100 | 2.51 | >100 | |
| K-562 | 0.783 | >100 | 2.33 | >100 | 2.83 | >100 | 3.98 | >100 | |
| MOLT-4 | 1.19 | >100 | 2.45 | >100 | 3.56 | >100 | 0.32 | >100 | |
| RPMI-8226 | 0.612 | — | 2.07 | >100 | 1.56 | >100 | 0.05 | >100 | |
| SR | 0.422 | >100 | 3.1 | >100 | 1.95 | >100 | 0.03 | >100 | |
| Non-small cell lung cancer | A549/ATCC | 2.18 | >100 | 2.74 | >100 | 3.48 | 40.2 | 0.20 | >100 |
| EKVX | 2.83 | >100 | 2.44 | 32 | 3.41 | 41.8 | 63.10 | >100 | |
| HOP-62 | 2.72 | >100 | 2.22 | 78.6 | 8.55 | 50.5 | 0.40 | >100 | |
| HOP-92 | 1.61 | >100 | 2.48 | >100 | 5.39 | >100 | 79.43 | >100 | |
| NCI-H226 | 2.15 | >100 | 2.32 | >100 | 4.29 | 62.7 | 50.12 | >100 | |
| NCI-H23 | 2.03 | >100 | 2.55 | 29.4 | 10.7 | 64.1 | 0.32 | >100 | |
| NCI-H322M | 2.18 | >100 | 2.63 | 36.2 | 14.9 | >100 | 0.20 | >100 | |
| NCI-H460 | 1.58 | 7.64 | 2.15 | 9.18 | 4 | 36.9 | 0.06 | >100 | |
| NCI-H522 | 1.59 | >100 | 1.85 | 57.9 | 7.82 | 59.4 | 7.94 | >100 | |
| Colon cancer | COLO 205 | 1.8 | — | 2.09 | 7.94 | 4.8 | 41.7 | 0.16 | >100 |
| HCC-2998 | 1.8 | 6.03 | 2.34 | 19.2 | 2.93 | 28.5 | 0.05 | >100 | |
| HCT-116 | 1.3 | 6.91 | 1.43 | 5.23 | 1.48 | 5.41 | 0.25 | 25.12 | |
| HCT-15 | 1.43 | >100 | 1.89 | 19.5 | 2.08 | 26.2 | 0.10 | >100 | |
| HT29 | 1.7 | — | 2 | 11.6 | 2.32 | 43.2 | 0.16 | >100 | |
| KM12 | 1.66 | — | 1.8 | 6.05 | 2.13 | 12.7 | 0.20 | >100 | |
| SW-620 | 1.81 | — | 1.92 | 8.57 | 2.17 | 14.4 | 1.00 | >100 | |
| CNS cancer | SF-268 | 1.88 | >100 | 2.68 | 40.6 | 5.05 | 52.2 | 1.58 | >100 |
| SF-295 | 3.03 | >100 | 2.67 | 31.5 | 3.98 | 38.3 | 0.25 | >100 | |
| SF-539 | 1.63 | 6.11 | 1.64 | 5.7 | 2.39 | 23.6 | 0.06 | >100 | |
| SNB-75 | 1.07 | — | 1.84 | 28.6 | 2.05 | 30.6 | 3.98 | >100 | |
| U251 | 1.38 | 6.41 | 1.63 | 7.02 | 2.18 | 18.8 | 1.00 | >100 | |
| Melanoma | LOX IMVI | 1.48 | — | 1.67 | 5.79 | 1.66 | 6.27 | 0.25 | 79.43 |
| MALME-3M | 1.45 | — | 2.91 | 32.6 | 13.9 | 58.3 | 0.05 | >100 | |
| M14 | 1.79 | — | 2.01 | 9.08 | 3.5 | 37.4 | 1.00 | >100 | |
| MDA-MB-435 | 1.66 | — | 2.23 | 16.8 | 3.46 | 35.8 | 0.08 | >100 | |
| SK-MEL-2 | 1.71 | 8.14 | 8.66 | 48.3 | 11.7 | 51 | 63.10 | >100 | |
| SK-MEL-28 | 1.78 | — | 3.25 | 34 | 5.88 | 42.8 | 1.00 | >100 | |
| SK-MEL-5 | 1.76 | 6.12 | 3.16 | 32 | 14 | 62.7 | 0.50 | 79.43 | |
| UACC-257 | 1.44 | — | 3.98 | 70.1 | 10.1 | 96.5 | 3.98 | >100 | |
| UACC-62 | 1.54 | 6.67 | 2.76 | 30.2 | 6.32 | 44.1 | 0.50 | >100 | |
| Ovarian cancer | IGROV1 | 1.9 | >100 | 2 | 9.77 | 4.92 | 40.2 | 1.26 | >100 |
| OVCAR-3 | 1.69 | >100 | 1.96 | 7.34 | 2.75 | 28 | 0.02 | 50.12 | |
| OVCAR-4 | 1.63 | — | 3.05 | 54.2 | 4.14 | 39.3 | 3.98 | >100 | |
| OVCAR-5 | 2.02 | >100 | 1.74 | 9.07 | 5.93 | 43.4 | 10.00 | >100 | |
| OVCAR-8 | 2.2 | >100 | 3.25 | 86.5 | 5.66 | >100 | 1.58 | >100 | |
| NCI/ADR-RES | 2.32 | >100 | 2.87 | >100 | 5.58 | 98.8 | 0.32 | >100 | |
| SK-OV-3 | 3.05 | >100 | 3.82 | >100 | 13.4 | 87.8 | 19.95 | >100 | |
| Renal cancer | 786-0 | 1.76 | — | 1.65 | 5.82 | 2.25 | 16.6 | 0.79 | >100 |
| A498 | 1.32 | >100 | 1.4 | 35.6 | 1.67 | 35.5 | 0.40 | >100 | |
| ACHN | 1.95 | >100 | 1.86 | 8.03 | 4.81 | 41.2 | 0.32 | >100 | |
| CAKI-1 | 2.09 | >100 | 2.54 | 69.6 | 5.12 | 43.4 | 0.08 | >100 | |
| RXF 393 | 1.46 | 7.14 | 1.63 | 6.53 | 1.82 | 9.5 | 2.51 | >100 | |
| SN12C | 1.78 | >100 | 2.22 | 21.6 | 5.53 | 41.9 | 0.50 | >100 | |
| UO-31 | 1.24 | — | 1.31 | 5.08 | 1.75 | 12.5 | 1.26 | >100 | |
| Prostate cancer | PC-3 | 2.14 | >100 | 2.24 | >100 | 2.99 | >100 | 1.58 | >100 |
| DU-145 | 1.35 | — | 2.19 | 9.56 | 6.08 | 43.2 | 2.51 | >100 | |
| Breast cancer | MCF7 | 1.17 | 6.55 | 1.25 | 79.4 | 1.49 | 53.7 | 0.40 | >100 |
| MDA-MB-231/ATCC | 2.2 | >100 | 2.69 | >100 | 4.21 | 40.8 | 0.08 | >100 | |
| HS 578T | 1.65 | >100 | 2.61 | >100 | 2.82 | >100 | 6.31 | >100 | |
| BT-549 | 1.35 | 6.06 | 1.56 | 6.11 | 3.07 | 30.5 | 10.00 | >100 | |
| T-47D | 1.8 | >100 | 2.28 | >100 | 3.4 | >100 | 10.00 | >100 | |
| MDA-MB-468 | 1.34 | >100 | 1.8 | 8.3 | 5.54 | 64.8 | 7.94 | >100 | |
Additionally, mean GI50 values (per panel) of chalcones 7d,g and 8g in comparison with the standard anticancer agent 5-fluorouracil (5-FU) were determined and drawn in Fig. 2 for a better understanding. Based on these data, chalcone 7d was more active against all panels of cancer cell lines than compounds 7g and 8g, exhibiting lower activity in only two panels of cancer cell lines (i.e. colon and CNS) compared to the standard drug 5-FU. Remarkably, chalcone 7d exhibited the lowest mean GI50 value against leukemia panel (0.96 μM), while the lowest mean GI50 values for compounds 7g and 8g were obtained for renal (1.80 μM) and colon (2.56 μM) cancer panels, respectively. These finding indicates that compound 7d might be used as promising lead molecule for discovering a new class of anticancer agents.
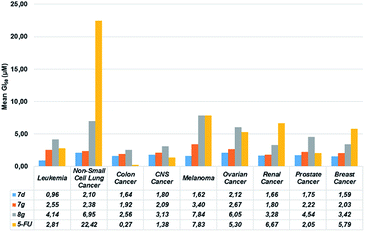 | ||
| Fig. 2 Comparison of the mean GI50 values, per panel, displayed by chalcones 7d,g, 8g and the standard drug 5-fluorouracil (5-FU) against the 60 human cancer cell lines. | ||
2.3. In silico insights
The antiproliferative activities showed by the chalcone and pyrazoline derivatives, lead us to investigate on the suitable biological target that can play a crucial role in the biological response. With this aim molecular modeling protocols, well established in previous works,30–33 were applied by using a mixed ligand- and structure-based approach.The features of the ligand-based approaches supported by molecular descriptors allowed the evaluation of the topological, thermodynamic, and charge-related characteristics of the ligands. Thus, two complementary standpoints in the evaluation of the binding capability (ligand- and structure-based) covered all the interaction aspects in the ligand–target complex.
In the first step the structures were submitted to the web-server DRUDIT (DRUgs Discovery Tools), an open access virtual screening platform, recently developed by us (https://www.drudit.com) based on molecular descriptors,34 useful in the identification of suitable biological targets for small molecules.
In particular, by the Biotarget Predictor tool, the DRUDIT Affinity Score (DAS) is assigned to each input structure versus the biological targets database implemented in the tool. The DAS values of the molecules under investigation against the best biological targets are reported in Table 3.
| Biological target/cmd | Thymidylate synthase | Serine threonine protein kinase4 | Voltage gated sodium channel subunit Nav1-5 | Proto-oncogene tyrosine protein kinase Src | Tyrosine protein kinase ZAP-70 | Epidermal growth factor receptor EGFR | Nociceptin receptor |
|---|---|---|---|---|---|---|---|
| 5 | 0.778 | 0.762 | 0.812 | 0.84 | 0.712 | 0.702 | 0.716 |
| 6 | 0.822 | 0.796 | 0.81 | 0.808 | 0.742 | 0.786 | 0.748 |
| 7a | 0.882 | 0.846 | 0.898 | 0.896 | 0.886 | 0.87 | 0.854 |
| 7b | 0.894 | 0.852 | 0.89 | 0.9 | 0.894 | 0.87 | 0.88 |
| 7c | 0.89 | 0.84 | 0.9 | 0.89 | 0.898 | 0.874 | 0.858 |
| 7d | 0.804 | 0.754 | 0.768 | 0.794 | 0.85 | 0.824 | 0.736 |
| 7e | 0.882 | 0.848 | 0.882 | 0.872 | 0.888 | 0.892 | 0.834 |
| 7f | 0.896 | 0.834 | 0.888 | 0.876 | 0.854 | 0.874 | 0.824 |
| 7g | 0.886 | 0.768 | 0.84 | 0.764 | 0.776 | 0.798 | 0.754 |
| 8a | 0.874 | 0.848 | 0.818 | 0.846 | 0.83 | 0.858 | 0.8 |
| 8b | 0.858 | 0.866 | 0.83 | 0.854 | 0.858 | 0.876 | 0.832 |
| 8c | 0.898 | 0.856 | 0.84 | 0.864 | 0.872 | 0.884 | 0.83 |
| 8d | 0.84 | 0.774 | 0.78 | 0.782 | 0.782 | 0.834 | 0.734 |
| 8e | 0.856 | 0.832 | 0.766 | 0.802 | 0.798 | 0.832 | 0.794 |
| 8f | 0.888 | 0.836 | 0.804 | 0.826 | 0.8 | 0.868 | 0.778 |
| 8g | 0.878 | 0.774 | 0.738 | 0.724 | 0.724 | 0.778 | 0.74 |
| 9a | 0.794 | 0.822 | 0.81 | 0.816 | 0.804 | 0.816 | 0.792 |
| 9b | 0.8 | 0.822 | 0.802 | 0.824 | 0.788 | 0.782 | 0.794 |
| 9c | 0.786 | 0.83 | 0.814 | 0.81 | 0.818 | 0.744 | 0.798 |
| 9d | 0.762 | 0.79 | 0.748 | 0.718 | 0.756 | 0.668 | 0.716 |
| 9e | 0.788 | 0.82 | 0.778 | 0.784 | 0.788 | 0.778 | 0.772 |
| 9f | 0.808 | 0.826 | 0.804 | 0.81 | 0.792 | 0.804 | 0.802 |
| 9g | 0.818 | 0.79 | 0.736 | 0.724 | 0.7 | 0.646 | 0.734 |
| 10a | 0.776 | 0.822 | 0.708 | 0.728 | 0.754 | 0.712 | 0.738 |
| 10b | 0.768 | 0.812 | 0.714 | 0.728 | 0.728 | 0.678 | 0.748 |
| 10c | 0.77 | 0.804 | 0.714 | 0.722 | 0.76 | 0.68 | 0.746 |
| 10d | 0.746 | 0.764 | 0.692 | 0.652 | 0.704 | 0.604 | 0.682 |
| 10e | 0.756 | 0.788 | 0.704 | 0.694 | 0.732 | 0.66 | 0.724 |
| 10f | 0.78 | 0.808 | 0.722 | 0.718 | 0.746 | 0.69 | 0.744 |
| 10g | 0.77 | 0.756 | 0.678 | 0.62 | 0.616 | 0.574 | 0.678 |
| Average | 0.825 | 0.811 | 0.790 | 0.790 | 0.788 | 0.775 | 0.773 |
The obtained results showed as the molecules under investigation have a quite good affinity against thymidylate synthase (TS) (DAS averaged 0.825, which reaches 0.87 for the derivatives 7 and 8), thus to get information on which molecular features are involved in the binding, structure-based studies were performed.
The monomer of TS consists of an α/β-fold containing 7α-helices and 10β-strands, arranged in three layers: a six stranded mixed β-sheet, a long α-helix across the sheet flanked by two shorter helices, and a mixed layer containing two antiparallel two stranded β-sheets and the remaining four helices. The large β-sheets from the monomers stack against each other to form the dimer interface (Fig. 3). The dimer has two active sites, one within each monomer. In this study, the monomer was extracted from the high-resolution crystal structure of TS (PDB ID: 5X5Q)35 downloaded from the Protein Databank (https://www.rcsb.org).36
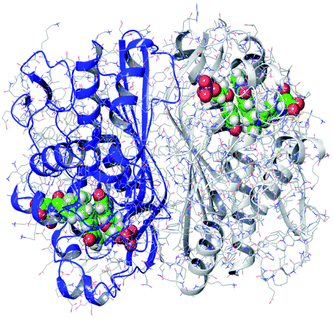 | ||
| Fig. 3 Thymidylate synthase dimer (from PDB ID 5X5Q). | ||
With the aim to confirm the ligand-based results, to investigate the structural interactions, and to predict the binding poses within the human thymidylate synthase binding site, all the synthesized molecules were processed by Induced Fit Docking (IFD) calculations.
The IFD results on chalcone- and pyrazoline-based 1,3,5-triazine derivatives confirmed the ligand-based evidence. In particular, the most active compounds (7d,g and 8g) showed excellent docking scores, comparable to reference compound raltitrexed (Table 4). The derivatives 9e and 10c showed the higher scores, although the in wet results and the ligand based outputs not classified them as TS modulators. It is conceivable that, although the molecular structures well fit TS active site, other chemical–physical properties do not contribute to overall affinity.
| Cmd | Docking XP score | IFD score |
|---|---|---|
| 5 | −9.485 | −589.383 |
| 6 | −12.496 | −591.632 |
| 7a | −9.645 | −594.712 |
| 7b | −13.681 | −597.86 |
| 7c | −13.301 | −595.368 |
| 7d | −12.161 | −598.947 |
| 7e | −12.382 | −596.397 |
| 7f | −10.889 | −594.444 |
| 7g | −12.672 | −597.106 |
| 8a | −12.765 | −592.985 |
| 8b | −13.461 | −598.765 |
| 8c | −11.157 | −597.44 |
| 8d | −11.701 | −596.742 |
| 8e | −12.406 | −594.546 |
| 8f | −12.018 | −593.787 |
| 8g | −15.116 | −598.53 |
| 9a | −12.401 | −597.581 |
| 9b | −11.993 | −597.761 |
| 9c | −11.898 | −597.398 |
| 9d | −9.545 | −593.173 |
| 9e | −14.838 | −601.736 |
| 9f | −13.206 | −599.526 |
| 9g | −12.101 | −598.997 |
| 10a | −13.657 | −600.72 |
| 10b | −9.9 | −600.609 |
| 10c | −15.252 | −599.967 |
| 10d | −11.00 | −589.16 |
| 10e | −11.842 | −598.052 |
| 10f | −13.633 | −598.584 |
| 10g | −11.598 | −591.941 |
| Raltitrexed | −13.828 | −584.621 |
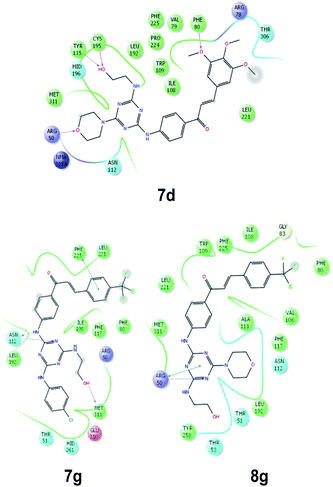
The analysis of the amino acid maps of the most active compounds (7d,g and 8g), confirmed as they interact with the pivotal amino acid residues (Arg50, Leu 108, Asn112, Leu192, Leu221, Phe225, and Met311), which have a central role in the catalytic enzyme process of the TS (Fig. 4).
As seen from 3D docking representation of the 8g derivative the core occupies the same binding site as does the reference ligand (Fig. 5, left) and makes stacking interaction with pyrimidine ring of UMP (Fig. 5, right). This stacking interaction is crucial and has been conserved in all the thymidylate synthases for with crystal structures that have been solved in complex with the cofactor and the inhibitor.
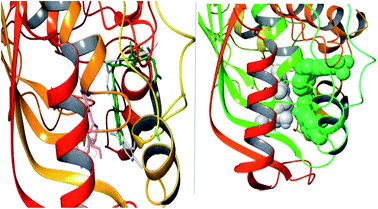 | ||
| Fig. 5 Left: superposition of 8g (white) and raltitrexed (green). Right: stacking interaction between 8g (green) and UMP (white). | ||
The overall results are in agreement with the hypothesis that the antiproliferative activities are related to the capability of these structures to modulate the TS, although, they could involve other biological targets as suggested from the ligand-based results (Table 3).
3. Conclusions
In this study, novel chalcone- and pyrazoline-based 1,3,5-triazines were synthesized using a versatile methodology, achieving high yields and purity. These triazine derivatives were evaluated for their anticancer effects on nine panels of 60 human cancer cell lines. Compounds 7d,g and 8g exhibited significant antiproliferative activity on all cancer panels being specially effective against the SR leukemia cell line, MCF7 breast cancer cell line and HCT-116 colon cancer cell line, respectively, and furthermore showed better GI50 values than 5-FU against several cell lines. Compound 7d exhibited the lowest mean GI50 against all panels of cancer cell lines and showed excellent docking score, comparable to reference compound raltitrexed. According to these results, chalcone 7d (R1 = morpholine, R2 = 3,4,5-(OCH3)3) could be considered as promising leads for further development of more potent anticancer agents.4. Experimental section
4.1. General information
Reagents and solvents used were obtained from commercial sources and used without further purification. Melting points were measured using a Stuart SMP10 melting point device (Cole-Parmer Ltd, Stone, Staffordshire, UK) and are uncorrected. FTIR spectra were obtained with a IRAffinity-1 spectrophotometer (Shimadzu, Columbia, MD, USA). The 1H- and 13C-NMR spectra were run on a DPX 400 spectrometer (Bruker, Billerica, MA, USA) operating at 400 and 100 MHz respectively, using DMSO-d6 as solvent and TMS as internal standard. The mass spectra were obtained on a Shimadzu-GCMS-QP2010 spectrometer (Shimadzu, Kyoto, Honshu, Japan) operating at 70 eV. The elemental analyses were obtained using an Agilent CHNS elemental analyzer (Thermo Fischer Scientific Inc., Madison, WI, USA) and the values are within ±0.4% of the theoretical values. Thin layer chromatography (TLC) were performed on 0.2 mm pre-coated aluminium plates of silica gel 60 F254 (Merck, Darmstadt, Hesse, Germany).4.2. Chemistry
4.2.2.1. 1-(4-((4-Chloro-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)ethan-1-one (3). White solid; 70% yield; mp 218–220 °C. FT-IR (ATR) ν (cm−1) 3316 (N–H), 3117 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1659 (C
C–H), 1659 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1578 and 1516 (C
O), 1578 and 1516 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.50 (s, 3H, CH3), 3.44–3.81 (m, 8H, CH2), 7.76 (d, J = 8.8 Hz, 2H, Ar–H), 7.90 (d, J = 8.8 Hz, 2H, Ar–H), 10.36 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.4 (CH3), 43.4 (CH2), 59.15 (CH2), 119.1 (CH), 129.3 (CH), 131.1 (Cq), 143.5 (Cq), 143.7 (Cq), 165.5 (Cq), 167.9 (Cq), 196.4 (Cq). MS (70 eV) m/z (%): 333 (11), 282 (23), 267 (100), 170 (26), 145 (44), 90 (32). Anal. calcd C15H16ClN5O2: C, 53.98; H, 4.83; N, 20.98; found: C, 54.01; H, 4.86; N, 20.70.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.50 (s, 3H, CH3), 3.44–3.81 (m, 8H, CH2), 7.76 (d, J = 8.8 Hz, 2H, Ar–H), 7.90 (d, J = 8.8 Hz, 2H, Ar–H), 10.36 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.4 (CH3), 43.4 (CH2), 59.15 (CH2), 119.1 (CH), 129.3 (CH), 131.1 (Cq), 143.5 (Cq), 143.7 (Cq), 165.5 (Cq), 167.9 (Cq), 196.4 (Cq). MS (70 eV) m/z (%): 333 (11), 282 (23), 267 (100), 170 (26), 145 (44), 90 (32). Anal. calcd C15H16ClN5O2: C, 53.98; H, 4.83; N, 20.98; found: C, 54.01; H, 4.86; N, 20.70.
4.2.3.1. 1-(4-((4-Chloro-6-((4-chlorophenyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)ethan-1-one (4). White solid; 80% yield; mp 258–260 °C. FT-IR (ATR) ν (cm−1) 3267 (N–H), 3101 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1666 (C
C–H), 1666 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1566 and 1512 (C
O), 1566 and 1512 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.54 (s, 3H, CH3), 7.42 (d, J = 8.8 Hz, 2H, Ar–H), 7.62–7.89 (m, 4H, Ar–H), 7.93 (d, J = 8.8 Hz, 2H, Ar–H), 10.48 (bs, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 119.8 (CH), 122.5 (CH), 127.4 (Cq), 128.5 (CH), 129.2 (CH), 131.6 (Cq), 137.3 (Cq), 143.0 (Cq), 163.8 (Cq), 168.3 (Cq), 168.4 (Cq), 196.5 (Cq). MS (70 eV) m/z (%): 373
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.54 (s, 3H, CH3), 7.42 (d, J = 8.8 Hz, 2H, Ar–H), 7.62–7.89 (m, 4H, Ar–H), 7.93 (d, J = 8.8 Hz, 2H, Ar–H), 10.48 (bs, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 119.8 (CH), 122.5 (CH), 127.4 (Cq), 128.5 (CH), 129.2 (CH), 131.6 (Cq), 137.3 (Cq), 143.0 (Cq), 163.8 (Cq), 168.3 (Cq), 168.4 (Cq), 196.5 (Cq). MS (70 eV) m/z (%): 373![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375
375![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 377 [M+]
377 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (90/59/11), 358 (75), 222 (10), 179 (16), 145 (100), 90 (27). Anal. calcd C17H13Cl2N5O: C, 54.56; H, 3.50; N, 18.71; found: C, 54.01; H, 3.55; N, 18.74.
[M + 4]+ (90/59/11), 358 (75), 222 (10), 179 (16), 145 (100), 90 (27). Anal. calcd C17H13Cl2N5O: C, 54.56; H, 3.50; N, 18.71; found: C, 54.01; H, 3.55; N, 18.74.
4.2.4.1. 1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)ethan-1-one (5). White solid. 76% yield; mp 191–193 °C. FT-IR (ATR): ν (cm−1) 3350 (N–H), 3279 (O–H), 3093 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1668 (C
C–H), 1668 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1583 and 1537 (C
O), 1583 and 1537 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.52 (s, 3H, CH3), 3.32–3.42 (m, 2H, CH2), 3.50–3.57 (m, 2H, CH2), 3.57–3.63 (m, 4H, CH2), 3.66–3.74 (m, 4H, CH2), 4.80 (s, 1H, OH), 6.88 (bs, 1H, NH), 7.83–7.90 (m, 4H, Ar–H), 9.39 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 43.0 (CH2), 43.6 (CH2), 60.2 (CH2), 66.2 (CH2), 118.6 (CH), 129.5 (CH), 130.1 (Cq), 145.4 (Cq), 165.0 (Cq), 165.9 (Cq), 166.1 (Cq), 196.9 (Cq). MS (70 eV) m/z (%): 358 [M+] (32), 328 (59), 313 (43), 69 (36), 55 (54), 43 (100). Anal. calcd C17H22N6O3: C, 56.97; H, 6.19; N, 23.45; found: C, 57.00; H, 6.25; N, 23.60.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.52 (s, 3H, CH3), 3.32–3.42 (m, 2H, CH2), 3.50–3.57 (m, 2H, CH2), 3.57–3.63 (m, 4H, CH2), 3.66–3.74 (m, 4H, CH2), 4.80 (s, 1H, OH), 6.88 (bs, 1H, NH), 7.83–7.90 (m, 4H, Ar–H), 9.39 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 43.0 (CH2), 43.6 (CH2), 60.2 (CH2), 66.2 (CH2), 118.6 (CH), 129.5 (CH), 130.1 (Cq), 145.4 (Cq), 165.0 (Cq), 165.9 (Cq), 166.1 (Cq), 196.9 (Cq). MS (70 eV) m/z (%): 358 [M+] (32), 328 (59), 313 (43), 69 (36), 55 (54), 43 (100). Anal. calcd C17H22N6O3: C, 56.97; H, 6.19; N, 23.45; found: C, 57.00; H, 6.25; N, 23.60.
4.2.5.1. 1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)ethan-1-one (6). White solid. 78% yield; mp 160–163 °C. FT-IR (ATR): ν (cm−1) 3398 (N–H), 3284 (O–H), 3113 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1666 (C
C–H), 1666 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 y 1566 (C
O), 1589 y 1566 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.54 (s, 3H, CH3), 3.45 (bs, 2H, CH2), 3.59 (t, J = 5.6 Hz, 2H, CH2), 4.12 (s, 1H, OH), 7.39 (d, J = 8.2 Hz, 2H, Ar–H), 7.75 (d, J = 8.2 Hz, 2H, Ar–H), 7.84–7.94 (m, 4H, Ar–H), 8.24 (bs, 1H, NH), 10.15–10.55 (m, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 43.3 (CH2), 59.4 (CH2), 119.9 (CH), 122.4 (CH), 122.9 (Cq), 128.5 (CH), 129.2 (CH), 131.4 (Cq), 131.5 (Cq), 143.2 (Cq), 158.7 (Cq), 161.0 (Cq), 161.2 (Cq), 196.5 (Cq). MS (70 eV) m/z (%): 398
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.54 (s, 3H, CH3), 3.45 (bs, 2H, CH2), 3.59 (t, J = 5.6 Hz, 2H, CH2), 4.12 (s, 1H, OH), 7.39 (d, J = 8.2 Hz, 2H, Ar–H), 7.75 (d, J = 8.2 Hz, 2H, Ar–H), 7.84–7.94 (m, 4H, Ar–H), 8.24 (bs, 1H, NH), 10.15–10.55 (m, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 26.5 (CH3), 43.3 (CH2), 59.4 (CH2), 119.9 (CH), 122.4 (CH), 122.9 (Cq), 128.5 (CH), 129.2 (CH), 131.4 (Cq), 131.5 (Cq), 143.2 (Cq), 158.7 (Cq), 161.0 (Cq), 161.2 (Cq), 196.5 (Cq). MS (70 eV) m/z (%): 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 [M+]
400 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (65/23), 367 (98), 353 (92), 296 (36), 176 (42), 145 (45), 57 (38), 43 (100). Anal. calcd C19H19ClN6O2: C, 57.22; H, 4.80; N, 21.07; found: C, 57.19; H, 4.83; N, 21.10.
[M + 2]+ (65/23), 367 (98), 353 (92), 296 (36), 176 (42), 145 (45), 57 (38), 43 (100). Anal. calcd C19H19ClN6O2: C, 57.22; H, 4.80; N, 21.07; found: C, 57.19; H, 4.83; N, 21.10.
4.2.6.1. (E)-1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-phenylprop-2-en-1-one (7a). Yellow solid. 75% yield; mp 175–177 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3315 (O–H), 3120 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1657 (C
C–H), 1657 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1576 y 1533 (C
O), 1576 y 1533 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.31–3.47 (m, 2H, CH2), 3.49–3.57 (m, 2H, CH2), 3.59–3.67 (m, 4H, CH2), 3.68–3.76 (m, 4H, CH2), 4.76 (s, 1H, OH), 6.99 (t, J = 5.0 Hz, 1H, NH), 7.42–7.48 (m, 3H, Ar–H), 7.70 (d, J = 15.6 Hz, 1H, CH), 7.87 (d, J = 4.8 Hz, 2H, Ar–H), 7.91–8.01 (m, 3H, Ar–H, CH), 8.11 (t, J = 10.0 Hz, 2H, Ar–H), 9.53 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 60.0 (CH2), 66.1 (CH2), 114.6 (Cq), 118.4 (CH), 122.1 (CH), 128.8 (CH), 128.9 (CH), 129.8 (Cq), 129.9 (CH), 130.4 (CH), 134.9 (Cq), 142.9 (CH), 164.1 (Cq), 164.8 (Cq), 165.6 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 446 [M+] (19), 416 (17), 401 (12), 131 (27), 103 (37), 69 (60), 55 (86). Anal. calcd C24H26N6O3: C, 64.56; H, 5.87; N, 18.82; found: C, 64.60; H, 5.90; N, 18.79.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.31–3.47 (m, 2H, CH2), 3.49–3.57 (m, 2H, CH2), 3.59–3.67 (m, 4H, CH2), 3.68–3.76 (m, 4H, CH2), 4.76 (s, 1H, OH), 6.99 (t, J = 5.0 Hz, 1H, NH), 7.42–7.48 (m, 3H, Ar–H), 7.70 (d, J = 15.6 Hz, 1H, CH), 7.87 (d, J = 4.8 Hz, 2H, Ar–H), 7.91–8.01 (m, 3H, Ar–H, CH), 8.11 (t, J = 10.0 Hz, 2H, Ar–H), 9.53 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 60.0 (CH2), 66.1 (CH2), 114.6 (Cq), 118.4 (CH), 122.1 (CH), 128.8 (CH), 128.9 (CH), 129.8 (Cq), 129.9 (CH), 130.4 (CH), 134.9 (Cq), 142.9 (CH), 164.1 (Cq), 164.8 (Cq), 165.6 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 446 [M+] (19), 416 (17), 401 (12), 131 (27), 103 (37), 69 (60), 55 (86). Anal. calcd C24H26N6O3: C, 64.56; H, 5.87; N, 18.82; found: C, 64.60; H, 5.90; N, 18.79.
4.2.6.2. (E)-1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-(p-tolyl)prop-2-en-1-one (7b). Yellow solid. 68% yield; mp 185–191 °C. FT-IR (ATR): ν (cm−1) 3419 (N–H), 3291 (O–H), 3110 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1651 (C
C–H), 1651 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1585 y 1537 (C
O), 1585 y 1537 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.35 (s, 3H, CH3), 3.26–3.47 (m, 2H, CH2), 3.54 (q, J = 6.1 Hz, 2H, CH2), 3.59–3.66 (m, 4H, CH2), 3.67–3.75 (m, 4H, CH2), 4.74 (s, 1H, OH), 6.98 (t, J = 5.6 Hz, 1H, NH), 7.26 (d, J = 8.0 Hz, 2H, Ar–H), 7.67 (d, J = 15.2 Hz, 1H, CH), 7.77 (d, J = 8.0 Hz, 2H, Ar–H), 7.85–7.98 (m, 3H, Ar–H, CH), 8.10 (t, J = 9.8 Hz, 2H, Ar–H), 9.51 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 21.1 (CH3), 42.9 (CH2), 43.4 (CH2), 59.9 (CH2), 66.1 (CH2), 118.4 (CH), 121.0 (CH), 128.8 (CH), 128.9 (Cq), 129.2 (Cq), 129.6 (CH), 129.8 (CH), 132.2 (Cq), 140.4 (Cq), 143.0 (CH), 164.6 (Cq), 164.8 (Cq), 165.9 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 460 [M+] (12), 430 (10), 145 (23), 117 (15), 69 (58), 55 (78), 43 (100). Anal. calcd C25H28N6O3: C, 65.20; H, 6.13; N, 18.25; found: C, 65.18; H, 6.18; N, 18.19.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.35 (s, 3H, CH3), 3.26–3.47 (m, 2H, CH2), 3.54 (q, J = 6.1 Hz, 2H, CH2), 3.59–3.66 (m, 4H, CH2), 3.67–3.75 (m, 4H, CH2), 4.74 (s, 1H, OH), 6.98 (t, J = 5.6 Hz, 1H, NH), 7.26 (d, J = 8.0 Hz, 2H, Ar–H), 7.67 (d, J = 15.2 Hz, 1H, CH), 7.77 (d, J = 8.0 Hz, 2H, Ar–H), 7.85–7.98 (m, 3H, Ar–H, CH), 8.10 (t, J = 9.8 Hz, 2H, Ar–H), 9.51 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 21.1 (CH3), 42.9 (CH2), 43.4 (CH2), 59.9 (CH2), 66.1 (CH2), 118.4 (CH), 121.0 (CH), 128.8 (CH), 128.9 (Cq), 129.2 (Cq), 129.6 (CH), 129.8 (CH), 132.2 (Cq), 140.4 (Cq), 143.0 (CH), 164.6 (Cq), 164.8 (Cq), 165.9 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 460 [M+] (12), 430 (10), 145 (23), 117 (15), 69 (58), 55 (78), 43 (100). Anal. calcd C25H28N6O3: C, 65.20; H, 6.13; N, 18.25; found: C, 65.18; H, 6.18; N, 18.19.
4.2.6.3. (E)-1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (7c). Yellow solid. 83% yield; mp 179–183 °C. FT-IR (ATR): ν (cm−1) 3479 (N–H), 3277 (O–H), 3201 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1649 (C
C–H), 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1571 y 1506 (C
O), 1571 y 1506 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.41 (m, 2H, CH2), 3.53 (q, J = 6.2 Hz, 2H, CH2), 3.60–3.67 (m, 4H, CH2), 3.67–3.75 (m, 4H, CH2), 3.81 (s, 3H, OCH3), 4.76 (s, 1H, OH), 6.94–7.04 (m, 3H, NH, Ar–H), 7.67 (d, J = 15.6 Hz, 1H, CH), 7.77–7.86 (m, 3H, Ar–H, CH), 7.94 (d, J = 7.2 Hz, 2H, Ar–H), 8.09 (t, J = 9.8 Hz, 2H, Ar–H), 9.50 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.3 (CH2), 55.4 (CH3), 59.9 (CH2), 66.0 (CH2), 114.4 (CH), 118.4 (CH), 119.6 (CH), 127.5 (Cq), 129.6 (CH), 130.6 (CH), 142.8 (CH), 161.2 (Cq), 164.1 (Cq), 164.2 (Cq), 164.8 (Cq), 165.9 (Cq), 165.9 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 476 [M+] (6), 446 (5), 161 (7), 98 (12), 83 (19), 69 (40), 55 (61), 43 (100). Anal. calcd C25H28N6O4: C, 63.01; H, 5.92; N, 17.64; found: C, 62.99; H, 5.93; N, 17.60.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.41 (m, 2H, CH2), 3.53 (q, J = 6.2 Hz, 2H, CH2), 3.60–3.67 (m, 4H, CH2), 3.67–3.75 (m, 4H, CH2), 3.81 (s, 3H, OCH3), 4.76 (s, 1H, OH), 6.94–7.04 (m, 3H, NH, Ar–H), 7.67 (d, J = 15.6 Hz, 1H, CH), 7.77–7.86 (m, 3H, Ar–H, CH), 7.94 (d, J = 7.2 Hz, 2H, Ar–H), 8.09 (t, J = 9.8 Hz, 2H, Ar–H), 9.50 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.3 (CH2), 55.4 (CH3), 59.9 (CH2), 66.0 (CH2), 114.4 (CH), 118.4 (CH), 119.6 (CH), 127.5 (Cq), 129.6 (CH), 130.6 (CH), 142.8 (CH), 161.2 (Cq), 164.1 (Cq), 164.2 (Cq), 164.8 (Cq), 165.9 (Cq), 165.9 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 476 [M+] (6), 446 (5), 161 (7), 98 (12), 83 (19), 69 (40), 55 (61), 43 (100). Anal. calcd C25H28N6O4: C, 63.01; H, 5.92; N, 17.64; found: C, 62.99; H, 5.93; N, 17.60.
4.2.6.4. (E)-1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one (7d). Yellow solid. 87% yield; mp 218–224 °C. FT-IR (ATR): ν (cm−1) 3354 (N–H), 3267 (O–H), 3100 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1649 (C
C–H), 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 y 1537 (C
O), 1587 y 1537 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.36–3.40 (m, 2H, CH2), 3.53 (q, J = 5.7 Hz, 2H CH2), 3.60–3.67 (m, 4H, CH2), 3.68–3.75 (m, 7H, CH2, OCH3), 3.87 (s, 6H, OCH3), 4.73 (s, 1H, OH), 6.98 (t, J = 6.0 Hz, 1H, NH), 7.2 (s, 2H, Ar–H), 7.66 (d, J = 15.2 Hz, 1H, CH), 7.82–8.02 (m, 3H, Ar–H, CH), 8.13 (t, J = 9.6 Hz, 2H, Ar–H), 9.52 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 56.2 (CH3), 59.8 (CH2), 60.2 (CH3), 66.1 (CH2), 106.4 (CH), 118.4 (CH), 121.2 (CH), 129.7 (CH), 129.8 (CH), 130.5 (Cq), 139.5 (Cq), 143.3 (Cq), 145.4 (Cq), 153.1 (Cq), 165.6 (Cq), 165.7 (Cq), 165.8 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 536 [M+] (15), 505 (10), 491 (8), 475 (6), 461 (11), 145 (18), 69 (58), 55 (71). Anal. calcd C27H32N6O6: C, 60.47; H, 5.98; N, 15.68; found: C, 60.44; H, 6.01; N, 15.66.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.36–3.40 (m, 2H, CH2), 3.53 (q, J = 5.7 Hz, 2H CH2), 3.60–3.67 (m, 4H, CH2), 3.68–3.75 (m, 7H, CH2, OCH3), 3.87 (s, 6H, OCH3), 4.73 (s, 1H, OH), 6.98 (t, J = 6.0 Hz, 1H, NH), 7.2 (s, 2H, Ar–H), 7.66 (d, J = 15.2 Hz, 1H, CH), 7.82–8.02 (m, 3H, Ar–H, CH), 8.13 (t, J = 9.6 Hz, 2H, Ar–H), 9.52 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 56.2 (CH3), 59.8 (CH2), 60.2 (CH3), 66.1 (CH2), 106.4 (CH), 118.4 (CH), 121.2 (CH), 129.7 (CH), 129.8 (CH), 130.5 (Cq), 139.5 (Cq), 143.3 (Cq), 145.4 (Cq), 153.1 (Cq), 165.6 (Cq), 165.7 (Cq), 165.8 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 536 [M+] (15), 505 (10), 491 (8), 475 (6), 461 (11), 145 (18), 69 (58), 55 (71). Anal. calcd C27H32N6O6: C, 60.47; H, 5.98; N, 15.68; found: C, 60.44; H, 6.01; N, 15.66.
4.2.6.5. (E)-3-(4-Chlorophenyl)-1-(4-((4-((2-hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)prop-2-en-1-one (7e). Yellow solid. 92% yield; mp 200–205 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3294 (O–H), 3100 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1657 (C
C–H), 1657 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1572 y 1541 (C
O), 1572 y 1541 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.40 (m, 2H, CH2), 3.53 (q, J = 6.3 Hz, 2H, CH2), 3.60–3.66 (m, 4H, CH2), 3.76–3.67 (m, 4H, CH2), 4.76 (s, 1H, OH), 7.00 (t, J = 5.9 Hz, 1H, NH), 7.52 (d, J = 8.4 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.89–7.99 (m, 5H, Ar–H, CH), 8.11 (t, J = 10.0 Hz, 2H, Ar–H), 9.54 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 60.0 (CH2), 66.0 (CH2), 118.4 (CH), 122.9 (Cq), 129.0 (CH), 129.8 (CH), 129.9 (CH), 130.5 (CH), 130.9 (Cq), 133.9 (Cq), 134.9 (Cq), 141.4 (CH), 164.1 (Cq), 164.6 (Cq), 165.9 (Cq), 187.1 (Cq). MS (70 eV) m/z (%): 480
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.40 (m, 2H, CH2), 3.53 (q, J = 6.3 Hz, 2H, CH2), 3.60–3.66 (m, 4H, CH2), 3.76–3.67 (m, 4H, CH2), 4.76 (s, 1H, OH), 7.00 (t, J = 5.9 Hz, 1H, NH), 7.52 (d, J = 8.4 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.89–7.99 (m, 5H, Ar–H, CH), 8.11 (t, J = 10.0 Hz, 2H, Ar–H), 9.54 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 60.0 (CH2), 66.0 (CH2), 118.4 (CH), 122.9 (Cq), 129.0 (CH), 129.8 (CH), 129.9 (CH), 130.5 (CH), 130.9 (Cq), 133.9 (Cq), 134.9 (Cq), 141.4 (CH), 164.1 (Cq), 164.6 (Cq), 165.9 (Cq), 187.1 (Cq). MS (70 eV) m/z (%): 480![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 482 [M+]
482 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (12/4), 449 (14), 437 (12), 165 (16), 145 (16), 137 (18), 102 (17), 69 (61), 55 (92). Anal. calcd C24H25ClN6O3: C, 59.94; H, 5.24; N, 17.47; found: C, 60.01; H, 5.19; N, 17.53.
[M + 2]+ (12/4), 449 (14), 437 (12), 165 (16), 145 (16), 137 (18), 102 (17), 69 (61), 55 (92). Anal. calcd C24H25ClN6O3: C, 59.94; H, 5.24; N, 17.47; found: C, 60.01; H, 5.19; N, 17.53.
4.2.6.6. (E)-3-(4-Fluorophenyl)-1-(4-((4-((2-hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)prop-2-en-1-one (7f). Yellow solid. 80% yield; mp 198–204 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3278 (O–H), 3100 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1649 (C
C–H), 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1579 y 1541 (C
O), 1579 y 1541 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.31–3.38 (m, 2H, CH2), 3.48–3.58 (m, 2H, CH2), 3.60–3.67 (m, 4H, CH2), 3.68–3.76 (m, 4H, CH2), 4.77 (s, 1H, OH), 7.00 (t, J = 4.8 Hz, 1H, NH), 7.30 (t, JHF = 8.8 Hz, 2H, Ar–H), 7.70 (d, J = 15.6 Hz, 1H, CH), 7.84–8.01 (m, 5H, Ar–H, CH), 8.11 (t, J = 10.2 Hz, 2H, Ar–H), 9.53 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 60.1 (CH2), 66.0 (CH2), 115.9 (d, 2JCF = 22.0 Hz, (CH)), 118.4 (CH), 122.1 (CH), 129.8 (CH), 131.1 (d, 3JCF = 9.0 Hz, (CH)), 131.6 (d, 4JCF = 3.1 Hz, Cq), 139.2 (Cq), 141.6 (CH), 162.1 (Cq), 164.6 (Cq), 165.7 (Cq), 167.0 (Cq), 173.8 (d, 1JC–F = 241.6 Hz, Cq), 187.2 (Cq). MS (70 eV) m/z (%): 464 (1), 256 (1), 185 (1), 97 (8), 73 (33), 69 (57), 55 (56). Anal. calcd. C24H25FN6O3: C, 62.06; H, 5.43; N, 18.09; found: C, 62.10; H, 5.38; N, 18.09.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.31–3.38 (m, 2H, CH2), 3.48–3.58 (m, 2H, CH2), 3.60–3.67 (m, 4H, CH2), 3.68–3.76 (m, 4H, CH2), 4.77 (s, 1H, OH), 7.00 (t, J = 4.8 Hz, 1H, NH), 7.30 (t, JHF = 8.8 Hz, 2H, Ar–H), 7.70 (d, J = 15.6 Hz, 1H, CH), 7.84–8.01 (m, 5H, Ar–H, CH), 8.11 (t, J = 10.2 Hz, 2H, Ar–H), 9.53 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.4 (CH2), 60.1 (CH2), 66.0 (CH2), 115.9 (d, 2JCF = 22.0 Hz, (CH)), 118.4 (CH), 122.1 (CH), 129.8 (CH), 131.1 (d, 3JCF = 9.0 Hz, (CH)), 131.6 (d, 4JCF = 3.1 Hz, Cq), 139.2 (Cq), 141.6 (CH), 162.1 (Cq), 164.6 (Cq), 165.7 (Cq), 167.0 (Cq), 173.8 (d, 1JC–F = 241.6 Hz, Cq), 187.2 (Cq). MS (70 eV) m/z (%): 464 (1), 256 (1), 185 (1), 97 (8), 73 (33), 69 (57), 55 (56). Anal. calcd. C24H25FN6O3: C, 62.06; H, 5.43; N, 18.09; found: C, 62.10; H, 5.38; N, 18.09.
4.2.6.7. (E)-1-(4-((4-((2-Hydroxyethyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)phenyl)-3-(4-(trifluoromethyl)phenyl)prop-2-en-1-one (7g). Yellow solid. 76% yield; mp 227–230 °C. FT-IR (ATR): ν (cm−1) 3415 (N–H), 3277 (O–H), 3211 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1651 (C
C–H), 1651 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1577 y 1535 (C
O), 1577 y 1535 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.42–3.20 (m, 2H, CH2), 3.48–3.56 (m, 2H, CH2), 3.59–3.66 (m, 4H, CH2), 3.69–3.76 (m, 4H, CH2), 4.80 (s, 1H, OH), 7.04 (bs, 1H, NH), 7.75 (d, J = 14.0 Hz, 1H, CH), 7.80 (d, J = 8.4 Hz, 2H, Ar–H), 8.02–7.92 (m, 2H, Ar–H), 8.19–8.07 (m, 5H, Ar–H, CH), 9.58 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 59.9 (CH2), 66.1 (CH2), 118.5 (CH), 124.1 (q, 1JCF = 245.9 Hz, CF3), 124.9 (CH), 125.8 (CH), 129.5 (q, 3JCF = 16.7 Hz, (CH)), 130.1 (CH), 139.0 (Cq), 139.7 (Cq), 141.0 (CH), 145.8 (Cq), 146.2 (Cq), 164.0 (Cq), 164.2 (Cq), 165.3 (q, 2JCF = 24.6 Hz, Cq), 187.3 (Cq). MS (70 eV) m/z (%): 514 (6), 484 (8), 469 (6), 199 (8), 145 (13), 69 (58), 55 (83). Anal. calcd C25H25F3N6O3: C, 58.36; H, 4.90; N, 16.33; found: C, 58.40; H, 5.00; N, 16.30.
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.42–3.20 (m, 2H, CH2), 3.48–3.56 (m, 2H, CH2), 3.59–3.66 (m, 4H, CH2), 3.69–3.76 (m, 4H, CH2), 4.80 (s, 1H, OH), 7.04 (bs, 1H, NH), 7.75 (d, J = 14.0 Hz, 1H, CH), 7.80 (d, J = 8.4 Hz, 2H, Ar–H), 8.02–7.92 (m, 2H, Ar–H), 8.19–8.07 (m, 5H, Ar–H, CH), 9.58 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 59.9 (CH2), 66.1 (CH2), 118.5 (CH), 124.1 (q, 1JCF = 245.9 Hz, CF3), 124.9 (CH), 125.8 (CH), 129.5 (q, 3JCF = 16.7 Hz, (CH)), 130.1 (CH), 139.0 (Cq), 139.7 (Cq), 141.0 (CH), 145.8 (Cq), 146.2 (Cq), 164.0 (Cq), 164.2 (Cq), 165.3 (q, 2JCF = 24.6 Hz, Cq), 187.3 (Cq). MS (70 eV) m/z (%): 514 (6), 484 (8), 469 (6), 199 (8), 145 (13), 69 (58), 55 (83). Anal. calcd C25H25F3N6O3: C, 58.36; H, 4.90; N, 16.33; found: C, 58.40; H, 5.00; N, 16.30.
4.2.7.1. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-phenylprop-2-en-1-one (8a). Yellow solid. 88% yield; mp 192–206 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3292 (O–H), 3119 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1637 (C
C–H), 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1572 y 1558 (C
O), 1572 y 1558 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.36–3.48 (m, 2H, CH2), 3.53–3.63 (m, 2H, CH2), 4.78 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.28–7.34 (m, 2H, Ar–H), 7.46 (d, J = 5.2 Hz, 2H, Ar–H), 7.71 (d, J = 15.6 Hz, 1H, CH), 7.81–7.89 (m, 4H, Ar–H, CH), 7.90–7.98 (m, 2H, Ar–H), 8.03 (d, J = 8.4 Hz, 2H, Ar–H), 8.11 (t, J = 8.4 Hz, 2H, Ar–H), 9.33 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.8 (CH2), 118.8 (CH), 121.4 (CH), 122.2 (CH), 125.3 (Cq), 125.4 (Cq), 128.2 (CH), 128.7 (CH), 128.9 (CH), 129.6 (CH), 130.4 (CH), 134.0 (Cq), 134.9 (Cq), 139.2 (Cq), 142.9 (CH), 163.8 (Cq), 164.0 (Cq), 165.7 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 486
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.36–3.48 (m, 2H, CH2), 3.53–3.63 (m, 2H, CH2), 4.78 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.28–7.34 (m, 2H, Ar–H), 7.46 (d, J = 5.2 Hz, 2H, Ar–H), 7.71 (d, J = 15.6 Hz, 1H, CH), 7.81–7.89 (m, 4H, Ar–H, CH), 7.90–7.98 (m, 2H, Ar–H), 8.03 (d, J = 8.4 Hz, 2H, Ar–H), 8.11 (t, J = 8.4 Hz, 2H, Ar–H), 9.33 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.8 (CH2), 118.8 (CH), 121.4 (CH), 122.2 (CH), 125.3 (Cq), 125.4 (Cq), 128.2 (CH), 128.7 (CH), 128.9 (CH), 129.6 (CH), 130.4 (CH), 134.0 (Cq), 134.9 (Cq), 139.2 (Cq), 142.9 (CH), 163.8 (Cq), 164.0 (Cq), 165.7 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 486![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 [M+]
488 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (50/24), 456 (49), 443 (43), 178 (48), 145 (44), 131 (65), 103 (100), 77 (39). Anal. calcd C26H23ClN6O2: C, 64.13; H, 4.76; N, 17.26; found: C, 64.17; H, 4.80; N, 17.23.
[M + 2]+ (50/24), 456 (49), 443 (43), 178 (48), 145 (44), 131 (65), 103 (100), 77 (39). Anal. calcd C26H23ClN6O2: C, 64.13; H, 4.76; N, 17.26; found: C, 64.17; H, 4.80; N, 17.23.
4.2.7.2. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(p-tolyl)prop-2-en-1-one (8b). Yellow solid. 85% yield; mp 219–224 °C. FT-IR (ATR): ν (cm−1) 3543 (N–H), 3294 (O–H), 3119 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1616 (C
C–H), 1616 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1583 y 1531 (C
O), 1583 y 1531 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.35 (s, 3H, CH3), 3.41–3.48 (m, 2H, CH2), 3.54–3.62 (m, 2H, CH2), 4.70 (bs, 1H, OH), 7.13 (bs, 1H, NH), 7.35–7.22 (m, 4H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.76 (d, J = 7.6 Hz, 2H, Ar–H), 7.81–7.91 (m, 3H, Ar–H, CH), 8.00 (d, J = 8.4 Hz, 2H, Ar–H), 8.09 (t, J = 7.2 Hz, 2H, Ar–H), not observed NH. 13C-NMR (100 MHz, DMSO-d6) δ ppm 21.1 (CH3), 43.1 (CH2), 59.9 (CH2), 118.8 (CH), 121.2 (CH), 121.4 (CH), 125.3 (Cq), 128.2 (CH), 128.8 (CH), 129.6 (CH), 129.6 (CH), 130.7 (Cq), 132.2 (Cq), 139.3 (Cq), 140.4 (Cq), 143.0 (CH), 145.4 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 500
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.35 (s, 3H, CH3), 3.41–3.48 (m, 2H, CH2), 3.54–3.62 (m, 2H, CH2), 4.70 (bs, 1H, OH), 7.13 (bs, 1H, NH), 7.35–7.22 (m, 4H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.76 (d, J = 7.6 Hz, 2H, Ar–H), 7.81–7.91 (m, 3H, Ar–H, CH), 8.00 (d, J = 8.4 Hz, 2H, Ar–H), 8.09 (t, J = 7.2 Hz, 2H, Ar–H), not observed NH. 13C-NMR (100 MHz, DMSO-d6) δ ppm 21.1 (CH3), 43.1 (CH2), 59.9 (CH2), 118.8 (CH), 121.2 (CH), 121.4 (CH), 125.3 (Cq), 128.2 (CH), 128.8 (CH), 129.6 (CH), 129.6 (CH), 130.7 (Cq), 132.2 (Cq), 139.3 (Cq), 140.4 (Cq), 143.0 (CH), 145.4 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502 [M+]
502 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (5/3), 367 (10), 353 (9), 149 (12), 119 (22), 91 (33), 81 (31). Anal. calcd C27H25ClN6O2: C, 64.73; H, 5.03; N, 16.78; found: C, 64.70; H, 4.99; N, 16.80.
[M + 2]+ (5/3), 367 (10), 353 (9), 149 (12), 119 (22), 91 (33), 81 (31). Anal. calcd C27H25ClN6O2: C, 64.73; H, 5.03; N, 16.78; found: C, 64.70; H, 4.99; N, 16.80.
4.2.7.3. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (8c). Yellow solid. 75% yield; mp 210–214 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3277 (O–H), 3117 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1637 (C
C–H), 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1576 y 1558 (C
O), 1576 y 1558 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.46 (m, 2H, CH2), 3.54–3.62 (m, 2H, CH2), 3.82 (s, 3H, OCH3), 4.78 (bs, 1H, OH), 7.01 (d, J = 8.4 Hz, 2H, Ar–H), 7.16 (bs, 1H, NH), 7.31 (t, J = 6.0 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.81–7.89 (m, 5H, Ar–H, CH), 8.01 (d, J = 8.4 Hz, 2H, Ar–H), 8.09 (t, J = 8.2 Hz, 2H, Ar–H), 9.32 (bs, 1H, NH), 9.55 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 55.4 (CH3), 59.9 (CH2), 114.4 (CH), 118.8 (CH), 119.7 (CH), 121.4 (CH), 125.3 (Cq), 127.5 (Cq), 128.2 (CH), 129.5 (CH), 130.6 (CH), 130.9 (Cq), 139.3 (Cq), 142.9 (CH), 145.0 (Cq), 161.2 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 516
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.46 (m, 2H, CH2), 3.54–3.62 (m, 2H, CH2), 3.82 (s, 3H, OCH3), 4.78 (bs, 1H, OH), 7.01 (d, J = 8.4 Hz, 2H, Ar–H), 7.16 (bs, 1H, NH), 7.31 (t, J = 6.0 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.81–7.89 (m, 5H, Ar–H, CH), 8.01 (d, J = 8.4 Hz, 2H, Ar–H), 8.09 (t, J = 8.2 Hz, 2H, Ar–H), 9.32 (bs, 1H, NH), 9.55 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 55.4 (CH3), 59.9 (CH2), 114.4 (CH), 118.8 (CH), 119.7 (CH), 121.4 (CH), 125.3 (Cq), 127.5 (Cq), 128.2 (CH), 129.5 (CH), 130.6 (CH), 130.9 (Cq), 139.3 (Cq), 142.9 (CH), 145.0 (Cq), 161.2 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 516![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 [M+]
518 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (100/50), 486 (57), 473 (50), 356 (52), 236 (39), 161 (20), 121 (17), 77 (20). Anal. calcd C27H25ClN6O3: C, 62.73; H, 4.87; N, 16.26; found: C, 62.70; H, 4.90; N, 16.29.
[M + 2]+ (100/50), 486 (57), 473 (50), 356 (52), 236 (39), 161 (20), 121 (17), 77 (20). Anal. calcd C27H25ClN6O3: C, 62.73; H, 4.87; N, 16.26; found: C, 62.70; H, 4.90; N, 16.29.
4.2.7.4. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one (8d). Yellow solid. 91% yield; mp 242–246 °C. FT-IR (ATR): ν (cm−1) 3564 (N–H), 3290 (O–H), 3098 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1649 (C
C–H), 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1560 y 1516 (C
O), 1560 y 1516 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.65–3.52 (m, 4H), 3.72 (s, 3H, OCH3), 3.87 (s, 6H, OCH3), 4.79 (bs, 1H, OH), 7.16 (bs, 1H, NH), 7.22 (s, 2H, Ar–H), 7.31 (t, J = 6.0 Hz, 2H, Ar–H), 7.66 (d, J = 15.6 Hz, 1H, CH), 7.93–7.76 (m, 3H, Ar–H), 8.03 (d, J = 7.6 Hz, 2H, Ar–H), 8.13 (t, J = 8.2 Hz, 2H, Ar–H), 9.32 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 56.2 (CH3), 59.9 (CH2), 60.2 (CH3), 106.4 (CH), 107.1 (CH), 118.8 (CH), 121.5 (CH), 125.3 (Cq), 128.2 (CH), 129.7 (CH), 130.5 (Cq), 130.8 (Cq), 139.2 (Cq), 139.6 (Cq), 143.4 (CH), 152.4 (Cq), 153.2 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 576
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.65–3.52 (m, 4H), 3.72 (s, 3H, OCH3), 3.87 (s, 6H, OCH3), 4.79 (bs, 1H, OH), 7.16 (bs, 1H, NH), 7.22 (s, 2H, Ar–H), 7.31 (t, J = 6.0 Hz, 2H, Ar–H), 7.66 (d, J = 15.6 Hz, 1H, CH), 7.93–7.76 (m, 3H, Ar–H), 8.03 (d, J = 7.6 Hz, 2H, Ar–H), 8.13 (t, J = 8.2 Hz, 2H, Ar–H), 9.32 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 56.2 (CH3), 59.9 (CH2), 60.2 (CH3), 106.4 (CH), 107.1 (CH), 118.8 (CH), 121.5 (CH), 125.3 (Cq), 128.2 (CH), 129.7 (CH), 130.5 (Cq), 130.8 (Cq), 139.2 (Cq), 139.6 (Cq), 143.4 (CH), 152.4 (Cq), 153.2 (Cq), 163.9 (Cq), 164.1 (Cq), 165.8 (Cq), 187.4 (Cq). MS (70 eV) m/z (%): 576![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 578 [M+]
578 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (94/46), 561 (25), 545 (27), 501 (21), 356 (21), 195 (44), 98 (40), 84 (96). Anal. calcd C29H29ClN6O5: C, 60.36; H, 5.07; N, 14.56; found: C, 60.40; H, 5.03; N, 14.49.
[M + 2]+ (94/46), 561 (25), 545 (27), 501 (21), 356 (21), 195 (44), 98 (40), 84 (96). Anal. calcd C29H29ClN6O5: C, 60.36; H, 5.07; N, 14.56; found: C, 60.40; H, 5.03; N, 14.49.
4.2.7.5. (E)-3-(4-Chlorophenyl)-1-(4-((4-((4-chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)prop-2-en-1-one (8e). Yellow solid. 93% yield; mp 200–204 °C. FT-IR (ATR): ν (cm−1) 3400 (N–H), 3279 (O–H), 3117 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1641 (C
C–H), 1641 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1577 y 1529 (C
O), 1577 y 1529 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.47 (m, 2H, CH2), 3.54–3.61 (m, 2H, CH2), 4.77 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.31 (t, J = 7.5 Hz, 2H, Ar–H), 7.51 (d, J = 8.4 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.82–7.89 (m, 2H, Ar–H), 7.92 (d, J = 8.4 Hz, 2H, Ar–H), 7.94–8.05 (m, 3H, Ar–H, CH), 8.11 (t, J = 8.8 Hz, 2H, Ar–H), 9.41 (bs, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.9 (CH2), 118.7 (CH), 121.4 (CH), 123.0 (CH), 125.3 (Cq), 128.1 (CH), 128.9 (CH), 129.7 (CH), 130.5 (CH), 133.9 (Cq), 134.8 (Cq), 139.2 (Cq), 141.4 (CH), 143.6 (Cq), 145.3 (Cq), 163.8 (Cq), 164.0 (Cq), 165.8 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 520
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.47 (m, 2H, CH2), 3.54–3.61 (m, 2H, CH2), 4.77 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.31 (t, J = 7.5 Hz, 2H, Ar–H), 7.51 (d, J = 8.4 Hz, 2H, Ar–H), 7.68 (d, J = 15.6 Hz, 1H, CH), 7.82–7.89 (m, 2H, Ar–H), 7.92 (d, J = 8.4 Hz, 2H, Ar–H), 7.94–8.05 (m, 3H, Ar–H, CH), 8.11 (t, J = 8.8 Hz, 2H, Ar–H), 9.41 (bs, 2H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.9 (CH2), 118.7 (CH), 121.4 (CH), 123.0 (CH), 125.3 (Cq), 128.1 (CH), 128.9 (CH), 129.7 (CH), 130.5 (CH), 133.9 (Cq), 134.8 (Cq), 139.2 (Cq), 141.4 (CH), 143.6 (Cq), 145.3 (Cq), 163.8 (Cq), 164.0 (Cq), 165.8 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 520![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 522
522![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 [M+]
524 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (20/14/4), 491 (19), 477 (23), 475 (15), 367 (11), 339 (12), 178 (20), 145 (25), 135 (32). Anal. calcd C26H22Cl2N6O2: C, 59.89; H, 4.25; N, 16.12; found: C, 60.01; H, 4.25; N, 16.16.
[M + 4]+ (20/14/4), 491 (19), 477 (23), 475 (15), 367 (11), 339 (12), 178 (20), 145 (25), 135 (32). Anal. calcd C26H22Cl2N6O2: C, 59.89; H, 4.25; N, 16.12; found: C, 60.01; H, 4.25; N, 16.16.
4.2.7.6. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(4-fluorophenyl)prop-2-en-1-one (8f). Yellow solid. 85% yield; mp 196–200 °C. FT-IR (ATR): ν (cm−1) 3412 (N–H), 3279 (O–H), 3117 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1641 (C
C–H), 1641 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1576 y 1529 (C
O), 1576 y 1529 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.52 (m, 2H, CH2), 3.52–3.64 (m, 2H, CH2), 4.74 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.26–7.36 (m, 4H, Ar–H), 7.71 (d, J = 15.6 Hz, 1H, CH), 7.82–7.88 (m, 2H, Ar–H), 7.88–7.99 (m, 3H, Ar–H, CH), 8.03 (d, J = 8.4 Hz, 2H, Ar–H), 8.12 (t, J = 8.8 Hz, 2H, Ar–H), 9.33 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.9 (CH2), 115.9 (d, 2JCF = 21.8 Hz, (CH)), 118.7 (CH), 121.4 (CH), 122.1 (CH), 125.3 (Cq), 128.2 (CH), 129.7 (CH), 130.6 (Cq), 131.1 (d, 3JCF = 8.6 Hz, (CH)), 131.6 (d, 4JCF = 2.9 Hz, Cq), 139.2 (Cq), 141.7 (CH), 145.2 (Cq), 163.1 (d, 1JCF = 201.6 Hz, Cq), 163.8 (Cq), 164.5 (Cq), 165.7 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 504
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.52 (m, 2H, CH2), 3.52–3.64 (m, 2H, CH2), 4.74 (s, 1H, OH), 7.17 (bs, 1H, NH), 7.26–7.36 (m, 4H, Ar–H), 7.71 (d, J = 15.6 Hz, 1H, CH), 7.82–7.88 (m, 2H, Ar–H), 7.88–7.99 (m, 3H, Ar–H, CH), 8.03 (d, J = 8.4 Hz, 2H, Ar–H), 8.12 (t, J = 8.8 Hz, 2H, Ar–H), 9.33 (bs, 1H, NH), 9.57 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.9 (CH2), 115.9 (d, 2JCF = 21.8 Hz, (CH)), 118.7 (CH), 121.4 (CH), 122.1 (CH), 125.3 (Cq), 128.2 (CH), 129.7 (CH), 130.6 (Cq), 131.1 (d, 3JCF = 8.6 Hz, (CH)), 131.6 (d, 4JCF = 2.9 Hz, Cq), 139.2 (Cq), 141.7 (CH), 145.2 (Cq), 163.1 (d, 1JCF = 201.6 Hz, Cq), 163.8 (Cq), 164.5 (Cq), 165.7 (Cq), 187.3 (Cq). MS (70 eV) m/z (%): 504![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 506 [M+]
506 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (74/32), 474 (65), 461 (60), 313 (71), 264 (81), 236 (100), 98 (78), 84 (67). Anal. calcd C26H22ClFN6O2: C, 61.84; H, 4.39; N, 16.64; found: C, 61.86; H, 4.42; N, 16.70.
[M + 2]+ (74/32), 474 (65), 461 (60), 313 (71), 264 (81), 236 (100), 98 (78), 84 (67). Anal. calcd C26H22ClFN6O2: C, 61.84; H, 4.39; N, 16.64; found: C, 61.86; H, 4.42; N, 16.70.
4.2.7.7. (E)-1-(4-((4-((4-Chlorophenyl)amino)-6-((2-hydroxyethyl)amino)-1,3,5-triazin-2-yl)amino)phenyl)-3-(4-(trifluoromethyl)phenyl)prop-2-en-1-one (8g). Yellow solid. 83% yield; mp 235–240 °C. FT-IR (ATR): ν (cm−1) 3441 (N–H), 3281 (O–H), 3119 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1643 (C
C–H), 1643 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1574 y 1528 (C
O), 1574 y 1528 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.49 (m, 2H, CH2), 3.53–3.63 (m, 2H, CH2), 4.74 (s, 1H, OH), 7.18 (bs, 1H, NH), 7.25–7.37 (m, 2H, Ar–H), 7.76 (d, J = 16.0 Hz, 1H, CH), 7.81 (d, J = 8.0 Hz, 2H, Ar–H), 7.83–7.89 (m, 2H, Ar–H), 7.98–8.19 (m, 7H, Ar–H), 9.33 (bs, 1H, NH), 9.59 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.8 (CH2), 118.7 (CH), 121.3 (CH), 124.1 (q, 1JCF = 272.2 Hz, CF3), 124.9 (CH), 125.3 (Cq), 125.4 (Cq), 125.7 (q, 4JCF = 3.0 Hz, (CH)), 128.2 (CH), 129.3 (CH), 129.8 (q, 3JCF = 18.6 Hz, (CH)), 130.3 (Cq), 138.9 (Cq), 139.2 (Cq), 139.2 (Cq), 140.9 (CH), 145.4 (Cq), 163.9 (q, 2JCF = 23.2 Hz, Cq), 165.7 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 554
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.38–3.49 (m, 2H, CH2), 3.53–3.63 (m, 2H, CH2), 4.74 (s, 1H, OH), 7.18 (bs, 1H, NH), 7.25–7.37 (m, 2H, Ar–H), 7.76 (d, J = 16.0 Hz, 1H, CH), 7.81 (d, J = 8.0 Hz, 2H, Ar–H), 7.83–7.89 (m, 2H, Ar–H), 7.98–8.19 (m, 7H, Ar–H), 9.33 (bs, 1H, NH), 9.59 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 59.8 (CH2), 118.7 (CH), 121.3 (CH), 124.1 (q, 1JCF = 272.2 Hz, CF3), 124.9 (CH), 125.3 (Cq), 125.4 (Cq), 125.7 (q, 4JCF = 3.0 Hz, (CH)), 128.2 (CH), 129.3 (CH), 129.8 (q, 3JCF = 18.6 Hz, (CH)), 130.3 (Cq), 138.9 (Cq), 139.2 (Cq), 139.2 (Cq), 140.9 (CH), 145.4 (Cq), 163.9 (q, 2JCF = 23.2 Hz, Cq), 165.7 (Cq), 187.2 (Cq). MS (70 eV) m/z (%): 554![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556 [M+]
556 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+ (100/37), 535 (12), 524 (93), 511 (93), 452 (15), 199 (9), 127 (12), 98 (9). Anal. calcd C27H22ClF3N6O2: C, 58.44; H, 4.00; N, 15.14; found: C, 58.39; H, 4.09; N, 15.10.
[M + 2]+ (100/37), 535 (12), 524 (93), 511 (93), 452 (15), 199 (9), 127 (12), 98 (9). Anal. calcd C27H22ClF3N6O2: C, 58.44; H, 4.00; N, 15.14; found: C, 58.39; H, 4.09; N, 15.10.
4.2.8.1. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9a). Yellow solid. 78% yield; mp 279–281 °C. FT-IR (ATR): ν (cm−1) 3416 (N–H), 3202 (O–H), 3090 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1626 y 1599 (C
C–H), 1626 y 1599 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.16 (dd, J = 17.4, 4.6 Hz, 1H, H-4), 3.38–3.49 (m, 2H, CH2), 3.52–3.60 (m, 2H, CH2), 3.64–3.72 (m, 5H, CH2, OH), 3.74–3.84 (m, 4H, CH2), 3.94 (dd, J = 17.4, 11.8 Hz, 1H, H-4), 5.59 (dd, J = 11.8, 4.2 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.23–7.31 (m, 3H, Ar–H), 7.32–7.40 (m, 2H, Ar–H), 7.63–7.82 (m, 4H, Ar–H), 8.30 (bs, 1H, NH), 10.59 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 44.1 (CH2), 44.3 (CH2), 59.3 (CH2), 62.4 (CH), 65.7 (CH2), 110.8 (CH), 116.8 (Cq), 116.9 (CH), 120.3 (CH), 121.0 (Cq), 125.7 (CH), 126.8 (Cq), 126.9 (CH), 127.8 (CH), 129.2 (CH), 134.4 (Cq), 141.4 (Cq), 145.1 (Cq), 145.7 (Cq), 148.0 (Cq), 149.7 (Cq). MS (70 eV) m/z (%): 604
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.16 (dd, J = 17.4, 4.6 Hz, 1H, H-4), 3.38–3.49 (m, 2H, CH2), 3.52–3.60 (m, 2H, CH2), 3.64–3.72 (m, 5H, CH2, OH), 3.74–3.84 (m, 4H, CH2), 3.94 (dd, J = 17.4, 11.8 Hz, 1H, H-4), 5.59 (dd, J = 11.8, 4.2 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.23–7.31 (m, 3H, Ar–H), 7.32–7.40 (m, 2H, Ar–H), 7.63–7.82 (m, 4H, Ar–H), 8.30 (bs, 1H, NH), 10.59 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 44.1 (CH2), 44.3 (CH2), 59.3 (CH2), 62.4 (CH), 65.7 (CH2), 110.8 (CH), 116.8 (Cq), 116.9 (CH), 120.3 (CH), 121.0 (Cq), 125.7 (CH), 126.8 (Cq), 126.9 (CH), 127.8 (CH), 129.2 (CH), 134.4 (Cq), 141.4 (Cq), 145.1 (Cq), 145.7 (Cq), 148.0 (Cq), 149.7 (Cq). MS (70 eV) m/z (%): 604![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 606
606![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 608 [M+]
608 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/70/15), 588 (4), 574 (9), 527 (8), 310 (10), 287 (8), 252 (8). Anal. calcd C30H30Cl2N8O2: C, 59.51; H, 4.99; N, 18.51; found: C, 60.00; H, 4.89; N, 18.53.
[M + 4]+ (100/70/15), 588 (4), 574 (9), 527 (8), 310 (10), 287 (8), 252 (8). Anal. calcd C30H30Cl2N8O2: C, 59.51; H, 4.99; N, 18.51; found: C, 60.00; H, 4.89; N, 18.53.
4.2.8.2. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-(p-tolyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9b). Yellow solid. 83% yield; mp 262–265 °C. FT-IR (ATR): ν (cm−1) 3454 (N–H), 3248 (O–H), 3088 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1626 y 1599 (C
C–H), 1626 y 1599 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.24 (s, 3H, CH3), 3.10 (dd, J = 17.2, 3.8 Hz, 1H, H-4), 3.38–3.49 (m, 2H, CH2), 3.50–3.72 (m, 7H, CH2, OH), 3.82–3.74 (m, 4H, CH2), 3.89 (dd, J = 17.2, 11.6 Hz, 1H, H-4), 5.51 (dd, J = 11.6, 3.8 Hz, 1H, H-5), 6.77 (s, 1H, Ar–H), 6.91 (s, 2H, Ar–H), 7.18–7.09 (m, 4H, Ar–H), 7.62–7.80 (m, 4H, Ar–H), 8.36 (bs, 1H, NH), 10.65 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 20.7 (CH3), 43.1 (CH2), 43.3 (CH2), 44.4 (CH2), 59.1 (CH2), 62.3 (CH), 65.8 (CH2), 110.9 (CH), 116.9 (CH), 116.9 (Cq), 120.5 (CH), 121.2 (Cq), 125.8 (CH), 127.0 (CH), 127.3 (Cq), 129.8 (CH), 134.5 (Cq), 137.1 (Cq), 138.5 (Cq), 139.6 (Cq), 145.8 (Cq), 148.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 618
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.24 (s, 3H, CH3), 3.10 (dd, J = 17.2, 3.8 Hz, 1H, H-4), 3.38–3.49 (m, 2H, CH2), 3.50–3.72 (m, 7H, CH2, OH), 3.82–3.74 (m, 4H, CH2), 3.89 (dd, J = 17.2, 11.6 Hz, 1H, H-4), 5.51 (dd, J = 11.6, 3.8 Hz, 1H, H-5), 6.77 (s, 1H, Ar–H), 6.91 (s, 2H, Ar–H), 7.18–7.09 (m, 4H, Ar–H), 7.62–7.80 (m, 4H, Ar–H), 8.36 (bs, 1H, NH), 10.65 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 20.7 (CH3), 43.1 (CH2), 43.3 (CH2), 44.4 (CH2), 59.1 (CH2), 62.3 (CH), 65.8 (CH2), 110.9 (CH), 116.9 (CH), 116.9 (Cq), 120.5 (CH), 121.2 (Cq), 125.8 (CH), 127.0 (CH), 127.3 (Cq), 129.8 (CH), 134.5 (Cq), 137.1 (Cq), 138.5 (Cq), 139.6 (Cq), 145.8 (Cq), 148.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 618![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620
620![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 622 [M+]
622 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/68/14), 587 (8), 527 (14), 310 (12), 294 (7), 252 (6), 117 (9), 91 (9). Anal. calcd C31H32Cl2N8O2: C, 60.10; H, 5.21; N, 18.09; found: C, 60.15; H, 5.18; N, 18.10.
[M + 4]+ (100/68/14), 587 (8), 527 (14), 310 (12), 294 (7), 252 (6), 117 (9), 91 (9). Anal. calcd C31H32Cl2N8O2: C, 60.10; H, 5.21; N, 18.09; found: C, 60.15; H, 5.18; N, 18.10.
4.2.8.3. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9c). Yellow solid. 75% yield; mp 275–278 °C. FT-IR (ATR): ν (cm−1) 3327 (N–H), 3252 (O–H), 3094 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1661 y 1582 (C
C–H), 1661 y 1582 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.12 (dd, J = 17.2, 3.7 Hz, 1H, H-4), 3.44 (d, J = 18.5 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.61–3.74 (m, 7H, CH2, OCH3), 3.75–3.82 (m, 4H, CH2), 3.89 (dd, J = 17.2, 11.9 Hz, 1H, H-4), 4.50 (bs, 1H, OH), 5.52 (dd, J = 11.9, 3.7 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.82–6.98 (m, 4H, Ar–H), 7.18 (d, J = 8.2 Hz, 2H, Ar–H), 7.65–7.85 (m, 4H Ar–H), 8.46 (bs, 1H, NH), 10.76 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.2 (CH2), 44.4 (CH2), 55.0 (OCH3), 59.0 (CH2), 61.9 (CH), 65.7 (CH2), 110.8 (CH), 114.5 (CH), 116.8 (CH), 120.3 (CH), 120.4 (Cq), 121.0 (Cq), 126.7 (Cq), 126.9 (CH), 127.0 (CH), 127.2 (Cq), 133.2 (Cq), 134.3 (Cq), 145.7 (Cq), 149.6 (Cq), 149.7 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 634
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.12 (dd, J = 17.2, 3.7 Hz, 1H, H-4), 3.44 (d, J = 18.5 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.61–3.74 (m, 7H, CH2, OCH3), 3.75–3.82 (m, 4H, CH2), 3.89 (dd, J = 17.2, 11.9 Hz, 1H, H-4), 4.50 (bs, 1H, OH), 5.52 (dd, J = 11.9, 3.7 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.82–6.98 (m, 4H, Ar–H), 7.18 (d, J = 8.2 Hz, 2H, Ar–H), 7.65–7.85 (m, 4H Ar–H), 8.46 (bs, 1H, NH), 10.76 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.2 (CH2), 44.4 (CH2), 55.0 (OCH3), 59.0 (CH2), 61.9 (CH), 65.7 (CH2), 110.8 (CH), 114.5 (CH), 116.8 (CH), 120.3 (CH), 120.4 (Cq), 121.0 (Cq), 126.7 (Cq), 126.9 (CH), 127.0 (CH), 127.2 (Cq), 133.2 (Cq), 134.3 (Cq), 145.7 (Cq), 149.6 (Cq), 149.7 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 634![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636
636![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 638 [M+]
638 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/70/15), 604 (5), 527 (39), 310 (12), 280 (7), 252 (6), 134 (14), 121 (14). Anal. calcd C31H32Cl2N8O3: C, 58.59; H, 5.08; N, 17.63; found: C, 58.62; H, 5.12; N, 17.59.
[M + 4]+ (100/70/15), 604 (5), 527 (39), 310 (12), 280 (7), 252 (6), 134 (14), 121 (14). Anal. calcd C31H32Cl2N8O3: C, 58.59; H, 5.08; N, 17.63; found: C, 58.62; H, 5.12; N, 17.59.
4.2.8.4. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-(3,4,5-trimethoxyphenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9d). Yellow solid. 80% yield; mp 239–242 °C. FT-IR (ATR): ν (cm−1) 3410 (N–H), 3242 (O–H), 3098 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1659 y 1583 (C
C–H), 1659 y 1583 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.20 (dd, J = 17.7, 6.2 Hz, 1H, H-4), 3.36–3.43 (m, 2H, CH2), 3.49–3.54 (m, 2H, CH2), 3.59–3.66 (m, 7H, OCH3, CH2), 3.67–3.73 (m, 10H, OCH3, CH2), 3.90 (dd, J = 17.7, 11.7 Hz, 1H, H-4), 4.67 (s, 1H, OH), 5.39 (dd, J = 11.7, 6.2 Hz, 1H, H-5), 6.60 (s, 2H, Ar–H), 6.76–6.89 (m, 2H, Ar–H, NH), 6.96 (s, 2H, Ar–H), 7.68 (t, J = 7.2 Hz, 2H, Ar–H), 7.83 (d, J = 8.7 Hz, 2H, Ar–H), 9.24 (bs 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.0 (CH2), 43.3 (CH2), 56.0 (OCH3), 59.9 (CH2), 60.0 (OCH3), 63.0 (CH), 66.0 (CH2), 103.0 (CH), 109.7 (Cq), 110.9 (CH), 116.1 (CH), 119.1 (CH), 122.5 (Cq), 126.8 (CH), 131.2 (Cq), 134.4 (Cq), 136.8 (Cq), 137.4 (Cq), 149.0 (Cq), 150.5 (Cq), 153.2 (Cq), 153.4 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 694
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.20 (dd, J = 17.7, 6.2 Hz, 1H, H-4), 3.36–3.43 (m, 2H, CH2), 3.49–3.54 (m, 2H, CH2), 3.59–3.66 (m, 7H, OCH3, CH2), 3.67–3.73 (m, 10H, OCH3, CH2), 3.90 (dd, J = 17.7, 11.7 Hz, 1H, H-4), 4.67 (s, 1H, OH), 5.39 (dd, J = 11.7, 6.2 Hz, 1H, H-5), 6.60 (s, 2H, Ar–H), 6.76–6.89 (m, 2H, Ar–H, NH), 6.96 (s, 2H, Ar–H), 7.68 (t, J = 7.2 Hz, 2H, Ar–H), 7.83 (d, J = 8.7 Hz, 2H, Ar–H), 9.24 (bs 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.0 (CH2), 43.3 (CH2), 56.0 (OCH3), 59.9 (CH2), 60.0 (OCH3), 63.0 (CH), 66.0 (CH2), 103.0 (CH), 109.7 (Cq), 110.9 (CH), 116.1 (CH), 119.1 (CH), 122.5 (Cq), 126.8 (CH), 131.2 (Cq), 134.4 (Cq), 136.8 (Cq), 137.4 (Cq), 149.0 (Cq), 150.5 (Cq), 153.2 (Cq), 153.4 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 694![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 696
696![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 698 [M+]
698 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/72/15), 677 (6), 527 (49), 310 (15), 280 (7), 252 (14), 179 (15), 83 (27). Anal. calcd C33H36Cl2N8O5: C, 56.98; H, 5.22; N, 16.11; found: C, 57.01; H, 5.20; N, 16.15.
[M + 4]+ (100/72/15), 677 (6), 527 (49), 310 (15), 280 (7), 252 (14), 179 (15), 83 (27). Anal. calcd C33H36Cl2N8O5: C, 56.98; H, 5.22; N, 16.11; found: C, 57.01; H, 5.20; N, 16.15.
4.2.8.5. 2-((4-((4-(5-(4-Chlorophenyl)-1-(3,5-dichlorophenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9e). Yellow solid. 91% yield; mp 230–234 °C. FT-IR (ATR): ν (cm−1) 3520 (N–H), 3273 (O–H), 3105 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1653 y 1582 (C
C–H), 1653 y 1582 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.15 (dd, J = 17.7, 4.4 Hz, 1H, H-4), 3.44 (d, J = 18.5 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.62–3.74 (m, 4H, CH2), 3.74–3.83 (m, 5H, CH2, OH), 3.92 (dd, J = 17.7, 11.7 Hz, 1H, H-4), 5.63 (dd, J = 11.7, 4.4 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.92 (s, 2H, Ar–H), 7.27 (d, J = 8.4 Hz, 2H, Ar–H), 7.42 (d, J = 8.4 Hz, 2H, Ar–H), 7.63–7.81 (m, 4H Ar–H), 8.48 (bs, 1H, NH), 10.77 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.3 (CH2), 44.4 (CH2), 59.0 (CH2), 61.6 (CH), 65.7 (CH2), 110.9 (CH), 117.1 (CH), 117.1 (Cq), 120.4 (Cq), 120.4 (CH), 121.0 (Cq), 124.0 (Cq), 127.0 (CH), 127.8 (CH), 129.2 (CH), 132.3 (Cq), 134.5 (Cq), 140.3 (Cq), 145.5 (Cq), 149.5 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 638
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.15 (dd, J = 17.7, 4.4 Hz, 1H, H-4), 3.44 (d, J = 18.5 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.62–3.74 (m, 4H, CH2), 3.74–3.83 (m, 5H, CH2, OH), 3.92 (dd, J = 17.7, 11.7 Hz, 1H, H-4), 5.63 (dd, J = 11.7, 4.4 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.92 (s, 2H, Ar–H), 7.27 (d, J = 8.4 Hz, 2H, Ar–H), 7.42 (d, J = 8.4 Hz, 2H, Ar–H), 7.63–7.81 (m, 4H Ar–H), 8.48 (bs, 1H, NH), 10.77 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.3 (CH2), 44.4 (CH2), 59.0 (CH2), 61.6 (CH), 65.7 (CH2), 110.9 (CH), 117.1 (CH), 117.1 (Cq), 120.4 (Cq), 120.4 (CH), 121.0 (Cq), 124.0 (Cq), 127.0 (CH), 127.8 (CH), 129.2 (CH), 132.3 (Cq), 134.5 (Cq), 140.3 (Cq), 145.5 (Cq), 149.5 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 638![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640
640![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 642
642![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 [M+]
644 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (49/41/14/3), 607 (6), 527 (4), 310 (10), 239 (7), 185 (7), 123 (26), 97 (40). Anal. calcd C30H29Cl3N8O2: C, 56.30; H, 4.57; N, 17.51; found: C, 56.20; H, 4.60; N, 17.48.
[M + 6]+ (49/41/14/3), 607 (6), 527 (4), 310 (10), 239 (7), 185 (7), 123 (26), 97 (40). Anal. calcd C30H29Cl3N8O2: C, 56.30; H, 4.57; N, 17.51; found: C, 56.20; H, 4.60; N, 17.48.
4.2.8.6. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9f). Yellow solid. 86% yield; mp 249–252 °C. FT-IR (ATR): ν (cm−1) 3535 (N–H), 3244 (O–H), 3113 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1653 y 1587 (C
C–H), 1653 y 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.16 (dd, J = 17.4, 3.5 Hz, 1H, H-4), 3.44 (d, J = 18.4 Hz, 2H, CH2), 3.51–3.61 (m, 2H, CH2), 3.63–3.72 (m, 4H, CH2), 3.75–3.84 (m, 4H, CH2), 4.06 (bs, 1H, OH), 3.92 (dd, J = 17.4, 12.1 Hz, 1H, H-4), 5.62 (dd, J = 12.1, 3.5 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.18 (t, J = 8.2 Hz, 2H, Ar–H), 7.25–7.35 (m, 2H, Ar–H), 7.70 (t, J = 11.2 Hz, 2H, Ar–H), 7.78 (d, J = 8.4 Hz, 2H, Ar–H), 8.50 (bs, 1H, NH), 10.80 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.1 (CH2), 43.3 (CH2), 44.4 (CH2), 59.0 (CH2), 61.6 (CH), 65.7 (CH2), 110.9 (CH), 116.0 (d, 2JCF = 21.5 Hz, (CH)), 117.0 (CH), 117.1 (Cq), 120.4 (CH), 120.9 (Cq), 127.0 (CH), 127.9 (d, 3JCF = 8.3 Hz, (CH)), 134.4 (Cq), 137.5 (d, 4JCF = 3.2 Hz, Cq), 138.4 (Cq), 145.6 (Cq), 149.3 (Cq), 149.7 (Cq), 149.8 (Cq), 161.5 (d, 1JC–F = 244.0 Hz, Cq). MS (70 eV) m/z (%): 622
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.16 (dd, J = 17.4, 3.5 Hz, 1H, H-4), 3.44 (d, J = 18.4 Hz, 2H, CH2), 3.51–3.61 (m, 2H, CH2), 3.63–3.72 (m, 4H, CH2), 3.75–3.84 (m, 4H, CH2), 4.06 (bs, 1H, OH), 3.92 (dd, J = 17.4, 12.1 Hz, 1H, H-4), 5.62 (dd, J = 12.1, 3.5 Hz, 1H, H-5), 6.80 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.18 (t, J = 8.2 Hz, 2H, Ar–H), 7.25–7.35 (m, 2H, Ar–H), 7.70 (t, J = 11.2 Hz, 2H, Ar–H), 7.78 (d, J = 8.4 Hz, 2H, Ar–H), 8.50 (bs, 1H, NH), 10.80 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.1 (CH2), 43.3 (CH2), 44.4 (CH2), 59.0 (CH2), 61.6 (CH), 65.7 (CH2), 110.9 (CH), 116.0 (d, 2JCF = 21.5 Hz, (CH)), 117.0 (CH), 117.1 (Cq), 120.4 (CH), 120.9 (Cq), 127.0 (CH), 127.9 (d, 3JCF = 8.3 Hz, (CH)), 134.4 (Cq), 137.5 (d, 4JCF = 3.2 Hz, Cq), 138.4 (Cq), 145.6 (Cq), 149.3 (Cq), 149.7 (Cq), 149.8 (Cq), 161.5 (d, 1JC–F = 244.0 Hz, Cq). MS (70 eV) m/z (%): 622![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624
624![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626 [M+]
626 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/71/15), 592 (11), 527 (10), 310 (16), 266 (11), 124.69 (33). Anal. calcd C30H29Cl2FN8O2: C, 57.79; H, 4.69; N, 17.97; found: C, 57.90; H, 4.72; N, 18.00.
[M + 4]+ (100/71/15), 592 (11), 527 (10), 310 (16), 266 (11), 124.69 (33). Anal. calcd C30H29Cl2FN8O2: C, 57.79; H, 4.69; N, 17.97; found: C, 57.90; H, 4.72; N, 18.00.
4.2.8.7. 2-((4-((4-(1-(3,5-Dichlorophenyl)-5-(4-(trifluoromethyl)phenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)amino)ethan-1-ol (9g). Yellow solid. 84% yield; mp 258–263 °C. FT-IR (ATR): ν (cm−1) 3445 (N–H), 3242 (O–H), 3105 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1653 y 1584 (C
C–H), 1653 y 1584 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.21 (dd, J = 17.8, 4.2 Hz, 1H, H-4), 3.44 (d, J = 16.4 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.62–3.73 (m, 5H, CH2, OH), 3.74–3.85 (m, 4H, CH2), 3.96 (dd, J = 17.8, 11.8 Hz, 1H, H-4), 5.75 (dd, J = 11.8, 4.2 Hz, 1H, H-5), 6.82 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.48 (d J = 8.4 Hz, 2H, Ar–H), 7.62–7.86 (m, 6H, Ar–H), 8.40 (bs, 1H, NH), 10.72 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 44.1 (CH2), 44.4 (CH2), 59.0 (CH2), 61.7 (CH), 65.8 (CH2), 110.8 (CH), 110.9 (Cq), 113.0 (Cq), 114.1 (Cq), 117.2 (CH), 118.2 (Cq), 120.3 (Cq), 120.4 (Cq), 120.9 (CH), 121.0 (q, 1JCF = 244.51 Hz, CF3), 125.5 (Cq), 126.2 (d, 3JCF = 5.7 Hz, (CH)), 126.8 (CH), 127.0 (d, 4JCF = 1.2 Hz, (CH)), 134.6 (Cq), 145.8 (d, 2JCF = 46.76 Hz, Cq), 150.0 (Cq). MS (70 eV) m/z (%): 672
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.21 (dd, J = 17.8, 4.2 Hz, 1H, H-4), 3.44 (d, J = 16.4 Hz, 2H, CH2), 3.50–3.60 (m, 2H, CH2), 3.62–3.73 (m, 5H, CH2, OH), 3.74–3.85 (m, 4H, CH2), 3.96 (dd, J = 17.8, 11.8 Hz, 1H, H-4), 5.75 (dd, J = 11.8, 4.2 Hz, 1H, H-5), 6.82 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.48 (d J = 8.4 Hz, 2H, Ar–H), 7.62–7.86 (m, 6H, Ar–H), 8.40 (bs, 1H, NH), 10.72 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 44.1 (CH2), 44.4 (CH2), 59.0 (CH2), 61.7 (CH), 65.8 (CH2), 110.8 (CH), 110.9 (Cq), 113.0 (Cq), 114.1 (Cq), 117.2 (CH), 118.2 (Cq), 120.3 (Cq), 120.4 (Cq), 120.9 (CH), 121.0 (q, 1JCF = 244.51 Hz, CF3), 125.5 (Cq), 126.2 (d, 3JCF = 5.7 Hz, (CH)), 126.8 (CH), 127.0 (d, 4JCF = 1.2 Hz, (CH)), 134.6 (Cq), 145.8 (d, 2JCF = 46.76 Hz, Cq), 150.0 (Cq). MS (70 eV) m/z (%): 672![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 674
674![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 676 [M+]
676 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+ (100/66/13), 642 (12), 515 (7), 310 (14), 252 (8), 124 (25), 69 (32). Anal. calcd C31H29Cl2F3N8O2: C, 55.28; H, 4.34; N, 16.64; found: C, 55.32; H, 4.29; N, 16.58.
[M + 4]+ (100/66/13), 642 (12), 515 (7), 310 (14), 252 (8), 124 (25), 69 (32). Anal. calcd C31H29Cl2F3N8O2: C, 55.28; H, 4.34; N, 16.64; found: C, 55.32; H, 4.29; N, 16.58.
4.2.9.1. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10a). Yellow solid. 84% yield; mp 222–224 °C. FT-IR (ATR): ν (cm−1) 3419 (N–H), 3319 (O–H), 3113 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1622 y 1583 (C
C–H), 1622 y 1583 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 17.5, 4.6 Hz, 1H, H-4), 3.32–3.49 (m, 2H, CH2), 3.57–3.62 (m, 2H, CH2), 3.75–4.12 (m, 2H, H-4, OH), 5.58 (dd, J = 11.9, 4.6 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.24–7.31 (m, 3H, Ar–H), 7.33–7.44 (m, 4H, Ar–H), 7.68–7.83 (m, 6H, Ar–H), 8.50 (bs, 1H, NH), 10.43 (bs, 1H, NH), 10.65 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 43.4 (CH2), 59.2 (CH2), 62.5 (CH), 110.8 (CH), 116.9 (CH), 120.7 (CH), 121.2 (CH), 122.7 (Cq), 123.2 (Cq), 123.3 (Cq), 125.8 (CH), 126.6 (Cq), 126.7 (CH), 127.0 (Cq), 127.8 (CH), 128.6 (CH), 129.2 (CH), 134.4 (Cq), 136.8 (Cq), 137.2 (Cq), 141.4 (Cq), 145.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 644
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 17.5, 4.6 Hz, 1H, H-4), 3.32–3.49 (m, 2H, CH2), 3.57–3.62 (m, 2H, CH2), 3.75–4.12 (m, 2H, H-4, OH), 5.58 (dd, J = 11.9, 4.6 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.93 (s, 2H, Ar–H), 7.24–7.31 (m, 3H, Ar–H), 7.33–7.44 (m, 4H, Ar–H), 7.68–7.83 (m, 6H, Ar–H), 8.50 (bs, 1H, NH), 10.43 (bs, 1H, NH), 10.65 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 43.4 (CH2), 59.2 (CH2), 62.5 (CH), 110.8 (CH), 116.9 (CH), 120.7 (CH), 121.2 (CH), 122.7 (Cq), 123.2 (Cq), 123.3 (Cq), 125.8 (CH), 126.6 (Cq), 126.7 (CH), 127.0 (Cq), 127.8 (CH), 128.6 (CH), 129.2 (CH), 134.4 (Cq), 136.8 (Cq), 137.2 (Cq), 141.4 (Cq), 145.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 644![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 646
646![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648
648![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 [M+]
650 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (95/100/38/5), 628 (49), 567 (12), 350 (17), 221 (19), 178 (17), 124 (24), 91 (30). Anal. calcd C32H27Cl3N8O: C, 59.50; H, 4.21; N, 17.35; found: C, 59.48; H, 4.27; N, 17.40.
[M + 6]+ (95/100/38/5), 628 (49), 567 (12), 350 (17), 221 (19), 178 (17), 124 (24), 91 (30). Anal. calcd C32H27Cl3N8O: C, 59.50; H, 4.21; N, 17.35; found: C, 59.48; H, 4.27; N, 17.40.
4.2.9.2. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-(p-tolyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10b). Yellow solid. 90% yield; mp 277–279 °C. FT-IR (ATR): ν (cm−1) 3311 (N–H), 3252 (O–H), 3190 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1629 y 1583 (C
C–H), 1629 y 1583 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.25 (s, 3H, CH3), 3.14 (dd, J = 17.1, 4.4 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.85–4.00 (m, 2H, H-4, OH), 5.53 (dd, J = 12.1, 4.4 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.93 (d, J = 1.6 Hz, 2H, Ar–H), 7.13–7.17 (m, 4H, Ar–H), 7.35–7.45 (m, 2H, Ar–H), 7.67–7.83 (m, 6H, Ar–H), 8.47 (bs, 1H, NH), 10.41 (bs, 1H, NH), 10.62 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 20.7 (CH3), 43.2 (CH2), 43.4 (CH2), 59.2 (CH2), 62.3 (CH), 110.8 (CH), 116.8 (CH), 120.0 (Cq), 120.8 (CH), 122.7 (Cq), 123.2 (CH), 125.7 (CH), 125.9 (Cq), 126.7 (CH), 127.1 (Cq), 127.6 (Cq), 128.5 (CH), 128.6 (Cq), 129.7 (CH), 134.4 (Cq), 134.5 (Cq), 137.0 (Cq), 138.5 (Cq), 145.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 658
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 2.25 (s, 3H, CH3), 3.14 (dd, J = 17.1, 4.4 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.85–4.00 (m, 2H, H-4, OH), 5.53 (dd, J = 12.1, 4.4 Hz, 1H, H-5), 6.79 (s, 1H, Ar–H), 6.93 (d, J = 1.6 Hz, 2H, Ar–H), 7.13–7.17 (m, 4H, Ar–H), 7.35–7.45 (m, 2H, Ar–H), 7.67–7.83 (m, 6H, Ar–H), 8.47 (bs, 1H, NH), 10.41 (bs, 1H, NH), 10.62 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 20.7 (CH3), 43.2 (CH2), 43.4 (CH2), 59.2 (CH2), 62.3 (CH), 110.8 (CH), 116.8 (CH), 120.0 (Cq), 120.8 (CH), 122.7 (Cq), 123.2 (CH), 125.7 (CH), 125.9 (Cq), 126.7 (CH), 127.1 (Cq), 127.6 (Cq), 128.5 (CH), 128.6 (Cq), 129.7 (CH), 134.4 (Cq), 134.5 (Cq), 137.0 (Cq), 138.5 (Cq), 145.8 (Cq), 149.8 (Cq). MS (70 eV) m/z (%): 658![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 660
660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662
662![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 [M+]
664 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (100/98/37/5), 642 (23), 557 (36), 350 (31), 178 (27), 117 (33), 91 (34). Anal. calcd C33H29Cl3N8O: C, 60.05; H, 4.43; N, 16.98; found: C, 59.98; H, 4.40; N, 17.01.
[M + 6]+ (100/98/37/5), 642 (23), 557 (36), 350 (31), 178 (27), 117 (33), 91 (34). Anal. calcd C33H29Cl3N8O: C, 60.05; H, 4.43; N, 16.98; found: C, 59.98; H, 4.40; N, 17.01.
4.2.9.3. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10c). Yellow solid. 95% yield; mp 276–279 °C. FT-IR (ATR): ν (cm−1) 3308 (N–H), 3246 (O–H), 3188 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1634 y 1585 (C
C–H), 1634 y 1585 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.14 (dd, J = 17.0, 4.3 Hz, 1H, H-4), 3.42–3.50 (m, 2H, CH2), 3.55–3.63 (m, 2H, CH2), 3.71 (s, 3H, OCH3), 3.76–3.97 (m, 2H, H-4, OH), 5.52 (dd, J = 11.6, 4.3 Hz, 1H, H-5), 6.78 (s, 1H, Ar–H), 6.86–6.98 (m, 4H, Ar–H), 7.19 (d, J = 8.4, 2H, Ar–H), 7.36–7.44 (m, 2H, Ar–H), 7.65–7.85 (m, 6H, Ar–H), 8.59 (bs, 1H, NH), 10.50 (bs, 1H, NH), 10.76 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 43.4 (CH2), 55.1 (OCH3), 59.2 (CH2), 62.0 (CH), 110.9 (CH), 114.5 (CH), 116.8 (CH), 120.8 (CH), 121.3 (Cq), 122.7 (Cq), 123.3 (CH), 126.7 (CH), 126.8 (Cq), 127.1 (CH), 128.5 (Cq), 128.6 (CH), 133.3 (Cq), 134.4 (Cq), 136.6 (CH), 142.1 (Cq), 145.8 (Cq), 149.8 (Cq), 152.2 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 674
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.14 (dd, J = 17.0, 4.3 Hz, 1H, H-4), 3.42–3.50 (m, 2H, CH2), 3.55–3.63 (m, 2H, CH2), 3.71 (s, 3H, OCH3), 3.76–3.97 (m, 2H, H-4, OH), 5.52 (dd, J = 11.6, 4.3 Hz, 1H, H-5), 6.78 (s, 1H, Ar–H), 6.86–6.98 (m, 4H, Ar–H), 7.19 (d, J = 8.4, 2H, Ar–H), 7.36–7.44 (m, 2H, Ar–H), 7.65–7.85 (m, 6H, Ar–H), 8.59 (bs, 1H, NH), 10.50 (bs, 1H, NH), 10.76 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.2 (CH2), 43.4 (CH2), 55.1 (OCH3), 59.2 (CH2), 62.0 (CH), 110.9 (CH), 114.5 (CH), 116.8 (CH), 120.8 (CH), 121.3 (Cq), 122.7 (Cq), 123.3 (CH), 126.7 (CH), 126.8 (Cq), 127.1 (CH), 128.5 (Cq), 128.6 (CH), 133.3 (Cq), 134.4 (Cq), 136.6 (CH), 142.1 (Cq), 145.8 (Cq), 149.8 (Cq), 152.2 (Cq), 158.7 (Cq). MS (70 eV) m/z (%): 674![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 676
676![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 678
678![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 [M+]
680 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (30/29/11/2), 658 (9), 640 (8), 567 (13), 350 (12), 124 (25), 84 (100). Anal. calcd C33H29Cl3N8O2: C, 58.63; H, 4.32; N, 16.58; found: C, 58.66; H, 4.32; N, 16.60.
[M + 6]+ (30/29/11/2), 658 (9), 640 (8), 567 (13), 350 (12), 124 (25), 84 (100). Anal. calcd C33H29Cl3N8O2: C, 58.63; H, 4.32; N, 16.58; found: C, 58.66; H, 4.32; N, 16.60.
4.2.9.4. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-(3,4,5-trimethoxyphenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10d). Yellow solid. 86% yield; mp 226–229 °C. FT-IR (ATR): ν (cm−1) 3500 (N–H), 3192 (O–H), 3074 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1636 y 1583 (C
C–H), 1636 y 1583 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.24 (dd, J = 17.4, 5.8 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.64 (s, 3H, OCH3), 3.71 (s, 6H, OCH3), 3.79–3.99 (m, 2H, H-4, OH), 5.43 (dd, J = 12.2, 5.8 Hz, 1H, H-5), 6.62 (s, 2H, Ar–H), 6.83 (s, 1H, Ar–H), 6.97 (s, 2H, Ar–H), 7.40 (d, J = 6.8 Hz, 2H, Ar–H), 7.68–7.86 (m, 6H, Ar–H), 8.53 (bs, 1H, NH), 10.46 (bs, 1H, NH), 10.68 (s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.3 (CH2), 43.4 (CH2), 56.0 (OCH3), 59.2 (CH2), 60.0 (OCH3), 63.2 (CH), 103.1 (CH), 111.0 (CH), 117.1 (CH), 120.7 (CH), 120.8 (Cq), 121.2 (Cq), 122.7 (Cq), 123.1 (CH), 123.3 (Cq), 126.7 (Cq), 126.8 (CH), 126.9 (Cq), 128.6 (CH), 134.4 (Cq), 136.8 (Cq), 137.3 (Cq), 139.1 (Cq), 146.2 (Cq), 150.1 (Cq), 153.4 (Cq). MS (70 eV) m/z (%): 734
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.24 (dd, J = 17.4, 5.8 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.64 (s, 3H, OCH3), 3.71 (s, 6H, OCH3), 3.79–3.99 (m, 2H, H-4, OH), 5.43 (dd, J = 12.2, 5.8 Hz, 1H, H-5), 6.62 (s, 2H, Ar–H), 6.83 (s, 1H, Ar–H), 6.97 (s, 2H, Ar–H), 7.40 (d, J = 6.8 Hz, 2H, Ar–H), 7.68–7.86 (m, 6H, Ar–H), 8.53 (bs, 1H, NH), 10.46 (bs, 1H, NH), 10.68 (s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.3 (CH2), 43.4 (CH2), 56.0 (OCH3), 59.2 (CH2), 60.0 (OCH3), 63.2 (CH), 103.1 (CH), 111.0 (CH), 117.1 (CH), 120.7 (CH), 120.8 (Cq), 121.2 (Cq), 122.7 (Cq), 123.1 (CH), 123.3 (Cq), 126.7 (Cq), 126.8 (CH), 126.9 (Cq), 128.6 (CH), 134.4 (Cq), 136.8 (Cq), 137.3 (Cq), 139.1 (Cq), 146.2 (Cq), 150.1 (Cq), 153.4 (Cq). MS (70 eV) m/z (%): 734![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 736
736![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738
738![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 [M+]
740 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (18/15/6/1), 656 (18), 567 (9), 363 (12), 221 (18), 152 (15), 118 (18). Anal. calcd C35H33Cl3N8O4: C, 57.11; H, 4.52; N, 15.22; found: C, 57.15; H, 4.55; N, 15.32.
[M + 6]+ (18/15/6/1), 656 (18), 567 (9), 363 (12), 221 (18), 152 (15), 118 (18). Anal. calcd C35H33Cl3N8O4: C, 57.11; H, 4.52; N, 15.22; found: C, 57.15; H, 4.55; N, 15.32.
4.2.9.5. 2-((4-((4-(5-(4-Chlorophenyl)-1-(3,5-dichlorophenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-6-((4-chlorophenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10e). Yellow solid. 95% yield; mp 277–279 °C. FT-IR (ATR): ν (cm−1) 3309 (N–H), 3200 (O–H), 3192 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1632 y 1583 (C
C–H), 1632 y 1583 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 16.5, 4.0 Hz, 1H, H-4), 3.41–3.51 (m, 2H, CH2), 3.57–3.63 (m, 2H, CH2), 3.74–4.07 (m, 2H, H-4, OH), 5.63 (dd, J = 12.5, 4.0 Hz, 1H, H-5), 6.81 (s, 1H, Ar–H), 6.92 (s, 2H, Ar–H), 7.29 (d, J = 8.4 Hz, 2H, Ar–H), 7.36–7.45 (m, 4H, Ar–H), 7.66–7.86 (m, 6H, Ar–H), 8.54 (bs, 1H, NH), 10.46 (bs, 1H, NH), 10.69 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 59.2 (CH2), 61.7 (CH), 110.9 (CH), 117.1 (CH), 120.7 (CH), 121.2 (Cq), 121.3 (Cq), 122.7 (Cq), 123.1 (CH), 123.6 (Cq), 125.9 (Cq), 126.8 (CH), 126.9 (Cq), 127.8 (CH), 128.6 (CH), 128.9 (Cq), 129.2 (CH), 132.3 (Cq), 134.5 (Cq), 140.4 (Cq), 145.6 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 678
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 16.5, 4.0 Hz, 1H, H-4), 3.41–3.51 (m, 2H, CH2), 3.57–3.63 (m, 2H, CH2), 3.74–4.07 (m, 2H, H-4, OH), 5.63 (dd, J = 12.5, 4.0 Hz, 1H, H-5), 6.81 (s, 1H, Ar–H), 6.92 (s, 2H, Ar–H), 7.29 (d, J = 8.4 Hz, 2H, Ar–H), 7.36–7.45 (m, 4H, Ar–H), 7.66–7.86 (m, 6H, Ar–H), 8.54 (bs, 1H, NH), 10.46 (bs, 1H, NH), 10.69 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.0 (CH2), 43.4 (CH2), 59.2 (CH2), 61.7 (CH), 110.9 (CH), 117.1 (CH), 120.7 (CH), 121.2 (Cq), 121.3 (Cq), 122.7 (Cq), 123.1 (CH), 123.6 (Cq), 125.9 (Cq), 126.8 (CH), 126.9 (Cq), 127.8 (CH), 128.6 (CH), 128.9 (Cq), 129.2 (CH), 132.3 (Cq), 134.5 (Cq), 140.4 (Cq), 145.6 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 678![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680
680![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682
682![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 684
684![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 686 [M+]
686 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+
[M + 6]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 8]+ (70/89/47/11/1), 662 (25), 555 (18), 417 (20), 350 (25), 262 (31), 221 (27), 118 (40). Anal. calcd C32H26Cl4N8O: C, 56.49; H, 3.85; N, 16.47; found: C, 56.52; H, 3.87; N, 16.50.
[M + 8]+ (70/89/47/11/1), 662 (25), 555 (18), 417 (20), 350 (25), 262 (31), 221 (27), 118 (40). Anal. calcd C32H26Cl4N8O: C, 56.49; H, 3.85; N, 16.47; found: C, 56.52; H, 3.87; N, 16.50.
4.2.9.6. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10f). Yellow solid. 77% yield; mp 208–211 °C. FT-IR (ATR): ν (cm−1) 3319 (N–H), 3253 (O–H), 3095 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1620 y 1585 (C
C–H), 1620 y 1585 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 16.8, 4.8 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.86–4.01, (m, 2H, H-4, OH), 5.62 (dd, J = 12.3, 4.8 Hz, 1H, H-5), 6.81 (s, 1H, Ar–H), 6.93 (d, J = 1.3 Hz, 2H, Ar–H), 7.19 (t, J = 8.6 Hz, 2H, Ar–H), 7.28–7.35 (m, 2H, Ar–H), 7.36–7.44 (m, 2H, Ar–H), 7.66–7.86 (m, 6H, Ar–H), 8.50 (bs, 1H, NH), 10.43 (bs, 1H, NH), 10.63 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.1 (CH2), 43.4 (CH2), 59.2 (CH2), 61.7 (CH), 110.9 (CH), 116.0 (d, 2JCF = 21.5 Hz, (CH)), 117.0 (CH), 120.7 (CH), 120.8 (Cq), 121.3 (Cq), 122.3 (Cq), 122.8 (Cq), 123.2 (CH), 123.5 (Cq), 126.8 (CH), 126.9 (Cq), 127.9 (d, 3JCF = 7.8 Hz, (CH)), 128.5 (CH), 128.6 (Cq), 134.4 (Cq), 137.6 (d, 4JCF = 2.5 Hz, Cq), 145.6 (Cq), 149.8 (Cq), 161.5 (d, 1JC–F = 243.7 Hz, Cq). MS (70 eV) m/z (%): 662
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.18 (dd, J = 16.8, 4.8 Hz, 1H, H-4), 3.40–3.50 (m, 2H, CH2), 3.56–3.62 (m, 2H, CH2), 3.86–4.01, (m, 2H, H-4, OH), 5.62 (dd, J = 12.3, 4.8 Hz, 1H, H-5), 6.81 (s, 1H, Ar–H), 6.93 (d, J = 1.3 Hz, 2H, Ar–H), 7.19 (t, J = 8.6 Hz, 2H, Ar–H), 7.28–7.35 (m, 2H, Ar–H), 7.36–7.44 (m, 2H, Ar–H), 7.66–7.86 (m, 6H, Ar–H), 8.50 (bs, 1H, NH), 10.43 (bs, 1H, NH), 10.63 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 43.1 (CH2), 43.4 (CH2), 59.2 (CH2), 61.7 (CH), 110.9 (CH), 116.0 (d, 2JCF = 21.5 Hz, (CH)), 117.0 (CH), 120.7 (CH), 120.8 (Cq), 121.3 (Cq), 122.3 (Cq), 122.8 (Cq), 123.2 (CH), 123.5 (Cq), 126.8 (CH), 126.9 (Cq), 127.9 (d, 3JCF = 7.8 Hz, (CH)), 128.5 (CH), 128.6 (Cq), 134.4 (Cq), 137.6 (d, 4JCF = 2.5 Hz, Cq), 145.6 (Cq), 149.8 (Cq), 161.5 (d, 1JC–F = 243.7 Hz, Cq). MS (70 eV) m/z (%): 662![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664
664![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666
666![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 668 [M+]
668 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (65/51/17/3), 644 (25), 557 (9), 350 (13), 279 (13), 262 (17), 221 (27), 124 (25). Anal. calcd C32H26Cl3FN8O: C, 57.89; H, 3.95; 2.86; N, 16.88; found: C, 57.90; H, 4.00; N, 16.74.
[M + 6]+ (65/51/17/3), 644 (25), 557 (9), 350 (13), 279 (13), 262 (17), 221 (27), 124 (25). Anal. calcd C32H26Cl3FN8O: C, 57.89; H, 3.95; 2.86; N, 16.88; found: C, 57.90; H, 4.00; N, 16.74.
4.2.9.7. 2-((4-((4-Chlorophenyl)amino)-6-((4-(1-(3,5-dichlorophenyl)-5-(4-(trifluoromethyl)phenyl)-4,5-dihydro-1H-pyrazol-3-yl)phenyl)amino)-1,3,5-triazin-2-yl)amino)ethan-1-ol (10g). Yellow solid. 83% yield; mp 274–277 °C. FT-IR (ATR): ν (cm−1) 3310 (N–H), 3292 (O–H), 3190 (
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1632 y 1585 (C
C–H), 1632 y 1585 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.17–3.28 (m, 1H, H-4), 3.41–2.51 (m, 2H, CH2), 3.56–6.63 (m, 2H, CH2), 3.98 (dd, J = 16.6, 12.5 Hz, 1H, H-4), 4.23 (bs, 1H, OH), 5.74 (dd, J = 12.5, 4.8 Hz, 1H, H-5), 6.83 (s, 1H, Ar–H), 6.94 (s, 2H, Ar–H), 7.35–7.45 (m, 2H, Ar–H), 7.50 (d, J = 8.0 Hz, 2H, Ar–H), 7.68–7.84 (m, 8H, Ar–H), 8.48 (bs, 1H, NH), 10.42 (bs, 1H, NH), 10.61 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.3 (CH2), 59.2 (CH2), 61.8 (CH), 110.8 (CH), 117.2 (CH), 120.5 (Cq), 120.7 (CH), 121.3 (CH), 123.1 (CH), 123.4 (Cq), 124.1 (q, 1JCF = 272.2 Hz, CF3), 126.2 (Cq), 126.5 (d, 4JCF = 5.86 Hz, (CH)), 126.8 (d, 3JCF = 7.00 Hz, (CH)), 127.6 (Cq), 127.9 (Cq), 128.2 (Cq), 128.6 (CH), 134.5 (Cq), 136.8 (Cq), 137.1 (Cq), 145.8 (d, 2JCF = 46.58 Hz, Cq), 148.2 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 712
C). 1H-NMR (400 MHz, DMSO-d6) δ ppm 3.17–3.28 (m, 1H, H-4), 3.41–2.51 (m, 2H, CH2), 3.56–6.63 (m, 2H, CH2), 3.98 (dd, J = 16.6, 12.5 Hz, 1H, H-4), 4.23 (bs, 1H, OH), 5.74 (dd, J = 12.5, 4.8 Hz, 1H, H-5), 6.83 (s, 1H, Ar–H), 6.94 (s, 2H, Ar–H), 7.35–7.45 (m, 2H, Ar–H), 7.50 (d, J = 8.0 Hz, 2H, Ar–H), 7.68–7.84 (m, 8H, Ar–H), 8.48 (bs, 1H, NH), 10.42 (bs, 1H, NH), 10.61 (bs, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ ppm 42.9 (CH2), 43.3 (CH2), 59.2 (CH2), 61.8 (CH), 110.8 (CH), 117.2 (CH), 120.5 (Cq), 120.7 (CH), 121.3 (CH), 123.1 (CH), 123.4 (Cq), 124.1 (q, 1JCF = 272.2 Hz, CF3), 126.2 (Cq), 126.5 (d, 4JCF = 5.86 Hz, (CH)), 126.8 (d, 3JCF = 7.00 Hz, (CH)), 127.6 (Cq), 127.9 (Cq), 128.2 (Cq), 128.6 (CH), 134.5 (Cq), 136.8 (Cq), 137.1 (Cq), 145.8 (d, 2JCF = 46.58 Hz, Cq), 148.2 (Cq), 149.9 (Cq). MS (70 eV) m/z (%): 712![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 714
714![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 716
716![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 718 [M+]
718 [M+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 2]+
[M + 2]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 4]+
[M + 4]+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [M + 6]+ (100/94/35/5), 696 (11), 682 (12), 557 (10), 350 (23), 279 (19), 178 (20), 124 (29). Anal. calcd C33H26Cl3F3N8O: C, 55.52; H, 3.67; N, 15.69; found: C, 55.50; H, 3.69; N, 15.74.
[M + 6]+ (100/94/35/5), 696 (11), 682 (12), 557 (10), 350 (23), 279 (19), 178 (20), 124 (29). Anal. calcd C33H26Cl3F3N8O: C, 55.52; H, 3.67; N, 15.69; found: C, 55.50; H, 3.69; N, 15.74.
4.3. Anticancer activity
The human cancer cell lines of the cancer screening panel were grown in an RPMI-1640 medium containing 5% fetal bovine serum and 2 mM L-glutamine. For a typical screening experiment, cells were inoculated into 96-well microtiter plates. After cell inoculation, the microtiter plates were incubated at 37 °C, 5% CO2, 95% air, and 100% relative humidity for 24 h prior to the addition of the tested compounds. After 24 h, two plates of each cell line were fixed in situ with TCA, to represent a measurement of the cell population for each cell line at the time of sample addition (Tz). The samples were solubilized in dimethyl sulfoxide (DMSO) at 400–fold the desired final maximum test concentration and stored frozen prior to use. At the time of compound addition, an aliquot of frozen concentrate was thawed and diluted to twice the desired final maximum test concentration with complete medium containing 50 μg mL−1 gentamicin. An additional four 10-fold or 1/2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log serial dilutions were made to provide a total of five drug concentrations plus the control. Aliquots of 100 μL of these different sample dilutions were added to the appropriate microtiter wells already containing 100 μL of medium, resulting in the required final sample concentrations. After the tested compounds were added, the plates were incubated for an additional 48 h at 37 °C, 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay was terminated by the addition of cold TCA. Cells were fixed in situ by the gentle addition of 50 μL of cold 50% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The supernatant was discarded, and plates were washed five times with tap water and air dried. Sulforhodamine B (SRB) solution (100 μL) at 0.4% (w/v) in 1% acetic acid was added to each well, and plates were incubated for 10 min at room temperature. After staining, unbound dye was removed by washing five times with 1% acetic acid and the plates were air dried. Bound stain was subsequently solubilized with 10 mM trizma base, and the absorbance was read on an automated plate reader at a wavelength of 515 nm. Using the seven absorbance measurements [time zero (Tz), control growth in the absence of drug, and test growth in the presence of drug at the five concentration levels (Ti)], the percentage growth was calculated at each of the drug concentrations levels. Percentage growth inhibition was calculated as [(Ti − Tz)/(C − Tz)] × 100 for concentrations for which Ti > Tz, and [(Ti − Tz)/Tz] × 100 for concentrations for which Ti < Tz. Two dose–response parameters were calculated for each compound. Growth inhibition of 50% (GI50) was calculated from [(Ti − Tz)/(C − Tz)] × 100 = 50, which is the drug concentration resulting in a 50% lower net protein increase in the treated cells (measured by SRB staining) as compared to the net protein increase seen in the control cells and the LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared to that at the beginning), indicating a net loss of cells; calculated from [(Ti − Tz)/Tz] × 100 = −50. Values were calculated for each of these two parameters if the level of activity is reached; however, if the effect was not reached or was exceeded, the value for that parameter was expressed as greater or less than the maximum or minimum concentration tested.38
log serial dilutions were made to provide a total of five drug concentrations plus the control. Aliquots of 100 μL of these different sample dilutions were added to the appropriate microtiter wells already containing 100 μL of medium, resulting in the required final sample concentrations. After the tested compounds were added, the plates were incubated for an additional 48 h at 37 °C, 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay was terminated by the addition of cold TCA. Cells were fixed in situ by the gentle addition of 50 μL of cold 50% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The supernatant was discarded, and plates were washed five times with tap water and air dried. Sulforhodamine B (SRB) solution (100 μL) at 0.4% (w/v) in 1% acetic acid was added to each well, and plates were incubated for 10 min at room temperature. After staining, unbound dye was removed by washing five times with 1% acetic acid and the plates were air dried. Bound stain was subsequently solubilized with 10 mM trizma base, and the absorbance was read on an automated plate reader at a wavelength of 515 nm. Using the seven absorbance measurements [time zero (Tz), control growth in the absence of drug, and test growth in the presence of drug at the five concentration levels (Ti)], the percentage growth was calculated at each of the drug concentrations levels. Percentage growth inhibition was calculated as [(Ti − Tz)/(C − Tz)] × 100 for concentrations for which Ti > Tz, and [(Ti − Tz)/Tz] × 100 for concentrations for which Ti < Tz. Two dose–response parameters were calculated for each compound. Growth inhibition of 50% (GI50) was calculated from [(Ti − Tz)/(C − Tz)] × 100 = 50, which is the drug concentration resulting in a 50% lower net protein increase in the treated cells (measured by SRB staining) as compared to the net protein increase seen in the control cells and the LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared to that at the beginning), indicating a net loss of cells; calculated from [(Ti − Tz)/Tz] × 100 = −50. Values were calculated for each of these two parameters if the level of activity is reached; however, if the effect was not reached or was exceeded, the value for that parameter was expressed as greater or less than the maximum or minimum concentration tested.38
4.4. Molecular modeling
4.4.2.1. Ligand preparation. The default setting of the LigPrep tool implemented in Schrödinger's software (version 2017-1) has been used to prepare the ligands for docking.39 All possible tautomers and combination of stereoisomers have been generated for pH 7.0 ± 0.4, using the Epik ionization method.40 Energy minimization is subsequently done using the integrated OPLS 2005 force field.41
4.4.2.2. Protein preparation. The high-resolution (2.79 Å) X-ray structure of Human thymidylate synthase complexed with dUMP and raltitrexed (PDB ID: 5X5Q)35 is downloaded from the Protein Databank.36
The receptor grid preparation has been carried out with substrate dUMP and without water molecules, to elucidate the role of dUMP for the binding, by assigning the original ligand (raltitrexed) as the centroid of the grid box. Protein Preparation Wizard of Schrödinger software that was employed using the default settings.42 Bond orders have been assigned and hydrogen atoms added as well as protonation of the heteroatom states using Epik-tool (with the pH set at biologically relevant values, i.e. at 7.0 ± 0.4). The H-bond network has been then optimized. The structure is finally subjected to a restrained energy minimization step (rmsd of the atom displacement for terminating the minimization was 0.3 Å), using the Optimized Potentials for Liquid Simulations (OPLS) 2005 force field.41
Finally, IFD score (IFD score = 1.0 Glide Gscore + 0.05 prime energy), which accounts for both protein–ligand interaction energy and total energy of the system, is calculated and used to rank the IFD poses. The more negative is the IFDscore, the more favorable is the binding.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank The Developmental Therapeutics Program (DTP) of the National Cancer Institute of the United States for performing the anticancer screening of the compounds. This work was financially supported by COLCIENCIAS and Universidad del Valle, Colombia.References
- P. Martins, J. Jesus, S. Santos, L. Raposo, C. Roma, P. Baptista and A. Fernandes, Molecules, 2015, 20, 16852–16891 CrossRef CAS.
- G. Facchetti and I. Rimoldi, Bioorg. Med. Chem. Lett., 2019, 29, 1257–1263 CrossRef CAS.
- R. Kurukulasuriya, J. Rohde, B. Szczepankiewicz, F. Basha, C. Lai, H. Jae, M. Winn, K. Stewart, K. Longenecker, T. Lubben, S. Ballaron, H. Sham and T. von Geldern, Bioorg. Med. Chem. Lett., 2006, 16, 6226–6230 CrossRef CAS.
- R. Roskoski, Pharmacol. Res., 2019, 144, 19–50 CrossRef CAS.
- S. Cascioferro, B. Parrino, V. Spanò, A. Carbone, A. Montalbano, P. Barraja, P. Diana and G. Cirrincione, Eur. J. Med. Chem., 2017, 142, 523–549 CrossRef CAS.
- G. Riham, K. Manal, E. Dina and E. Ahmed, Bioorg. Chem., 2020, 99, 103780 CrossRef PubMed.
- S. Ranjbaria, M. Behzadib, S. Sepehric, M. Dadkhah, A. Jarrahpoura, M. Mohkame, Y. Ghasemib, A. Reza, S. Kianpourb, Z. Atioğluh, N. Özdemiri, M. Akkurtj, M. Nabavizadeha and E. Turos, Bioorg. Med. Chem., 2020, 28, 115408 CrossRef PubMed.
- S. Kuthyala, M. Hanumanthappa, S. Madan Kumar, S. Sheik, N. Gundibasappa Karikannar and A. Prabhu, J. Mol. Struct., 2019, 1197, 65–72 CrossRef CAS.
- H. Kothayer, S. Spencer, K. Tripathi, A. Westwell and K. Palle, Bioorg. Med. Chem. Lett., 2016, 26, 2030–2034 CrossRef CAS PubMed.
- Z. Nie, C. Perretta, P. Erickson, S. Margosiak, J. Lu, A. Averill, R. Almassy and S. Chu, Bioorg. Med. Chem. Lett., 2008, 18, 619–623 CrossRef CAS PubMed.
- B. Zhang, Q. Zhang, Z. Xiao, X. Sun, Z. Yang, Q. Gu, Z. Liu, T. Xie, Q. Jin, P. Zheng, S. Xu and W. Zhu, Bioorg. Chem., 2020, 95, 103525 CrossRef CAS PubMed.
- N. Lolak, S. Akocak, S. Bua, R. Sanku and C. Supuran, Bioorg. Med. Chem., 2019, 27, 1588–1594 CrossRef CAS PubMed.
- E. Havránková, J. Csöllei, D. Vullo, V. Garaj, P. Pazdera and C. Supuran, Bioorg. Chem., 2018, 77, 25–37 CrossRef.
- B. Żołnowska, J. Sławiński, K. Szafrański, A. Angeli, C. Supuran, A. Kawiak, M. Wieczór, J. Zielińska, T. Bączek and S. Bartoszewska, Eur. J. Med. Chem., 2018, 143, 1931–1941 CrossRef PubMed.
- K. Bergant, M. Janežič, K. Valjavec, I. Sosič, S. Pajk, M. Štampar, B. Žegura, S. Gobec, M. Filipič and A. Perdih, Eur. J. Med. Chem., 2019, 175, 330–348 CrossRef CAS PubMed.
- X. Zhou, K. Lin, X. Ma, W. K. Chui and W. Zhou, Eur. J. Med. Chem., 2017, 125, 1279–1288 CrossRef CAS.
- S. Narva, S. Chitti, S. Amaroju, D. Bhattacharjee, B. Rao, N. Jain, M. Alvala and K. Sekhar, Bioorganic Med. Chem. Lett., 2017, 27, 3794–3801 CrossRef CAS.
- B. Sever, M. Altıntop, M. Radwan, A. Özdemir, M. Otsuka, M. Fujita and H. Ciftci, Eur. J. Med. Chem., 2019, 182, 111648 CrossRef.
- Z. Brzozowski, F. Sa and M. Gdaniec, Eur. J. Med. Chem., 2000, 35, 1053–1064 CrossRef CAS.
- L. Moreno, J. Quiroga, R. Abonia, J. Ramírez-Prada and B. Insuasty, Molecules, 2018, 23, 1956 CrossRef PubMed.
- H. Dmytro, Z. Borys, V. Olexandr, G. Andrzej and L. Roman, J. Med. Chem., 2020, 55, 8630–8641 Search PubMed.
- G. Bagnolini, D. Milano, M. Manerba, F. Schipani, J. Ortega, D. Gioia, F. Falchi, A. Balboni, F. Farabegoli, F. De Franco, J. Robertson, R. Pellicciari, I. Pallavicini, S. Peri, S. Minucci, S. Girotto, G. Di Stefano, M. Roberti and A. Cavalli, J. Med. Chem., 2020, 63, 2588–2619 CrossRef CAS.
- S. Farooq and Z. Ngaini, Tetrahedron Lett., 2020, 61, 151416 CrossRef CAS.
- S. Mirzaei, F. Hadizadeh, F. Eisvand, F. Mosaffa and R. Ghodsi, J. Mol. Struct., 2020, 1202, 127310 CrossRef CAS.
- S. Burmaoglu, S. Ozcan, S. Balcioglu, M. Gencel, S. Noma, S. Essiz, B. Ates and O. Algul, Bioorg. Chem., 2019, 91, 103149 CrossRef PubMed.
- G. Wang, W. Liu, Z. Gong, Y. Huang, Y. Li and Z. Peng, Bioorg. Chem., 2020, 95, 103565 CrossRef CAS.
- L. Rahman, D. Voeller, R. Monzur, L. Stan, C. Allegra, C. Barrett, F. Kaye and M. Zajac-Kaye, Cancer Cell, 2004, 5, 341–351 CrossRef CAS.
- L. Taddia, D. D'Arca, S. Ferrari, C. Marraccini, L. Severi, G. Ponterini, Y. Assaraf, G. Marverti and M. Costi, Drug Resist. Updat., 2015, 23, 20–54 CrossRef PubMed.
- R. Xing, H. Zhang, J. Yuan, K. Zhang, L. Li, H. Guo, L. Zhao, C. Zhang, S. Li, T. Gao, Y. Liu and L. Wang, Eur. J. Med. Chem., 2017, 139, 531–541 CrossRef CAS PubMed.
- A. Lauria, M. Ippolito and A. M. Almerico, Comput. Biol. Chem., 2009, 33, 386–390 CrossRef CAS.
- A. Lauria, I. Abbate, C. Patella, A. Martorana, G. Dattolo and A. M. Almerico, Eur. J. Med. Chem., 2013, 62, 416–424 CrossRef CAS PubMed.
- A. Lauria, M. Ippolito and A. M. Almerico, J. Mol. Graph. Model., 2009, 27, 712–722 CrossRef CAS.
- A. Lauria, M. Ippolito and A. M. Almerico, QSAR Comb. Sci., 2009, 28, 387–395 CrossRef CAS.
- A. Lauria, S. Mannino, C. Gentile, G. Mannino, A. Martorana and D. Peri, Bioinformatics, 2020, 36, 1562–1569 CrossRef CAS PubMed.
- D. Chen, A. Jansson, D. Sim, A. Larsson and P. Nordlund, J. Biol. Chem., 2017, 292, 13449–13458 CrossRef CAS.
- H. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. Bhat, H. Weissig, I. Shindyalov and P. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS.
- P. Kathiriya, V. Patel and D. Purohit, J. Chem. Pharm. Res., 2013, 5, 103–107 CAS.
- NCI-60 Screening Methodology, https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm, accessed April 17, 2020 Search PubMed.
- Schrödinger Release 2017–2, LigPrep, Schrödinger, LLC, New York, 2017 Search PubMed.
- Schrödinger Release 2017-2: Protein Preparation Wizard, Epik, Schrödinger, LLC, New York, 2017 Search PubMed.
- J. Banks, H. Beard, Y. Cao, A. Cho, W. Damm, R. Farid, A. Felts, T. Halgren, D. Mainz, J. Maple, R. Murphy, D. Philipp, M. Repasky, L. Zhang, B. Berne, R. Friesner, E. Gallicchio and R. Levy, J. Comput. Chem., 2005, 26, 1752–1780 CrossRef CAS.
- G. M. Sastry, M. Adzhigirey and W. Sherman, J. Comput. Aided. Mol. Des., 2013, 27, 221–234 CrossRef.
- R. Friesner, R. Murphy, M. Repasky, L. Frye, J. Greenwood, T. Halgren, P. Sanschagrin and D. Mainz, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- T. Halgren, R. Murphy, R. Friesner, H. Beard, L. Frye, W. Pollard and J. Banks, J. Med. Chem., 2004, 2, 1750–1759 CrossRef.
- R. Friesner, J. Banks, R. Murphy, T. Halgren, J. Klicic, D. Mainz, M. Repasky, E. Knoll, M. Shelley, J. Perry, D. Shaw, P. Francis and P. Shenkin, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS PubMed.
- W. Sherman, T. Day, M. Jacobson, R. Friesner and R. Farid, J. Med. Chem., 2006, 49, 534–553 CrossRef CAS.
- W. Sherman, H. Beard and R. Farid, Chem. Biol. Drug Des., 2006, 67, 83–84 CrossRef CAS PubMed.
- Maestro, Version 9.3, Schrödinger, LLC. New York 2012 Search PubMed.
- H. Zhong, L. Tran and J. Stang, J. Mol. Graph. Model., 2009, 28, 336–346 CrossRef CAS.
- H. Luo, J. Wang, W. Deng and K. Zou, Med. Chem. Res., 2013, 22, 4970–4979 CrossRef CAS.
- H. Wang, R. Aslanian and V. Madison, J. Mol. Graph. Model., 2008, 27, 512–521 CrossRef CAS.
- M. Jacobson, R. Friesner, Z. Xiang and B. Honig, J. Mol. Biol., 2002, 320, 597–608 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1, spectra data and Fig. S1. See DOI: 10.1039/d0ra06799g |
| This journal is © The Royal Society of Chemistry 2020 |